Abstract
Cannabis sativa is a medicinal plant having a very complex matrix composed of mainly cannabinoids and terpenoids. The literature has numerous reports, which indicate that tetrahydrocannabinol (THC) is the only major psychoactive metabolite in C. sativa. It is important to explore other metabolites having the possibility of exhibiting the psychoactive character of various degrees and also to identify metabolites targeting other receptors such as opioid, γ amino butyric acid (GABA), glycine, serotonin, and nicotine present in C. sativa, the smoke of C. sativa, and other phytocannabinoid matrices. This article aims to achieve this goal by application of batteries of computational tools such as machine learning tools and multivariate methods on physiochemical and absorption, distribution, metabolism, excretion, and toxicity (ADMET) descriptors of 468 metabolites from C. sativa, its smoke and, other phytocannabinoids. The structure–activity relationship (SAR) showed that 54 metabolites from C. sativa have high scaffold homology with THC. Its implications on the route of administration and factors affecting the SAR are discussed. C. sativa smoke has metabolites that have possibility of interacting with GABA, and glycine receptors.
Introduction
Globally, mortality due to narcotic drugs increased by 60% from 2000 (105 000) to 2015 (168 000).1 World consumption of narcotic drugs was estimated to be 275 million in 2016, reporting the use at least once in the earlier year. Of 275 million, cannabis accounted for 192 million, and some large countries do not report the true statistics on the use of drugs.2 Many states in the US, Canada, and Uruguay have legalized cannabis for medical and recreational purposes. Over the last few decades, the exploration into the application of cannabis and the derived drugs to various disorders has been carried out by an increasing number of research groups in the world.3−6 It is shown to be effective in treatment for schizophrenia,7 epilepsy,8 Parkinson’s disease,9 brain tumor,10 brain injury,11 oxidative stress and inflammation,5 inflammatory bowel disease,12 and multiple sclerosis.13 Cannabis-based treatment for chronic neuropathic pain has been studied in detail14 by several groups. Apart from cannabis in natural form, a few synthetic drugs such as dronabinol and sativex have been approved by USFDA for controlling nausea15 and cancer pain,16 respectively. In spite of the use of cannabis and its products for therapeutic use, scientific evidence is yet to be generated for its widespread application for the general public.
Cannabis has three species such as Cannabis indica, Cannabis sativa, and Cannabis ruderalis.(17) Small and Beckstead classified three different chemophenotypes.18 Chemotype I contains >0.3% of tetrahydrocannabinol (THC) and <0.5% of cannabidiol (CBD) of the total dry plant material, whereas chemotype II has predominantly CBD and varying quantities of THC. Chemotype III has low THC content. The main difference between these two species is that indica has higher cannabidiol (CBD) content and sativa has a higher content of tetrahydrocannabinol (THC).19 THC has psychoactive properties resulting in “highness” on consumption, and CBD lacks any of the psychoactive characteristics. Commercially, C. sativa is cultivated for medical and recreational purposes. Cannabis is a very complex matrix of 568 compounds, and the final output depends on the route of administration such as ingestion, smoking, and vaporization. C. sativa has two phenotypes: one has a high content of THC and another has a high content of fibers called hemp and its THC content is less than 0.3%. Fiber-rich cannabis plants showed cannabionic acids such as cannabidiolic acid (CBDA) and cannabigerolic acid (CBGA) followed by their decarboxylated forms, namely, cannabidiol (CBD) and cannabigerol (CBG).20C. sativa has several natural product classes such as terpenes, carbohydrates, fatty acids, and their esters, amides, amines, phytosterols, phenolic compounds, and cannabinoids.21 Cannabinoids bind to cannabinoid receptors, CB1R and CB2R, which are similar to G-protein coupled receptor (GPCR) family of