Abstract
Examining the link between neural activity, transcriptional output, and synaptic function offers unique insights into how neurons adapt to changing environments and form memories. Epigenetic markers, such as DNA methylation and histone modifications, have been implicated in the formation of not only cellular memories such as cell fate, but also memories of experience at the organismal level. Here, we review recent advances in chromatin regulation that contribute to synaptic plasticity and drive adaptive behaviors through dynamic and precise regulation of transcription output in neurons. We discuss chromatin-associated proteins, histone variant proteins, the contribution of cis-regulatory elements and their interaction with histone modifications, and how these mechanisms are integrated into distinct behavior and environmental response paradigms.
Introduction
Dynamic regulation of transcription is a critical component of neuronal function. Transcription is regulated by many mechanisms in neurons, including through chromatin, the complex of DNA and histone proteins that package DNA into higher-order structures and control gene accessibility. The importance of chromatin in neuroscience is becoming increasingly appreciated, in both vertebrates and invertebrates, for its function in memory formation to its involvement in a wide range of behaviors (1–3). Here we review work on the role of chromatin in mature and healthy neurons from the last five years. Given the rapid growth of this field, we can address only a subset of this research, but aim to provide an overview of exciting new advances at the intersection of chromatin biology and neuroscience.
Reading, writing, and erasing the chromatin landscape of neurons
The N-terminal tails of core histones and the H1 linker histone are host to a striking number of post-translational modifications including acetylation, phosphorylation, methylation, and more. The classes of enzymes implicated in the deposition of histone modifications, termed ‘writers’, and the removal of modifications, termed ‘erasers’ are critical to these processes. Histone modifications regulate histone-DNA interactions and contacts between nucleosomes to facilitate selective unraveling or compaction of chromatin structure. In addition, these modifications interface with other proteins, termed ‘readers’, which can recruit additional factors that further modify chromatin and affect the transcriptional state of the cell. Together, the readers, writers, and erasers play important roles in the dynamic nature of chromatin and transcription regulation. In neurons, considerable recent work has illuminated how these proteins contribute to synaptic plasticity and memory formation.
Readers contain conserved domains such as bromodomains, chromodomains, and PHD domains. These proteins play a critical role in bridging neuronal activation, histone modifications, and transcriptional output that drives memory formation. Recent work demonstrated an important role for readers in regulation of AMPA-type glutamate receptors, the main excitatory receptor in neurons. L3mbtl1 is a polycomb group protein and recognizes mono- and dimethylated histone residues (4). L3mbtl1 is decreased upon neuronal activity and is required for AMPA receptor regulation in homeostatic downscaling, the process through which neurons modify their synapses to adjust to long-term changes in activity. Similarly, Brd4, a reader of acetylated lysines, regulates transcription of activity-dependent genes as well as genes encoding AMPA receptor subunits (5*, 6*). Small molecule inhibition of Brd4 results in impairments in memory consolidation and decreased seizure susceptibility, suggesting that Brd4-mediated transcription regulates neuronal firing.
Deposition of histone marks by ‘writers’ is another important epigenetic regulator of transcription, and considerable research in past decades has focused on the role of histone modifications in neuronal activity-dependent gene expression (7, 8). Exciting recent work has elegantly dissected the roles of acetyltransferases and methyltransferases in distinct memory processes. Kmt2a is H3K4 methyltransferase whose loss in the prefrontal cortex in mice impairs working memory and leads to increased anxiety (9). Conditional knockout of Kmt2a (10**) and the related histone methyltransferase Kmt2b (11) in the hippocampus both result in decreased activity-dependent gene expression. Interestingly, these enzymes regulate distinct memory processes, as Kmt2a deficient mice show deficits in working memory, and Kmt2b mice exhibit impairments in object recognition memory. Further, they affect H3K4 trimethylation at distinct genomic regions, and loss of Kmt2a and Kmt2b lead to unique gene expression patterns (10**).
The importance of methyltransferase activity in neurons is also demonstrated by the polycomb repressive complex 2 (PRC2). PRC2 catalyzes H3K27 methylation to repress gene expression and is required to maintain neuronal function and survival in the adult brain (12). Acetyltransferases such as KAT2a in the brain also affects specific memory processes, and KAT5 is implicated in hippocampal memory enhancement (13, 14).
Removal of methyl and acetyl marks by ‘erasers’ is a critical mechanism to gate the appropriate transcription of plasticity genes. Erasers are often regulated in concert with writers to facilitate a highly dynamic chromatin state in response to neuronal stimulation. Neuron-specific alternative splicing of Lysine-Specific Demethylase 1 (LSD1) results in the neuroLSD1 isoform (also called LSD1n). While LSD1 mainly exhibits H3K9 demethylation, neuroLSD1 functions more as a H4K20 demethylase (15*). This altered demethylation profile regulates activity-dependent gene expression including genes required for learning and memory. Loss of neuroLSD1 results in spatial learning deficiencies and changes in stress responses (discussed later).
