Abstract
The human cytochrome P450 1A1 (CYP1A1) is well known for chemical activation of procarcinogens and often has a substrate scope of small and highly planar compounds. Substrates deviating from these characteristics are certainly known, but how these larger and nonplanar substrates are accommodated and oriented within the CYP1A1 active site is not understood. Herein a new X-ray structure of CYP1A1 bound to the pan-Pim kinase inhibitor GDC-0339 reveals how the CYP1A1 active site cavity is reconfigured to bind larger and nonplanar compounds. The shape and size of the cavity are controlled by structural elements in the active site roof, with major changes in the conformation of the F helix break and relocation of Phe224 from the active site to the protein surface. This altered CYP1A1 active site architecture is consistent with the proposed mechanism for CYP1A1 generation of an unusual aminoazepane-rearranged metabolite for this substrate.
SIGNIFICANCE STATEMENT
Cytochrome P450 1A1 metabolizes drugs, procarcinogens, and toxins and although previous structures have revealed how its stereotypical planar, aromatic compounds are accommodated in the CYP1A1 active site, this is not the case for flexible and nonplanar compounds. The current work determines the X-ray structure of CYP1A1 with such a flexible, nonplanar Pim kinase inhibitor, revealing significant modification of the CYP1A1 roof that accommodate this preclinical candidate and support an unusual intramolecular rearrangement reaction.
Introduction
Many human cytochrome P450 enzymes are promiscuous monooxygenases capable of metabolizing chemically and morphologically diverse sets of drugs, environmental contaminants, and endogenous compounds. While some generalizations can be made about the substrate scope of an individual P450 enzyme based on known active site characteristics such as its size and features of the residues lining the cavity (Johnson and Stout, 2013; Scott and Godamudunage, 2018), there are also many examples of validated substrates and inhibitors that do not conform to known active site boundaries. This indicates that we do not yet understand the full range of active site malleability for different human P450 enzymes. These limitations in our knowledge of the conformational flexibility of a P450 enzyme significantly retard the utility of structure to predict drug metabolism, carcinogen/toxin activation, and to design selective P450 inhibitors.
Cytochrome P450 1A1 (CYP1A1) is a human enzyme most well known for its role in chemical carcinogenesis (Rendic and Guengerich, 2012). Until recently there was only a single structure of human CYP1A1 available (Walsh et al., 2013). This structure, solved with the compact, highly planar ligand α-naphthoflavone (molecular weight 272 g/mol) revealed a spatially complementary enclosed, highly planar active site. This cavity is compatible with a subset of known CYP1A1 ligands (Walsh et al., 2013) with low molecular weights (200–500 g/mol) and compact structures like benzo[a]pyrene (252 g/mol), but not larger and nonplanar CYP1A1 substrates. Although CYP1A1 has not historically been recognized for playing an extensive role in drug metabolism, Jungmann and Lang 2019 recently reported that screening a library of 650 drugs revealed ∼35% had appreciable metabolism by CYP1A1. This diverse library probably represents the most unbiased evaluation of CYP1A1 substrates. Those with appreciable metabolism typically have molecular weights from 200 to 500 g/mol, but surprising exceptions of larger compounds include the macrolide antibiotic troleandomycin (813 g/mol), the muscle relaxant atracurium (929 g/mol), the macrolide antibiotic tylosin (916 g/mol), and even the immunosuppressant cyclosporin A (1202 g/mol) and the decapeptide gonadotropin releasing hormone agonist gosrelin (1269 g/mol). Clearly, conformational changes must occur to accommodate such ligands. Recently, two additional structures of CYP1A1 were solved with two ligands larger than α-naphthoflavone that had both planar and nonplanar constituents, the natural product bergamottin (338 g/mol) and the tyrosine kinase inhibitor erlotinib (393 g/mol) (Bart and Scott, 2018). These structures revealed conformational alterations, allowing for a more extended substrate profile for CYP1A1. The structure with erlotinib was particularly important because CYP1A1 is known to bioactivate tyrosine kinase inhibitor drugs into reactive metabolites that have been implicated in patient toxicities (Li et al., 2009, 2010; Lin et al., 2017). However, all three structures of CYP1A1 demonstrated a planar active site and thus did not answer the question of how nonplanar ligands are accommodated.