receptors.22 CB1R encoded by the CNR1 gene consists of 472 amino acids in humans and displays a high degree of sequence homology (97–99%) with other species.23 CB1R has three isoforms showing differences in the N-terminal region due to alternative mRNA splicing patterns.24 The isoform CB1R is expressed predominantly in the brain and skeletal muscles, and CB1Rb is expressed in liver and pancreatic islet cells mainly involved in metabolic functions.25 CB2R is encoded by the CNR2 gene and comprises 360 amino acids in humans. It displays only 44% sequence homology with CB1R and shows higher diversity among the species.26 CB2R has two isoforms, and one isoform is expressed in testis and the other is highly expressed in spleen and the brain regions.27
Chemoinformatics plays a vital role in all stages of drug discovery. Drug target identification,28 virtual screening and structure–activity relationship (SAR) analysis,29 hit/lead identification and optimization,30 and absorption, distribution, metabolism, excretion, and toxicity (ADMET)31 prediction are different areas of drug discovery, where chemoinformatics is widely applied with high success. Analysis of drug-like space is important to filter the molecules from larger databases using chemoinformatic methods32,33 and further taking them to the next step of identification of lead-like compounds.34 Data mining using machine learning methods such as support vector machine, k-nearest neighbor (kNN), and neural network algorithms is used in the virtual screening stage of the drug discovery process.35 The goal of this article is to identify the THC- or opiate-like candidates from the database of metabolites from C. sativa, its smoke, and other phytocannabinoids, based on the physiochemical descriptors and predicted clinical ADMET properties using machine learning methods such as naïve-Bayes (NB), support vector machine, multilayer perceptron, and hierchirarchial clustering methods. Our results identified the possible candidates having similar physiochemical and ADMET properties to THC by machine learning, multivariate, and structure–activity relationship methods, and its consequences on the consumption of cannabis are also discussed in relation with receptor types in the brain.
Results and Discussion
Drug-like chemical space based on Lipinski’s rule of five for drug candidates is explored to improve its pharmacokinetic properties.36 Five physiochemical descriptors covered under Lipinski’s rule of five are shown for all of the metabolites along with reference psychoactive compounds in Figures 1−6. The molecular weights of the 468 metabolites from C. sativa, its smoke, and other phytocannabinoids are shown in Figure 1.
Figure 1.
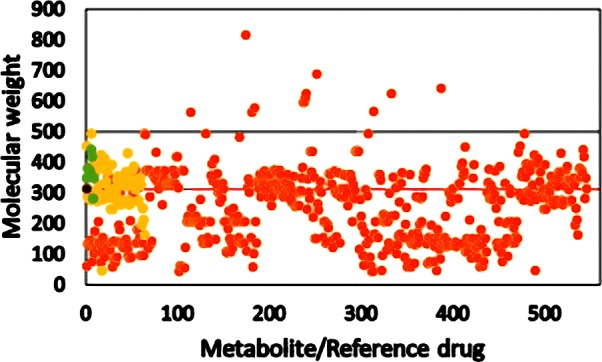
Molecular weights of metabolites from C. sativa, its smoke, and other phytocannabinoids. Orange circle solid represents metabolite, yellow circle solid reoresents psychoactive reference compound, green circle solid represents endocannabinoid, black circle solid represents THC, and black and red lines represent Lipinski’s and the central nervous system (CNS) cutoff, respectively.
Figure 6.
Nonmetric MDS map for CDK data type for 1–15 metabolites in 330 dataset. C, P, E, and T denote metabolite, opioid reference drug, endocannabinoid, and THC, respectively.
With Lipinski’s rule of five, only 13 metabolites violated the cutoff scale of 500 and all of the Schedule I−IV drugs were well below the Lipinski’s cutoff (black line) for molecular weight. Pajouhesh et al.37 in the analysis of physiochemical descriptors for drugs, which include CNS acting drugs, reported the new cutoff values for physiochemical descriptors for CNS-targeted drugs. The CNS cutoff (red line) for molecular weight was 310, and when it was applied to the 468 metabolites, it showed a significant number of outliers, which include 53% of Schedule I−IV substances. Endocannabinoids except oleamide, which are natural agonists for cannabinoid receptors, display molecular weight above the CNS cutoff value. It is important to mention that THC has the molecular weight (314) that is closer to CNS cutoff having a psychoactive property in C. sativa.