The histone deacetylases (HDACs) are generally described as suppressors of gene transcription and memory formation (7), and recent work has added new mechanistic insights into how blocking HDACs leads to memory enhancement. In a fear-induced learning model, HDAC3 inhibits memory by forming a complex with CREB to suppress transcription of activity-regulated genes implicated in synaptic plasticity (14). Upon fear training, HDAC3 is exchanged for the KAT5 acetyltransferase, which acetylates histones at the Fgf1b promoter, and promotes learning. HDAC3 has also been implicated in regulating synaptic plasticity in the primary auditory cortex during reward learning (16). However, histone tails are not the only substrates for HDACs which also deacetylate critical synaptic proteins, producing memory deficits in knockout animals (17, 18). The cytoplasmic and synaptic activities of HDACs underlie the importance of careful dissection of mechanism without bias towards well-studied histone functions.
While transcription regulation of synaptic function has been explored, in only some cases is it clear how the synapse signals to chromatin through specific kinases or signaling cascades (5, 19). It will be intriguing to further investigate how neuronal activity results in altered activity or expression of histone-associated proteins. Together, these exciting studies illuminate the carefully orchestrated interplay between histone readers, writers, and erasers and their important links to synaptic function and memory formation (Fig. 1). This recent work also demonstrates the remarkable degree to which closely related histone-modifying enzymes regulate distinct transcriptional programs in an activity-dependent manner to allow the brain to perform complex tasks such as memory formation.
Figure 1. Chromatin-associated proteins in learning and memory formation.
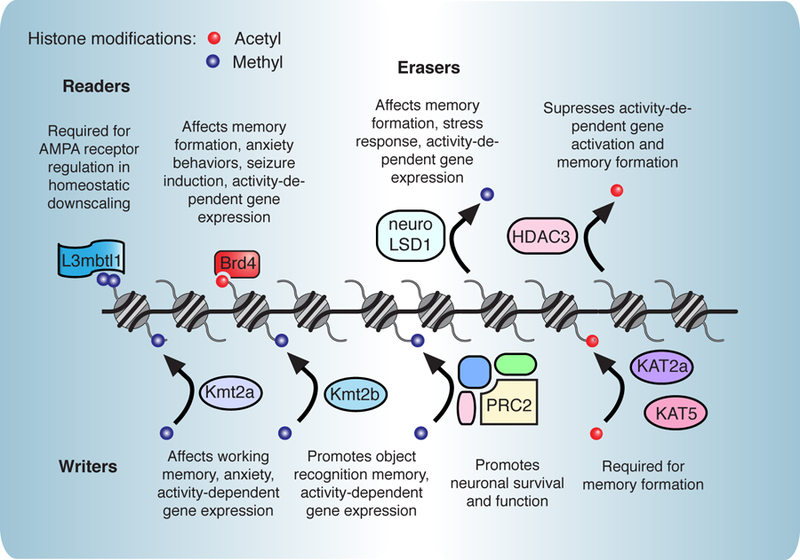
Numerous reader [4,5*,6*], writer [9,10**,11–14], and eraser [15*,16, 41] proteins have recently been implicated in neuronal function. In particular, these proteins regulate activity-dependent gene expression and key synaptic receptor genes. In addition, many have been implicated in various forms of synaptic plasticity and various types of learning and memory formation.
Histone variants in modulating neuronal plasticity
In addition to regulation by post-translational modifications, histone proteins themselves can also come in different ‘flavors’ called histone variants. These variants are encoded by separate genes and have distinct amino acid sequences from their canonical counterparts. Differences range from a few scattered amino acids to large-scale changes that modify the charge of critical regions of the histone. Unlike most histone proteins, several of these variants do not require cell replication to be incorporated into chromatin so are potentially highly important in postmitotic cells like neurons. Recent work has demonstrated that histone variants play a particularly critical role in the brain and are closely linked to transcriptional changes underlying learning and memory.
Histone variants H3.3, H2A.Z, and H2BE contribute to various aspects of neuronal function, from neuronal development to neuronal cell death (Fig. 2). One of the first histone variants studied in the brain was H2BE, a variant of the H2B histone (20). H2BE is expressed in the mouse olfactory bulb in neurons that receive low levels of input. Unlike much of the brain, the olfactory system uses cell turnover to continually adapt to new environments through a process controlled in part by H2BE-mediated cell death. While this extraordinary mechanism may be specific to the unique neurons of the olfactory system, this work sparked the wider study of histone variants throughout the brain.