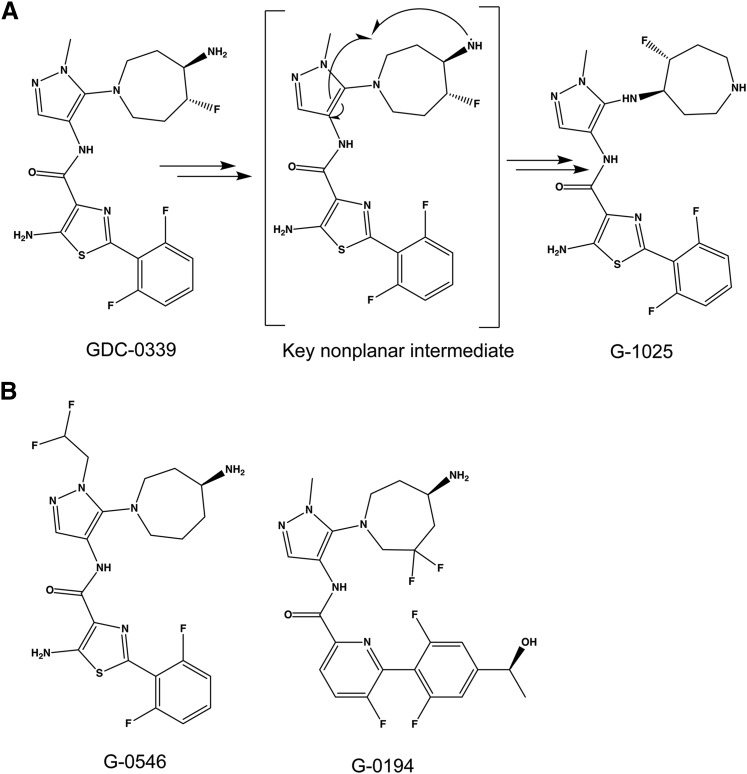
Genentech researchers recently demonstrated that CYP1A1 could efficiently metabolize the pan Pim kinase inhibitor GDC-0339 [5-amino-N-(5-((4R,5R)-4-amino-5-fluoroazepan-1-yl)-1-methyl-1H-pyrazol-4-yl)-2-(2,6-difluorophenyl)thiazole-4-carboxamide] (Takahashi et al., 2017). CYP1A1 converted the flexible, nonplanar GDC-0339 into an unusual aminoazepane-rearranged metabolite (G-1025, Fig. 1A), with the proposed mechanism requiring a highly nonplanar intermediate (Takahashi et al., 2017). This intramolecular rearrangement was difficult to rationalize based on any of the currently known CYP1A1 structures with highly planar active sites. To help understand how CYP1A1 is able to accommodate this larger and potentially non-planar substrate, CYP1A1 was co-crystallized with GDC-0339. The observed orientation of GDC-0339 in CYP1A1 is consistent with initiation of the proposed rearrangement reaction mechanism (Takahashi et al., 2017) and also reveals the ability of CYP1A1 to deviate from the previously observed planar active site through conformational flexibility involving the F helix portion of the active site roof. This new structure furthers our understanding of the plasticity of this human enzyme, revealing how CYP1A1 accommodates and metabolizes a more diverse substrate profile than could be previously rationalized.
Fig. 1.
Pim kinase inhibitor GDC-0339, its metabolite G-1025, and analogs. (A) GDC-0339 is reported to undergo a CYP1A1-mediated rearrangement to G-1025 via a key nonplanar intermediate. (B) Structures of close-in analogs G-0546 and G-0194 used for spectral binding experiments.
Materials and Methods
Materials.
GDC-0339 and two structurally related compounds, G-0546 and G-0194 (Fig. 1B), were synthesized at Genentech Inc. (South San Francisco, CA) and provided for research purposes.
Protein Expression and Purification.
Human CYP1A1 was generated using a construct containing a synthetic, codon-optimized gene with an N-terminal truncation and C-terminal 6x histidine tag, as described previously (Walsh et al., 2013). Expression and purification of the CYP1A1 construct was performed without modification as described (Bart and Scott, 2018). Protein purity was assessed by SDS-PAGE. The reduced carbon monoxide-difference spectrum was used to evaluate protein quality (Omura and Sato, 1964). Protein concentration was quantified by UV-visible spectroscopy from the absorbance of the Soret peak with an extinction coefficient of 100 mM−1cm−1 in a buffer containing 50 mM potassium phosphate, 500 mM NaCl, 20% (v/v) glycerol, 250 mM imidazole, 0.5% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid, 1 mM EDTA, pH 7.4.
Ligand Binding Assays.
Assays to measure ligand binding to CYP1A1 were performed in tandem cuvettes as described previously (Bart and Scott, 2018). Ligand binding data were determined in duplicate. Each of the ligands examined herein bound tightly enough (Kd values < 400 nM) that significant ligand is depleted from solution, and the assumption that ligand concentration does not substantially change is not valid. Thus the quadratic equation for tight binding ligands was used to fit the data (DeVore et al., 2009).
Crystallization, Data Collection, and Structure Determination.
Co-crystallization of purified CYP1A1 with the pan Pim kinase inhibitor GDC-0339 was performed as previously described (Bart and Scott, 2018), with a few modifications as listed below. Purified CYP1A1 was saturated with ligand by addition of 100 μM GDC-0339 (stock in 100% dimethylsulfoxide) to dilute protein and subsequently concentrated by centrifugal ultrafiltration to 20 mg/ml, measured by the Soret peak in the buffer described above. Crystals of CYP1A1 with GDC-0339 were grown using the sitting-drop vapor diffusion method in 96-well plates by mixing 0.75 μl CYP1A1 saturated with GDC-0339 with 0.75 μl crystallization solution (0.2 M potassium phosphate dibasic, 20% PEG 3350, 10% glycerol, 1 mM hexadecyltrimethylammonium bromide). Triangular prism-shaped crystals grew over 3–5 days and were cryoprotected using the crystallization solution plus 20% glycerol before being flash cooled in liquid nitrogen. A dataset was collected on a single crystal at Advanced Photon Source LS-CAT beamline 21-ID-G (Table 1). Data processing was performed using HKL2000 (Otwinowski and Minor, 1997), and an initial structure solution was obtained by molecular replacement via Phaser (McCoy et al., 2007) with the CYP1A1/bergamottin structure (Bart and Scott, 2018) (PDB 6DWM, molecule A), with a log likelihood score of 1244. This solution placed two copies of CYP1A1 in the asymmetric unit, chains A and B. This partial structure, along with the PDB 6DWM, molecule A, was used in Phaser to find a third copy, chain C, with a log likelihood score of 6549. Additional electron density and packing indicated the presence of a fourth copy of CYP1A1 in the asymmetric unit, chain D, which was modeled by partially building helices and then using the secondary structure matching algorithm in COOT to superpose the competely built chain A onto the partial model for chain D. This was followed by a round of simulated annealing using torsion angles in PHENIX. Manual model building and refinement were performed using COOT (Emsley et al., 2010) and PHENIX (Adams et al., 2010), respectively (Table 1). Coordinates and restraints for GDC-0339 were generated using PHENIX eLBOW (Moriarty et al., 2009) with AM1 geometry optimization. Calculation of the active site void volume was performed using VOIDOO (Kleywegt and Jones, 1994), with probe radius set to 1.4 Å and grid spacing of 1.0. Crystal structure figures were prepared using PyMOL (DeLano, 2017).
TABLE 1.
Data collection and refinement statistics
| CYP1A1/GDC-0339 (6O5Y) | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions (Å) | 65.15, 195.90, 236.98 |
| Molecules/a.u. | 4 |
| Resolution (Å)a | 50.00–3.16 (3.21–3.16) |
| Total reflectionsa | 343,766 (13,457) |
| Unique reflectionsa | 52,336 (2539) |
| Redundancya | 6.6 (5.3) |
| Rpima | 0.102 (0.786) |
| <I/σ(I)>a | 16.5 (1.8) |
| CC1/2a | 0.991 (0.490) |
| Completeness (%)a | 99.6 (97.6) |
| Refinement | |
| Resolution (Å) | 49.32–3.17 |
| No. reflections | 52,207 |
| R/Rfree (%) | 24.0/26.7 |
| Ramachandran (%) Favored/Allowed/Outliers | 92.39/6.90/0.71 |
| No. non-H atoms/B factors (Å2) | |
| Protein | 14,918/81.27 |
| Ligand | 32/87.42 |
| Heme | 172/68.31 |
| NO3 | 4/64.02 |
| Water | 10/61.61 |
| RMSD bond (Å) | 0.005 |
| RMSD angle (°) | 0.706 |
| Coordinate error (max. Likelihood based) (Å) | 0.43 |
Statistics for highest resolution shell shown in parentheses.
Results
Binding of GDC-0339 and Analogs to CYP1A1.
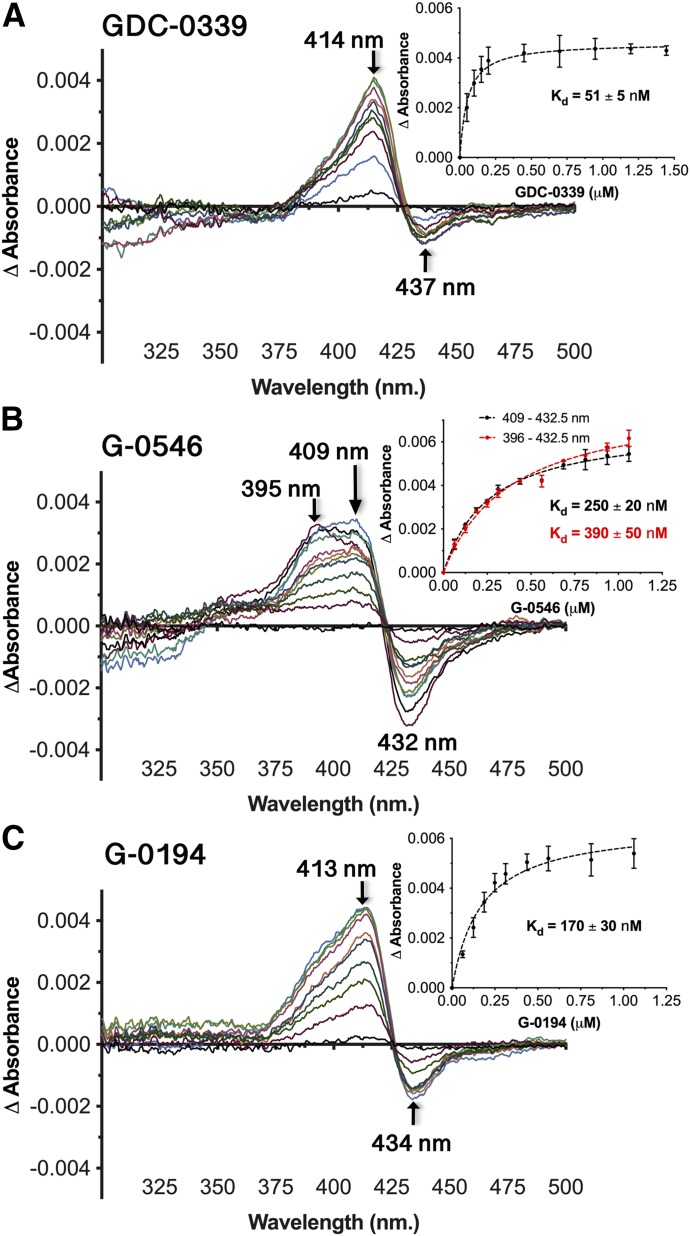
Spectral binding experiments were employed to examine further the binding mode and affinity of GDC-0339 toward human CYP1A1. Titrations of GDC-0339 to CYP1A1 resulted in unusual spectral shifts, with the difference spectra revealing an absorbance peak emerging at approximately 414–415 nm and a shallow trough at 437 nm (Fig. 2A). These changes do not correspond to classic P450 ligand binding modes of Type I, Type II, or reverse Type I. However, fitting these spectral changes required the tight-binding equation and revealed that CYP1A1 has very high-affinity for GDC-0339 at 51 ± 5 nM (Fig. 2A, inset). By comparison, bergamottin and erlotinib, two other good CYP1A1 substrates, bind with affinities of 370 nM and 1 µM, respectively (Bart and Scott, 2018).
Fig. 2.
CYP1A1 (1 μM) spectral binding assays with GDC-0339 (A), G-0546 (B), and G-0194 (C) result in atypical binding response. Insets, the peak-trough absorbance change vs. compound concentration fit by nonlinear regression to a tight-binding equation. Kd is given ±S.E. (error bars).