Figure 2. The role of histone variants in neurons.
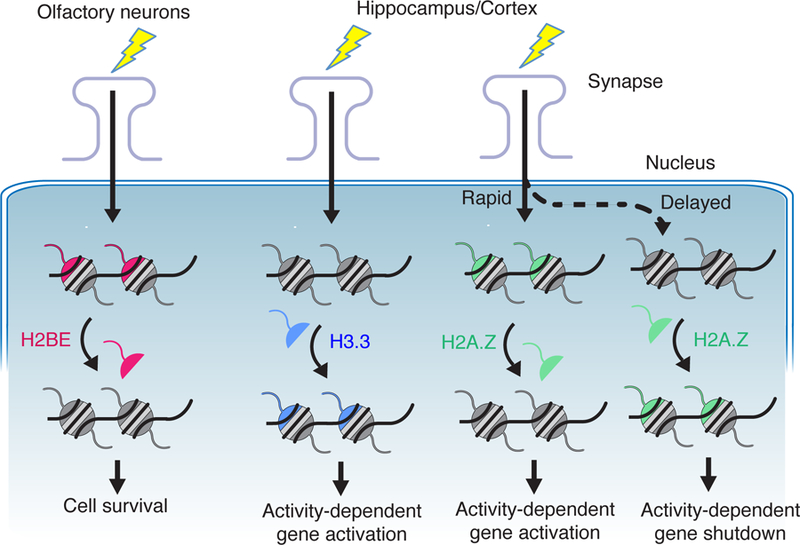
The histone variants H2BE, H3.3 and H2A.Z are involved in various aspects of neuronal function. In olfactory epithelial neurons, the variant H2BE is expressed at low levels in active neurons and at high levels in inactive neurons and regulates cell survival [20]. The variants H3.3 and H2A.Z are expressed throughout the brain and have opposing effects on the neuronal response to activity. Increased activity results in greater expression of H3.3, which contributes to activity-dependent gene expression [24**, 29]. H2A.Z is initially evicted from chromatin in response to activity which promotes gene activation and later is deposited in chromatin to shut down gene expression [21**, 22, 23, 24**,25,26**,27].
Exciting new research has explored the role of H2A.Z, a variant of H2A, in neuronal function and in regulation of activity-dependent gene expression. H2A.Z is actively exchanged with other histones in chromatin in response to learning paradigms. Intriguingly, knockdown of H2A.Z promotes stronger fear conditioning suggesting that H2A.Z restrains memory formation in the brain (21**), while brain-specific knockout early in development impairs neurogenesis and memory formation (22). As animals age, H2A.Z accumulates in the brain (21**), similar to other histone variants (24**). However, in both young and old mice, H2A.Z is evicted from genes during learning, though the genes regulated by H2A.Z change with aging (23). As an added layer of complexity, two different genes encode distinct versions of H2A.Z and these ‘hypervariants’ also appear to regulate different subsets of genes (25).
H2A.Z is deposited at the promoters of activity-dependent genes by the nucleosome remodeling and deacetylase (NuRD) complex (26**). Knockdown of one component of this complex, Chd4, decreases deposition of H2A.Z at specific genes, promotes their expression, and blocks dendritic pruning. This results in increased responsivity of neurons to sensorimotor inputs causing procedural learning deficits. The link between decreased H2A.Z deposition and increased recruitment of neurons that respond to specific stimuli could explain how H2A.Z loss could also lead to enhanced memory formation in other tests (21**). H2A.Z deposition is also regulated by the Tip60- p400 complex and inhibition of this complex also improves memory formation (27).
Similar to H2A.Z deposition, H2A.Z eviction is likely a highly regulated and activity-dependent process (21**) though is less well studied in neurons. However, monoubiquitination of histone H2B blocks access to the H2A.Z evictor INO80 at inducible enhancers in HEK293- T cells (28). Such a mechanism could apply to inducible genes in neurons as well.
The histone variant H3.3 is also incorporated into chromatin in response to increased neuronal activity (24**). H3.3 is incorporated throughout gene bodies while H2A.Z is primarily at promoters and H3.3 appears to function in a manner opposite to H2A.Z. Knockdown of H3.3 in postmitotic neurons decreased activity-dependent gene induction and caused deficits in learning and memory. More recently, this work has been extended to show that depression in humans and chronic stress in rodents can affect H3.3 levels (29).