Binding assays were also performed with two analogs of GDC-0339 (Fig. 1B) to determine if various extensions on this ligand framework could also be accommodated by CYP1A1. G-0546 was selected to probe the impact of an increased bulk such as a difluoromethyl group on the pyrazole moiety relatively close to the center of rearrangement. The CYP1A1 catalyzed rearrangement was observed for the methyl (in place of difluoromethyl) analog of G-0546 (Takahashi et al., 2017). Titrations of G-0546 resulted in a broad peak, which has approximate λmax values of 409 nm and 395–396 nm, and a pronounced trough at 432–433 nm (Fig. 2B). This spectral response is consistent with G-0546 binding in the active site and furthermore suggests multiple binding modes, with the λmax of 409 nm most similar to the observations with GDC-0339 and the λmax at 395–396 more representative of a typical Type I binding profile where the ligand displaces a water from the heme iron. If the amino-azepane moiety of G-0546 is directed toward the heme, as the metabolite G-1025 suggests, addition of the bulky difluoromethane group on the pyrazole could logically alter the ligand’s spatial relationship to the heme iron, giving rise to the classic Type I character. The binding affinity of G-0546 was 250 ± 20 nM (using the absorbance at 409 nm) or 390 ± 50 nM (using the absorbance at 396 nm) (Fig. 2B inset). Both Kd values indicate that the substitutions in G-0546 (Fig. 1B) weaken the binding in comparison with the 1A1/GDC-0339 complex (Kd ∼50 nM).
The second analog tested was G-0194 (Fig. 1B), which differs from GDC-0339 in three areas: 1) G-0194 replaces the central aminothiazole with a slightly larger fluoropyridine and removes the aniline NH2 group; 2) G-0194 adds a second fluorine and both fluorine groups are at the γ-position of the amine group on the amino-azepane ring in contrast to the β-position in GDC-0339; 3) G-0194 adds a hydroxyethyl group extending from the difluorophenyl moiety. Knowing that the γ-difluoro-amino-azepane moiety could still rearrange, we were interested to know how the increased diversity away from the center of rearrangement could impact CYP1A1’s binding and catalysis. Spectral changes from G-0194 titrations to CYP1A1 resulted in responses similar to GDC-0339, where difference spectra evolved an absorbance peak at 413–414 nm and a smaller trough at ∼434 nm (Fig. 2C). The broader, bifurcated peak observed for G-0546 was not present. This is consistent with the major changes in structure of G-0194 are farther from the amino-azepane epicenter. The Kd for G-0194 binding to CYP1A1 was 170 ± 30 nM (Fig. 2C, inset), which is intermediate between the other two compounds examined herein. Due to multiple changes to the analogs, it is unknown which specific alterations are attributed to effects on binding mode and/or affinity.
CYP1A1 Structure with GDC-0339.
Since CYP1A1 produced a unique rearranged GDC-0339 metabolite and unusual spectral behavior upon binding, an X-ray structure was determined using co-crystallization (3.17 Å). The space group and unit cell dimensions were similar to previously determined CYP1A1 structures (Walsh et al., 2013; Bart and Scott, 2018). There were four copies of the protein within the asymmetric unit, with weaker electron density for chains C and D.
Among these four copies of the CYP1A1 protein, there are three types of structures. Molecule B has some density for only the ligand azepane ring, located low in the active site just above the heme, consistent with partial occupancy of GDC-0339, and the conformation of the F helix forming part of the active site roof is similar to that observed in previous structures, most similar to the CYP1A1 structure with α-naphthoflavone bound. Molecules C and D have essentially no active site density, consistent with low occupancy for the ligand in the active site and the poor density for side chains in the F helix, precluding definition of their orientation either into or out of the active site. However, the active site of molecule A contains electron density clearly defining an elongated conformation of the GDC-0339 ligand in concert with significant corresponding differences in the F helix and upper active site (Fig. 3).
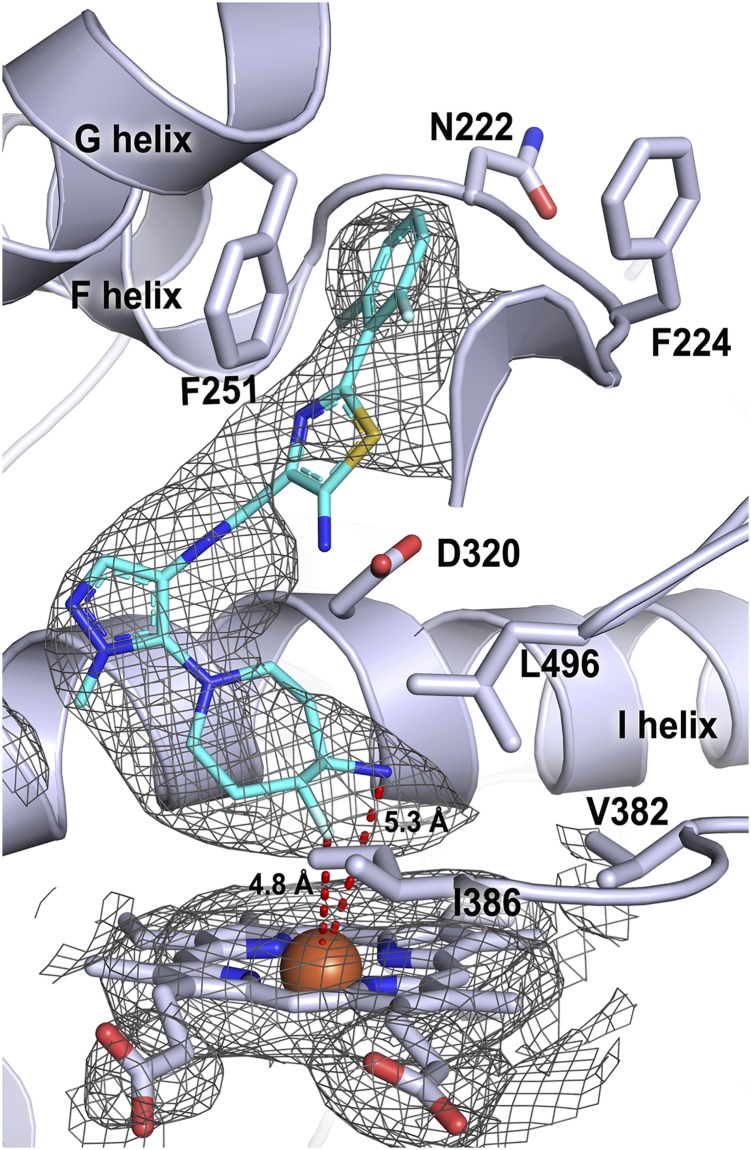
Fig. 3.