Research on H2BE, H3.3 and H2A.Z has demonstrated that our environment and experiences control not just histone modifications but also the makeup of nucleosomes in our neurons. It is particularly intriguing to consider how these variants may work together to regulate gene expression. The opposing effects on gene expression of H3.3 and H2A.Z, particularly in response to neuronal activity, suggest a model in which neurons make use of multiple variants to balance their response to neuronal activity. Also intriguing is whether more replication-independent histones remain to be discovered. Whether other specific cell types or brain regions also have unusual histone variants like H2BE that allow neurons to perform their unique functions has yet to be seen.
Interplay of chromatin and neuronal activity-dependent enhancers
Enhancers are short (100–500 bp) regions in the genome that act as regulatory elements and recruit sequence-specific transcription factors (TFs). The binding of TFs to the correct enhancer is crucial in allowing any given cell type, such as neurons, to precisely activate cell type-specific genes at the right time and in response to the right signaling pathways (30). However, for a given enhancer to be activated, nucleosomes may need to be evicted or moved (31). Active enhancers are also marked by specific histone modifications on the surrounding nucleosomes, particularly H3K27ac and H3K4me1 (32).
A subset of enhancers control stimulus-dependent genes. In neurons, these genes, termed Immediate Early Genes (IEGs), are rapidly transcribed in response to neuronal activity and are critical for numerous neuronal functions (33). Such enhancers are marked by dynamic regulation of H3K27ac (34, 35) making this mark particularly important in ensuring that IEGs are activated as animals learn and store information. In addition, enhancer regions themselves are actually transcribed in the process of recruitment of TFs and polymerase, generating a class of RNAs termed enhancer RNAs (eRNAs) (36). eRNAs can then affect transcription through mechanisms such as regulating chromatin looping (37). The process of inducible acetylation, TF recruitment, eviction of nucleosomes, and eRNA transcription are all critical points of regulation in neuronal gene induction.
Recent work has shed light on how these processes are interconnected and converge in neurons to allow for highly specific transcriptional responses to synaptic activity. The enhancers regulating the IEG Fos can distinguish between different types of neuronal stimuli as seen by distinct patters of eRNA synthesis at different Fos enhancers (38*). This could allow for different stimuli to recruit TFs and chromatin-regulating proteins to distinct enhancer regions. Different durations of neuronal activity also result in different patterns of IEG activation with the most rapidly induced genes having the most open chromatin state indicated by various histone marks around gene promoters (39*). IEGs Fos and Jun are TFs that, once induced, can recruit BAF remodeling complexes to establish open chromatin domains (40). Interestingly, enhancer regions that recruit these TFs differ markedly between mouse strains implying that DNA sequence heterogeneity could affect chromatin remodeling in IEG activation. Enhancers can also counteract large heterochromatin domains to allow neurons to activate a single olfactory receptor gene in the midst of large regions of silenced genes (41). Taken together, these findings begin to hint at the complexity involved in the transcriptional response to any given neuronal stimuli, at least some of which is highly influenced by interplay of enhancers and the surrounding chromatin (Fig. 3).
Figure 3. Chromatin and enhancers co-regulate activity-dependent gene activation in neurons.
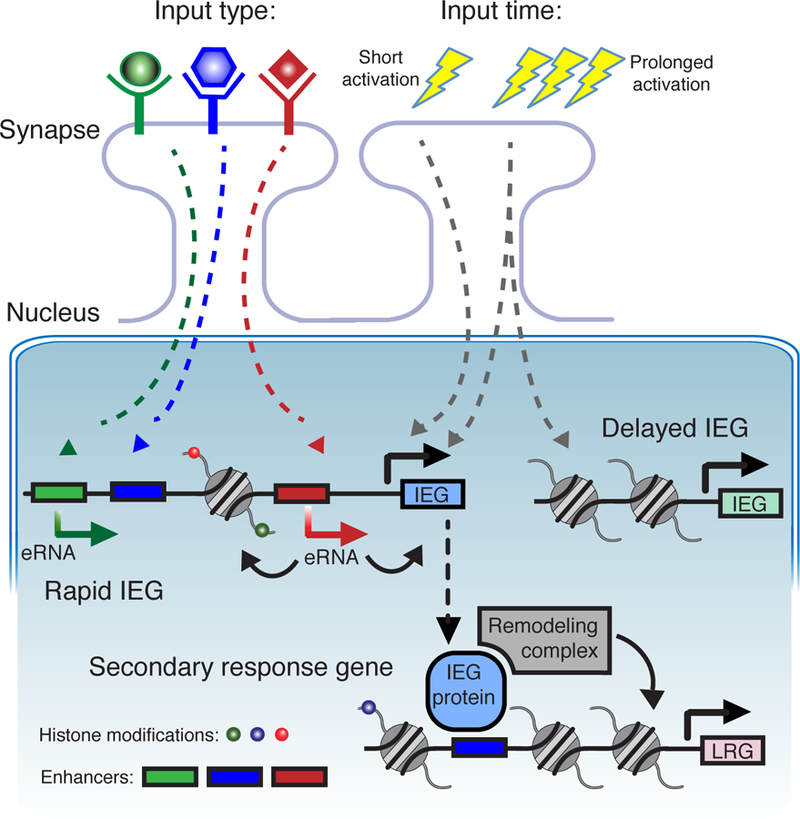
The combinatorial effects of enhancers and chromatin allow neurons to distinguish between different types of stimuli [38*] and different lengths of stimuli [39*]. Different signaling pathways activate different enhancers. This generates specific eRNAs [34] which in turn affect promoter activity and the surrounding chromatin. The most rapidly induced IEGs have more open chromatin with marks such as H3K27ac near gene promoters [34, 35]. The gene products of IEGs include transcription factors which can then regulate late response genes (LGRs) and recruit chromatin regulators and remodelers such as the BAF complex [40]. IEG, immediate early gene. eRNA, enhancer RNA.