CYP1A1 (light blue ribbons and sticks) active site with GDC-0339 (cyan sticks) bound. Heteroatoms are as indicated: nitrogen, royal blue; oxygen, red; sulfur, yellow; fluorine, light blue; iron, rust. Electron density is shown as 2Fo – Fc composite omit map contoured at 1.0 σ (gray mesh) for ligand and heme. Distances from the heme iron to the closest GDC-0339 atoms are shown by red dashed lines and labeled with the respective distances.
Correct orientation of GDC-0339 within the active site density was aided by the large, seven-membered fluoro-amino-azepane ring. This ring system is located closest to the heme with the fluorine and primary amine approximately 4.8 and 5.3 Å from the iron, respectively (Fig. 3). This binding orientation is consistent with the mechanism proposed by Takahashi et al. where production of the rearranged metabolite was initiated by hydrogen abstraction at the primary amine on the fluoro-amino-azepane (Takahashi et al., 2017). Moving away from the heme-oriented fluoro-amino-azepane ring, the ligand methyl-pyrazole is directed toward the central I helix (Fig. 3). The subsequent amide linkage directs both the aminothiazole and difluorophenyl moieties of ligand toward the roof of the active site and the F helix. The aminothiazole has less electron density, suggesting that it may adopt multiple conformations. Regardless, the ligand aminothiazole and difluorophenyl rings occupy the space where Phe224 and the protein backbone between the two halves of the F helix normally reside in structures with other ligands.
Despite the high affinity, as measured in spectral assays here and reflected in efficient GDC-0339 turnover by and potent competitive inhibition for CYP1A1 (Takahashi et al., 2017), there are relatively few specific interactions between GDC-0339 and CYP1A1. The fluorine group on the fluoro-amino-azepane is confined by hydrophobic residues such as Val382 and Leu496 from the K/β1-4 and β4 loops near the bottom of the active site (Fig. 3). The aromatic thiazole ring has favorable pi-pi edge-to-face interaction with Phe251 on the G helix (Fig. 3). Overall, slightly weaker density is observed for the aminothiazole and fluoropyridine moieties of GDC-0339 as it begins to extend toward the roof and edge of the active site bounds. This suggests less complete immobilization of the ligand in this area, whereas the methyl-pyrazole and fluoro-amino-azepane moieties lie in a more confined environment near the heme.
Discussion
Many CYP1A1 substrates share physicochemical features of high planarity, aromaticity, small size, and compact shape. The structural basis for this profile was evident when the first CYP1A1 structure, bound to α-naphthoflavone, was solved (Walsh et al., 2013). This structural architecture was also observed in the other family 1 human P450 enzymes, 1A2 (Sansen et al., 2007) and 1B1 (Wang et al., 2011). Synthetic derivatives based on the flavonoid core like 4′-propargyloxy-β-naphthoflavone (Liu et al., 2013) and tyrosine kinase inhibitors (Li et al., 2010; Lin et al., 2017; Bart and Scott, 2018) suggested the capacity of CYP1A1 to accommodate more elongated ligands, which still maintain a predominately planar shape. A recent report revealed CYP1A1-mediated metabolism of the flexible, nonplanar Pim-kinase inhibitor GDC-0339. This compound inhibited CYP1A1 competitively and with greater potency than α-naphthoflavone when phenacetin was used as a substrate (Takahashi et al., 2017), indicating GDC-0339 binds in close proximity to the heme and with high affinity. Based on the rearranged metabolite and structural analogs of GDC-0339, Takahashi et al. (2017), proposed that CYP1A1 initiated the reaction through hydrogen abstraction from the primary amine on the fluoro-amino-azepane moiety, indicating this group on GDC-0339 would be close to the heme iron. This rearrangement mechanism involves the compound folding on itself in the active site with interactions between this primary amine radical and the adjacent methylpyrazole (Takahashi et al., 2017), also strongly suggesting the CYP1A1 active site would have to be substantially reconfigured to allow such a reaction.
Binding of GDC-0339 and Analogs to CYP1A1.
In general, the bulkier analogs had decreased affinity to CYP1A1 compared with GDC-0339, although binding was still in the nanomolar range. The main spectral binding changes induced by all three of these compounds do not correspond to common P450 binding modes. They are most similar to observations reported for CYP46A1 binding bicalutamide (Mast et al., 2013). In this case there is crystallographic evidence of the bicalutamide cyano group hydrogen bonding to a water molecule, which is in turn coordinated to the heme iron. While it was possible that the primary amine of the fluoro-amino-azepane also hydrogen bonds to a water coordinated to the heme iron, this is not consistent with the X-ray structure herein. Addition of difluoromethane on the pyrazole adjacent to the fluoro-amino-azepane ring in G-0546 might reposition the ligand, consistent with the addition of a classic Type I response for displacement of the heme-coordinated water. For all three ligands, the net absorbance change in the spectral binding experiments is low. This suggests a low proportion of CYP1A1 is binding GDC-0339 despite high affinity and is consistent with ligand occupancy in only one of the four CYP1A1 molecules in the asymmetric unit.