The influence of the environment on neuronal chromatin
The external environment and experiences of an organism are tightly linked to chromatin state and modulation of neuronal transcription. Because most neurons persist for the full lifetime of an animal, this connection also raises the questions of whether our environment can have enduring effects on the chromatin landscape (Fig. 4). Recent elegant work demonstrated that the level of gene expression early in life has lasting consequences for gene expression in the adult brain (42). This research focused on DNA methylation but similar effects could occur as a result of histone modifications, particularly those that are more stable.
Figure 4. The effect of environment on neuronal chromatin.
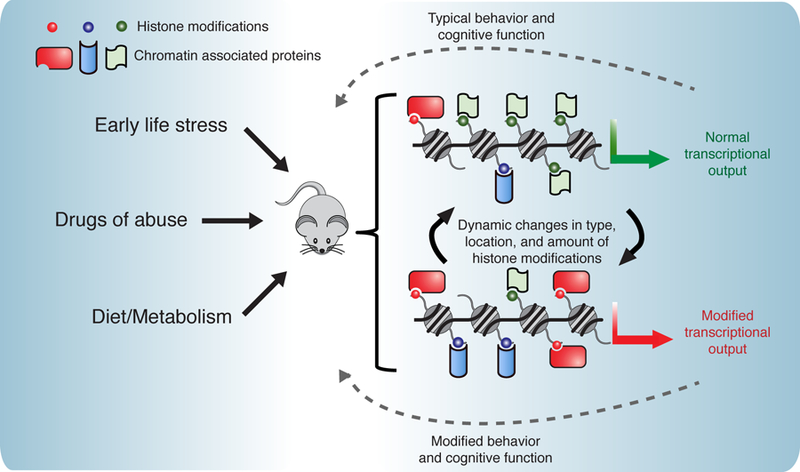
The external environment and experience of an animal can have lasting effects on chromatin, leading to changes in transcriptional output and animal behavior. These external influences include exposure to drugs and alcohol [47–52], diet and metabolism [53–58], and adverse experiences such as social stressors [43–46]. Such influences can modify histones in numerous ways including the type of modification deposited on histones (such as acyl groups regulated by metabolic enzymes), the location of histone modifications, and the activity of the enzymes that deposit and remove modifications. These changes can in turn affect the transcriptional output and function of the neuron.
One such external influence that has long-term consequences for the epigenome of neurons is stress. In particular, early life stress causes lasting transcriptional changes and leads to future behavioral changes such as increased susceptibility to depression (43). Newly uncovered mechanisms demonstrate how chromatin is involved in the response to stress. Histone demethylase LSD1, described above, as well as its neuronal-specific splicing isoform neuroLSD1, are induced by social stress and function as corepressors of IEGs (44*). Mice lacking neuroLSD1 show altered histone methylation and acetylation and have a low-anxiety phenotype. In addition, HDAC2 is required for the resilience to chronic stress observed in TRPV1 knockout mouse (45). Modifying histones at key genes can also induce behavioral changes in response to stress. Using zinc finger proteins to modify H3 acetylation at the Cdk5 locus promotes resilience in response to social stress (46) demonstrating that the chromatin landscape at individual genes can dictate whether specific experiences have long-lasting adverse consequences. These exciting studies provide examples of how chromatin-associated proteins can link past experiences to future transcriptional output.
In addition to stress, external substances we are exposed to throughout our lives have clear effects on our chromatin landscape. Drugs of abuse have long been known to modulate the epigenome and new work has expanded on our knowledge of such mechanisms (47). Histone deacetylase HDAC5 (48), histone dimethyltransferase G9a (49), and many other histone modifiers and histone readers (50) have been implicated in the response to drugs. Alcohol also alters HDAC expression, resulting in enhanced vulnerability to subsequent exposure to cocaine (50). Less well-studied modifications such as histone arginine methylation are also linked to addiction (52) suggesting that more mechanisms tying chromatin to the drugs of abuse have yet to be discovered.
Our diet and metabolism also play an important role in regulating chromatin and many chemical modifications deposited on histones are generated from metabolites (53–56). For example, the metabolic enzyme acetyl-CoA synthetase 2 (ACSS2) generates acetyl-CoA for histone acetylation. Recently, exciting work found that ACSS2 binds near memory-related genes providing local generation of acetyl-CoA (57**). Loss of ACSS2 impairs the activation of these genes and the formation of long-term memory. Other recent work demonstrated that metabolites generated during digestion that modify histones and are also expressed at very high levels in the brain (58). Together, these findings hint at the possibility that our diet may affect our neuronal epigenome by changing the availability of key metabolites needed to modify histones and regulate key neuronal genes.
Conclusion
The last several years of research into the role of chromatin in neurons has helped to advance our understanding of many critical mechanisms of transcriptional regulation in the brain. In particular, the complex interplay of histone modifications, histone variants, enhancers, and environmental factors has emerged as an exciting new forefront in neuroscience. Yet much remains to be discovered and the next phase of research in this field promises to be at least as thrilling as these recent discoveries.
Highlights.
Histone modifications are highly dynamic in the brain and drive changes in many behaviors including learning and memory.
Chromatin-associated proteins that interact with and regulate histone modifications control expression of genes critical for synaptic function.
Exchange and accumulation of specific histone variants affect neuronal function and survival.
Enhancer elements are marked by histone modifications and regulate transcription of activity-dependent genes.
Short and long-term changes to the neuronal epigenome by external influences from stress, drugs of abuse, and diet drive dynamic changes in gene expression and behavior.
Acknowledgements
This work was supported by the National Institutes of Health (grant numbers F30DC017658, R00MH111836 and P50MH096890). We thank Drs. C. David Allis, Ian Maze, Azad Bonni, Iva Zovkic, David Sweatt, Elizabeth Heller, and Cate Peña for feedback, and J.J.G. and S.A.A for editing.
References and recommended reading
Publications of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Zovkic IB, Guzman-Karlsson MC, Sweatt JD, Epigenetic regulation of memory formation and maintenance. Learn. Mem. Cold Spring Harb. N 20, 61–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Opachaloemphan C, Yan H, Leibholz A, Desplan C, Reinberg D, Recent Advances in Behavioral (Epi)Genetics in Eusocial Insects. Annu. Rev. Genet (2018), doi: 10.1146/annurev-genet-120116-024456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levenson JM, Sweatt JD, Epigenetic mechanisms: a common theme in vertebrate and invertebrate memory formation. Cell. Mol. Life Sci. CMLS 63, 1009–1016 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao W et al. , Activity-Induced Regulation of Synaptic Strength through the Chromatin Reader L3mbtl1. Cell Rep 23, 3209–3222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korb E, Herre M, Zucker-Scharff I, Darnell RB, Allis CD, BET protein Brd4 activates transcription in neurons and BET inhibitor Jq1 blocks memory in mice. Nat. Neurosci 18, 1464–1473 (2015).* The authors demonstrate the critical role of a reader of acetylated lysines, Brd4, in the transcription regulation of memory-related genes and utilize pharmacologic manipulation to demonstrate its importance in memory consolidation.
- 6.Sullivan JM et al. , Autism-like syndrome is induced by pharmacological suppression of BET proteins in young mice. J. Exp. Med 212, 1771–1781 (2015).* This study identifies BET chromatin reader proteins as critical regulators for neuronal development and function and demonstrates that treatment of mice with a BET protein inhibitor causes behavioral changes.
- 7.Penney J, Tsai L-H, Histone deacetylases in memory and cognition. Sci. Signal 7, re12–re12 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Gupta S et al. , Histone Methylation Regulates Memory Formation. J. Neurosci 30, 3589–3599 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jakovcevski M et al. , Neuronal Kmt2a/Mll1 Histone Methyltransferase Is Essential for Prefrontal Synaptic Plasticity and Working Memory. J. Neurosci 35, 5097–5108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerimoglu C et al. , KMT2A and KMT2B Mediate Memory Function by Affecting Distinct Genomic Regions. Cell Rep 20, 538–548 (2017).** This work dissects the roles of two closely related methyltransferases, Kmt2a and Kmt2b. By investigating how these enzymes affect distinct memory processes and genomic signatures, the authors demonstrate remarkable specificity of these chromatin modifying enzymes in their transcriptional activity and behavioral output.