CYP1A1 Structure with GDC-0339.
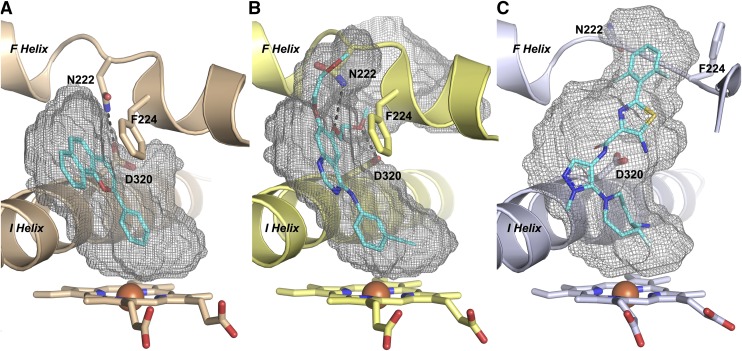
Since the top of the GDC-0339 ligand occupies the space where the F helix normally resides in structures with other ligands, this region is substantially reconfigured (Fig. 4). As in most human P450 enzymes, this F helix forms the roof of the active site. In CYP1 enzymes there is typically a break in this helix as it passes over the active site. The positioning of this break is key in determining whether the CYP1A1 active site is enclosed or open to the surface and its preference for planar ligands. In the α-naphthoflavone complex (Walsh et al., 2013), F helix residue Asn222 hydrogen bonds to I helix Asp320 to close off access to the protein exterior between these two structural elements (Fig. 4A). In the bergamottin complex, these two side chains torsion so that there is no longer a hydrogen bond between them and a little additional space is made available to accommodate the furanocoumarin ring (Bart and Scott, 2018). In the erlotinib complex, both Asn222 and Asp320 torsion farther apart, opening up a channel between them all the way to the protein surface (Fig. 4B). In each of these latter two structures, Asn222 instead interacts with various oxygens in the ligands and the torsions of the side chains occur without changing the backbone conformation of the F helix break (Fig. 4B). However, in this new GDC-0339 structure, the protein backbone between the two halves of the F helix adopts an entirely new conformation, with the Asn222 side chain disordered on the protein surface and not interacting with the ligand at all (Fig. 4C). The second important impact of F helix break reconfiguration is positioning of Phe224. In all other known CYP1A1 structures, the F helix backbone conformation positions Phe224 to project into the active site, pi-pi stacking with the planar portions of ligands (Fig. 4, A and B). This is a key feature reinforcing planarity in the middle and top portions of the CYP1A1 active site. With nonplanar GDC-0339, however, the new conformation of the F helix break instead projects Phe224 in the opposite direction, projecting from the protein surface toward solvent (Fig. 4C). Distances between the nearest side chain atoms of Asp320 and Asn222 vary from 3.5 Å in the hydrogen bond in the α-naphthoflavone structure to 6.2 Å in the erlotinib structure to 10 Å in the GDC-0339 structure.
Fig. 4.
Comparison of active-site cavities for CYP1A1 structures with α-naphthoflavone (A), erlotinib (B), and GDC-0339 (C). Heteroatoms are as indicated: nitrogen, blue; oxygen, red; sulfur, yellow; fluorine, light blue; iron, rust. The F helix break conformation in the CYP1A1/GDC-0339 structure results in both Phe224 and Asn222 positioned outside of the active site, with increased width of the cavity volume compared with the much more planar nature of the CYP1A1 structures and active sites shown in (A and B). Black dashed lines indicate hydrogen-bonds.
Restructuring of the F helix break as observed in the CYP1A1/GDC-0339 structure and repositioning Phe224 outside the active site significantly expand the active site cavity. Comparisons to other CYP1A1 co-crystal structures (Fig. 4, gray mesh) reveal that the volume of the CYP1A1/GDC-0339 active site at 820 Å3 is much larger than structures with α-naphthoflavone (408 Å3, Fig. 4A) or bergamottin (456 Å3) and similar to erlotinib (806 Å3). Although the CYP1A1 structures with GDC-0339 and erlotinib structures have similar cavity volumes, the identity of the repositioned residues and structural elements differed and the shape of the upper active site/channel is different. Expansions in the CYP1A1/erlotinib active site cavity were largely a result of torsioning of Asn222 (F helix break), Asp320 (I helix), and Leu254 (G helix), with an additional offset of the G helix away for the active site (Fig. 4B). In contrast, the CYP1A1/GDC-0339 active site increased substantially more in width at about the level of the aminothiazole ring (Fig. 4C).
This increased width of the middle of the active-site cavity is interesting in the context of the proposed GDC-0339 rearrangement mechanism (Takahashi et al., 2017). The mechanism that was proposed to form the rearranged metabolite would proceed through an intramolecular tetrahedral intermediate occurring between the pyrazole and primary amine radical (Fig. 1A), which likely would have to be accommodated within the active site. The reconfiguration of the F helix break, to swing Phe224 outside the active site cavity may not only accommodate the nonplanar substrate, but also permit the formation of this three-dimensional intermediate. Therefore, the F helix break conformation observed in this crystal structure might help prime CYP1A1 for accommodating large deviations to the normally highly planar active site, such as the GDC-0339 rearrangement intermediate. Notably none of the five residues in the middle of this F helix break are conserved in human CYP1A2, which could correlate with the selectivity for GDC-0339 metabolism by CYP1A1 over CYP1A2 (Takahashi et al., 2017). The F helix break in CYP1A2 may not be able to adopt a conformation consistent with the binding of GDC-0339 in this orientation.