- 11.Kerimoglu C et al. , Histone-Methyltransferase MLL2 (KMT2B) Is Required for Memory Formation in Mice. J. Neurosci 33, 3452–3464 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Schimmelmann M et al. , Polycomb repressive complex 2 (PRC2) silences genes responsible for neurodegeneration. Nat. Neurosci 19, 1321–1330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stilling RM et al. , K‐Lysine acetyltransferase 2a regulates a hippocampal gene expression network linked to memory formation. EMBO J 33, 1912–1927 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uchida S et al. , CRTC1 Nuclear Translocation Following Learning Modulates Memory Strength via Exchange of Chromatin Remodeling Complexes on the Fgf1 Gene. Cell Rep 18, 352–366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J et al. , LSD1n is an H4K20 demethylase regulating memory formation via transcriptional elongation control. Nat. Neurosci 18, 1256–1264 (2015).* This work shows that an alternative splicing event of a histone lysine demethylase results in distinct chromatin-regulatory activity in neurons and affects activity-dependent gene expression and spatial learning.
- 16.Bieszczad KM et al. , Histone Deacetylase Inhibition via RGFP966 Releases the Brakes on Sensory Cortical Plasticity and the Specificity of Memory Formation. J. Neurosci 35, 13124–13132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang G et al. , Crucial Roles for SIRT2 and AMPA Receptor Acetylation in Synaptic Plasticity and Memory. Cell Rep 20, 1335–1347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry S, Kiragasi B, Dickman D, Ray A, The Role of Histone Deacetylase 6 in Synaptic Plasticity and Memory. Cell Rep 18, 1337–1345 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Backs J, CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J. Clin. Invest 116, 1853–1864 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santoro SW, Dulac C, The activity-dependent histone variant H2BE modulates the life span of olfactory neurons. eLife 1, e00070 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zovkic IB, Paulukaitis BS, Day JJ, Etikala DM, Sweatt JD, Histone H2A.Z subunit exchange controls consolidation of recent and remote memory. Nature 515, 582–586 (2014).** This work addresses a critical question related to activity-induced transcription in neurons: how is transcription inactivated after stimulation? This study demonstrated that the histone variant exchange of H2A.Z is a suppressor of activity-induced transcription and acts as a critical mediator of memory formation.
- 22.Shen T et al. , Brain-specific deletion of histone variant H2A.z results in cortical neurogenesis defects and neurodevelopmental disorder. Nucleic Acids Res 46, 2290–2307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stefanelli G et al. , Learning and Age-Related Changes in Genome-wide H2A.Z Binding in the Mouse Hippocampus. Cell Rep 22, 1124–1131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maze I et al. , Critical Role of Histone Turnover in Neuronal Transcription and Plasticity. Neuron 87, 77–94 (2015).** This work links incorporation of H3.3 at activity-dependent neuronal genes to transcription, synaptic connectivity, and cognition demonstrating the importance of histone turnover in neuronal function.
- 25.Dunn CJ et al. , Histone Hypervariants H2A.Z.1 and H2A.Z.2 Play Independent and Context-Specific Roles in Neuronal Activity-Induced Transcription of Arc/Arg3.1 and Other Immediate Early Genes. eNeuro 4 (2017), doi: 10.1523/ENEURO.0040-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y et al. , Chromatin remodeling inactivates activity genes and regulates neural coding. Science 353, 300–305 (2016).** This study demonstrates that the NuRD nucleosome remodeling complex inactivates activity-dependent transcription by depositing the histone variant H2A.Z at promoters of neuronal genes. Further, the authors show the importance of this process for proper dendritic pruning in the cerebellum.
- 27.Narkaj K et al. , Blocking H2A.Z Incorporation via Tip60 Inhibition Promotes Systems Consolidation of Fear Memory in Mice. eNeuro 5 (2018), doi: 10.1523/ENEURO.0378-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segala G, Bennesch MA, Pandey DP, Hulo N, Picard D, Monoubiquitination of Histone H2B Blocks Eviction of Histone Variant H2A.Z from Inducible Enhancers. Mol. Cell 64, 334–346 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Lepack AE et al. , Aberrant H3.3 dynamics in NAc promote vulnerability to depressive-like behavior. Proc. Natl. Acad. Sci. U. S. A 113, 12562–12567 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray JM et al. , Genomic Views of Transcriptional Enhancers: Essential Determinants of Cellular Identity and Activity-Dependent Responses in the CNS. J. Neurosci. Off. J. Soc. Neurosci 35, 13819–13826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cirillo LA et al. , Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9, 279–289 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Rada-Iglesias A et al. , A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yap E-L, Greenberg ME, Activity-Regulated Transcription: Bridging the Gap between Neural Activity and Behavior. Neuron 100, 330–348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malik AN et al. , Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat. Neurosci 17, 1330–1339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ostuni R et al. , Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Kim T-K et al. , Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng H, Bartholomew B, Emerging roles of transcriptional enhancers in chromatin looping and promoter-proximal pausing of RNA polymerase II. J. Biol. Chem 293, 13786–13794 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joo J-Y, Schaukowitch K, Farbiak L, Kilaru G, Kim T-K, Stimulus-specific combinatorial functionality of neuronal c-fos enhancers. Nat. Neurosci 19, 75–83 (2016).* This study details the ways in which multiple enhancers act in concert to drive c-fos expression in response to various and specific stimuli.
- 39.Tyssowski KM et al. , Different Neuronal Activity Patterns Induce Different Gene Expression Programs. Neuron 98, 530–546.e11 (2018).* This work mechanistically dissects how different neuronal stimulation paradigms effect distinct expression outputs, and demonstrates differential roles for histone acetylation and enhancer RNA transcription in enhancer activation.
- 40.Vierbuchen T et al. , AP-1 Transcription Factors and the BAF Complex Mediate Signal-Dependent Enhancer Selection. Mol. Cell 68, 1067–1082.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monahan K et al. , Cooperative interactions enable singular olfactory receptor expression in mouse olfactory neurons. eLife 6 (2017), doi: 10.7554/eLife.28620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stroud H et al. , Early-Life Gene Expression in Neurons Modulates Lasting Epigenetic States. Cell 171, 1151–1164.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peña CJ et al. , Early life stress confers lifelong stress susceptibility in mice via ventral tegmental area OTX2. Science 356, 1185–1188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rusconi F et al. , LSD1 modulates stress-evoked transcription of immediate early genes and emotional behavior. Proc. Natl. Acad. Sci. U. S. A 113, 3651–3656 (2016).* This study implicates a neuronal splicing variant of a histone demethylase in the regulation of immediate early gene transcription in response to stress.
- 45.Wang SE et al. , TRPV1 Regulates Stress Responses through HDAC2. Cell Rep 19, 401–412 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Heller EA et al. , Targeted Epigenetic Remodeling of the Cdk5 Gene in Nucleus Accumbens Regulates Cocaine- and Stress-Evoked Behavior. J. Neurosci. Off. J. Soc. Neurosci 36, 4690–4697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker DM, Cates HM, Heller EA, Nestler EJ, Regulation of chromatin states by drugs of abuse. Curr. Opin. Neurobiol 30, 112–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taniguchi M et al. , HDAC5 and Its Target Gene, Npas4, Function in the Nucleus Accumbens to Regulate Cocaine-Conditioned Behaviors. Neuron 96, 130–144.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson EM et al. , Overexpression of the Histone Dimethyltransferase G9a in Nucleus Accumbens Shell Increases Cocaine Self-Administration, Stress-Induced Reinstatement, and Anxiety. J. Neurosci. Off. J. Soc. Neurosci 38, 803–813 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sartor GC, Powell SK, Brothers SP, Wahlestedt C, Epigenetic Readers of Lysine Acetylation Regulate Cocaine-Induced Plasticity. J. Neurosci. Off. J. Soc. Neurosci 35, 15062–15072 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Griffin EA et al. , Prior alcohol use enhances vulnerability to compulsive cocaine self-administration by promoting degradation of HDAC4 and HDAC5. Sci. Adv 3, e1701682 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Damez-Werno DM et al. , Histone arginine methylation in cocaine action in the nucleus accumbens. Proc. Natl. Acad. Sci. U. S. A 113, 9623–9628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simithy J et al. , Characterization of histone acylations links chromatin modifications with metabolism. Nat. Commun 8, 1141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M, The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol 15, 536–550 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Dutta A, Abmayr SM, Workman JL, Diverse Activities of Histone Acylations Connect Metabolism to Chromatin Function. Mol. Cell 63, 547–552 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sabari BR, Zhang D, Allis CD, Zhao Y, Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol 18, 90–101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mews P et al. , Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 546, 381–386 (2017).** This study demonstrates that a metabolic enzyme that generates acetyl moieties for histone acetylation associates with chromatin upon neuronal differentiation and is localized near memory-related genes. This work shows exciting links between cellular metabolism, chromatin regulation, and neuronal plasticity.
- 58.Fellows R et al. , Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun 9, 105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]