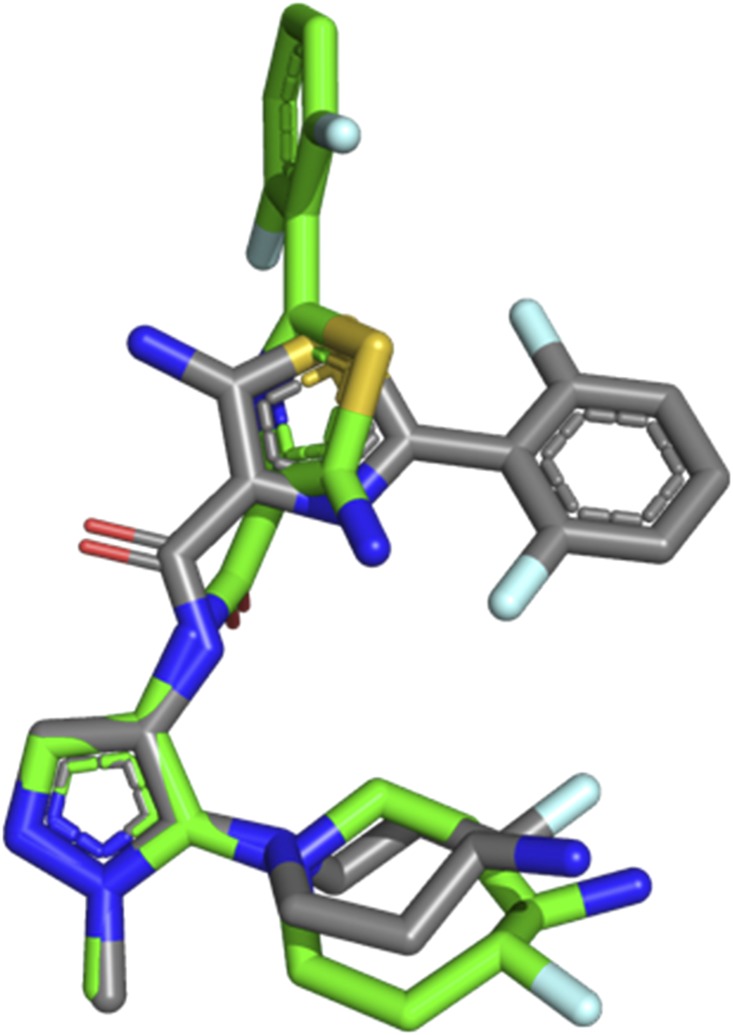
The therapeutic target for GDC-0339 are the Pim kinases, where the ligand is a potent pan-inhibitor designed to interact with the ATP binding pocket. Recently a structure of GDC-0339 bound to Pim-1 was reported (Wang et al., 2019). When bound to Pim-1, GDC-0339 adopts a compact U-shaped conformation (Fig. 5, gray sticks). In contrast when GDC-0339 is bound to CYP1A1, the conformation is much more extended (Fig. 5, green sticks). GDC-0339 is metabolized efficiently by CYP1A1 to form the rearranged metabolite G-1025, which displays 10- to 20-fold reduction in potency toward Pim isoforms (Takahashi et al., 2017). This likely results from loss of key salt-bridges formed between the primary amine on the fluoro-amino-azepane with an acidic region in the Pim kinase (Wang et al., 2019). Engineering conformational constraints into GDC-0339 to favor the U-shaped Pim-compatible conformation may be desirable to reduce its binding to and metabolic deactivation by CYP1A1, as a strategy to sustain its efficacy and avoid multiple myeloma drug resistance. CYP1A1 expression levels and activity have previously been shown to have a resistance role in for the multiple myeloma drug bortezomib (Xu et al., 2012).
Fig. 5.
Comparison of the GDC-0339 conformation when bound to CYP1A1 (green sticks) vs. the conformation bound to Pim-1 (gray sticks, PDB Code: 6NO9) reveals significant conformational differences that could be used to decrease CYP1A1 metabolism. Heteroatoms are as indicated: nitrogen, blue; oxygen, red; sulfur, yellow; fluorine, light blue.
In conclusion, our structural investigations into CYP1A1 revealed that this enzyme has greater degrees of conformational malleability than previously observed, permitting the binding and metabolism of nonplanar compounds and their likely intermediates. Specifically, this structure demonstrates the CYP1A1 active site can increase approximately twice in volume and rearrange both polar and aromatic residues. Even larger nonplanar CYP1A1 substrates such as the macrolide antibiotics troleandomycin and tylosin, the immunosuppressant cyclosporin A, and the decapeptide gosrelin may take advantage of this nonplanarity and/or protrude outside of the active site through the channel observed. The roles observed for CYP1A1 in chemical carcinogenesis and the generation of reactive metabolites underscore the importance in predicting chemical liabilities of various dietary compounds, pollutants, and drugs. This more complete understanding of the structural elements contributing to flexibility within the CYP1A1 active site should help in predicting a broader range of substrates potentially susceptible to CYP1A1 bioactivation, and in some cases aid in engineering drugs to remove CYP1A1-mediated metabolic liabilities.
Acknowledgments
GDC-0339, G-0546, and G-0194 were kindly provided as gifts from Genentech. Crystallization employed instrumentation in the University of Michigan Center for Structural Biology (CSB). The CSB is grateful for support from the U-M Life Sciences Institute, the U-M Rogel Cancer Center, the U-M Medical School Endowment for the Basic Sciences, and grants from the National Institutes of Health. X-ray diffraction data were collected at Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Abbreviations
- CYP1A1
cytochrome P450 1A1
- GDC-0339
[5-amino-N-(5-((4R,5R)-4-amino-5-fluoroazepan-1-yl)-1-methyl-1H-pyr-azol-4-yl)-2-(2,6-difluorophenyl)thiazole-4-carboxamide
Authorship Contributions
Participated in research design: Bart, Takahashi, Wang, Scott.
Conducted experiments: Bart.
Contributed new reagents or analytical tools: Wang.
Performed data analysis: Bart, Scott.
Wrote or contributed to the writing of the manuscript: Bart, Takahashi, Wang, Scott.
Footnotes
This work was supported by National Institutes of Health (NIH) [Grant R37 GM076343 to E.E.S.]. A.G.B.’s training was partially supported by the NIH [T32 GM008545]. The content of this publication is solely the responsibility of the authors and do not necessarily represent the official views of NIH.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart AG, Scott EE. (2018) Structures of human cytochrome P450 1A1 with bergamottin and erlotinib reveal active-site modifications for binding of diverse ligands. J Biol Chem 293:19201–19210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. (2017) The PyMOL Molecular Graphics System, Schrodeinger, LLC, New York. [Google Scholar]
- DeVore NM, Smith BD, Wang JL, Lushington GH, Scott EE. (2009) Key residues controlling binding of diverse ligands to human cytochrome P450 2A enzymes. Drug Metab Dispos 37:1319–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EF, Stout CD. (2013) Structural diversity of eukaryotic membrane cytochrome p450s. J Biol Chem 288:17082–17090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann N, Lang D. (2019) Drug library screening for human CYP1A1 substrates, in 12th International Society for the Study of Xenobiotics; 2019 July 28–31; Portland, OR, pp 55. [Google Scholar]
- Kleywegt GJ, Jones TA. (1994) Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr 50:178–185. [DOI] [PubMed] [Google Scholar]
- Li X, Kamenecka TM, Cameron MD. (2009) Bioactivation of the epidermal growth factor receptor inhibitor gefitinib: implications for pulmonary and hepatic toxicities. Chem Res Toxicol 22:1736–1742. [DOI] [PubMed] [Google Scholar]
- Li X, Kamenecka TM, Cameron MD. (2010) Cytochrome P450-mediated bioactivation of the epidermal growth factor receptor inhibitor erlotinib to a reactive electrophile. Drug Metab Dispos 38:1238–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Kostov R, Huang JTJ, Henderson CJ, Wolf CR. (2017) Novel pathways of ponatinib disposition catalyzed by CYP1A1 involving generation of potentially toxic metabolites. J Pharmacol Exp Ther 363:12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Taylor SF, Dupart PS, Arnold CL, Sridhar J, Jiang Q, Wang Y, Skripnikova EV, Zhao M, Foroozesh M. (2013) Pyranoflavones: a group of small-molecule probes for exploring the active site cavities of cytochrome P450 enzymes 1A1, 1A2, and 1B1. J Med Chem 56:4082–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast N, Zheng W, Stout CD, Pikuleva IA. (2013) Binding of a cyano- and fluoro-containing drug bicalutamide to cytochrome P450 46A1: unusual features and spectral response. J Biol Chem 288:4613–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. (2007) Phaser crystallographic software. J Appl Cryst 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty NW, Grosse-Kunstleve RW, Adams PD. (2009) electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr 65:1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura T, Sato R. (1964) The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239:2370–2378. [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. [DOI] [PubMed] [Google Scholar]
- Rendic S, Guengerich FP. (2012) Contributions of human enzymes in carcinogen metabolism. Chem Res Toxicol 25:1316–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, Johnson EF. (2007) Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J Biol Chem 282:14348–14355. [DOI] [PubMed] [Google Scholar]
- Scott EE, Godamudunage MP. (2018) Structures of human cytochrome P450 enzymes: variations on a theme, in Dioxygen-Dependent Heme Enzymes (Ikeda-Saito M, Raven E. eds), pp 249–273, Royal Society of Chemistry, London. [Google Scholar]
- Takahashi RH, Wang X, Segraves NL, Wang J, Chang JH, Khojasteh SC, Ma S. (2017) CYP1A1-mediated intramolecular rearrangement of aminoazepane in GDC-0339. Drug Metab Dispos 45:1084–1092. [DOI] [PubMed] [Google Scholar]
- Walsh AA, Szklarz GD, Scott EE. (2013) Human cytochrome P450 1A1 structure and utility in understanding drug and xenobiotic metabolism. J Biol Chem 288:12932–12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Savas U, Stout CD, Johnson EF. (2011) Structural characterization of the complex between alpha-naphthoflavone and human cytochrome P450 1B1. J Biol Chem 286:5736–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Blackaby W, Allen V, Chan GKY, Chang JH, Chiang PC, Diène C, Drummond J, Do S, Fan E, et al. (2019) Optimization of pan-Pim kinase activity and oral bioavailability leading to diaminopyrazole (GDC-0339) for the treatment of multiple myeloma. J Med Chem 62:2140–2153. [DOI] [PubMed] [Google Scholar]
- Xu D, Hu J, De Bruyne E, Menu E, Schots R, Vanderkerken K, Van Valckenborgh E. (2012) Dll1/Notch activation contributes to bortezomib resistance by upregulating CYP1A1 in multiple myeloma. Biochem Biophys Res Commun 428:518–524. [DOI] [PubMed] [Google Scholar]