In the case of M log P data shown in Figure 2, Lipinski’s rule of five cutoff is 4.15 (the black horizontal line in Figure 2) and CNS drug cutoff is 2.08 (red line).
Figure 2.
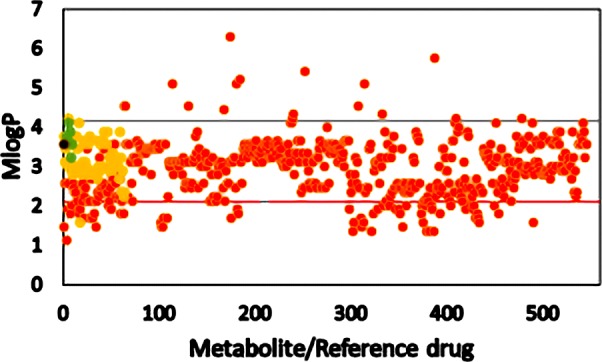
M log P of metabolites from C. sativa, its smoke, and other phytocannabinoids. Orange circle solid represents metabolite, yellow circle solid represents psychoactive reference compound, green circle solid represents endocannabinoid, black circle solid represents THC, and black and red lines represent Lipinski’s and the CNS cutoff, respectively.
Most of the metabolites along with Schedule I−IV drugs obey Lipinski’s cutoff and showed deviation from the CNS drug cutoff value of 2.08. Cannabinoids from C. sativa, phytocannabinoids, and the volatile products derived from C. sativa showed moderate hydrophobicity.
Total polar surface area (TPSA) controls the absorption property of the drugs,38−40 and Lipinski’s cutoff is 140 Å2. Any drug above this value shows poor absorption into the intestines. TPSA for all of the metabolites, endocannabinoids, and Schedule I−IV substances is shown in Figure 3.
Figure 3.
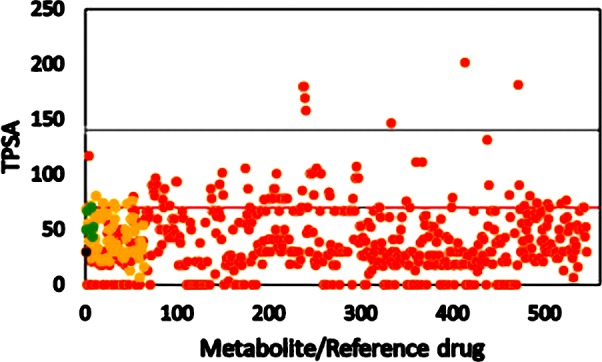
TPSA of metabolites from C. sativa, its smoke, and other phytocannabinoids. Orange circle solid represents metabolite, yellow circle solid represents psychoactive reference compound, green circle solid represents endocannabinoid, black circle solid represents THC, and black and red lines represent Lipinski’s and the CNS cutoff, respectively.
The cutoff value of TPSA for CNS drugs is 70 Å2, and surprisingly the majority of metabolites from C. sativa have TPSA lower than 70 Å2. THC showed a TPSA value of 29 Å2, and the highest TPSA value among endocannabinoids is 69 Å2. Cannabidiol (CBD), which does not have a psychoactive property, showed a TPSA value of 78 Å2, and lower TPSA is pronounced in the case of cannabinoids having a psychoactive property. TPSA may be one of the best filters to identity the metabolites from cannabinoids having psychoactive properties.
H-bond donor and H-bond acceptor properties determine the mode and strength of ligand–receptor interaction through hydrogen bonds. As per Lipinski’s rule of five, the permissible number of H-bond donors is 5 and H-bond acceptors is 10 for a successful drug. However, in the case of CNS drugs, the cutoff is reduced to 1.5 for a H-bond donor and 4.32 for a H-bond acceptor. As shown in Figure 4a, the majority of metabolites and endocannabinoids have maximum of 3 H-bond donors. In the case of Schedule I−IV substances, 95% are shown to have only 2 H-bond donors.
Figure 4.
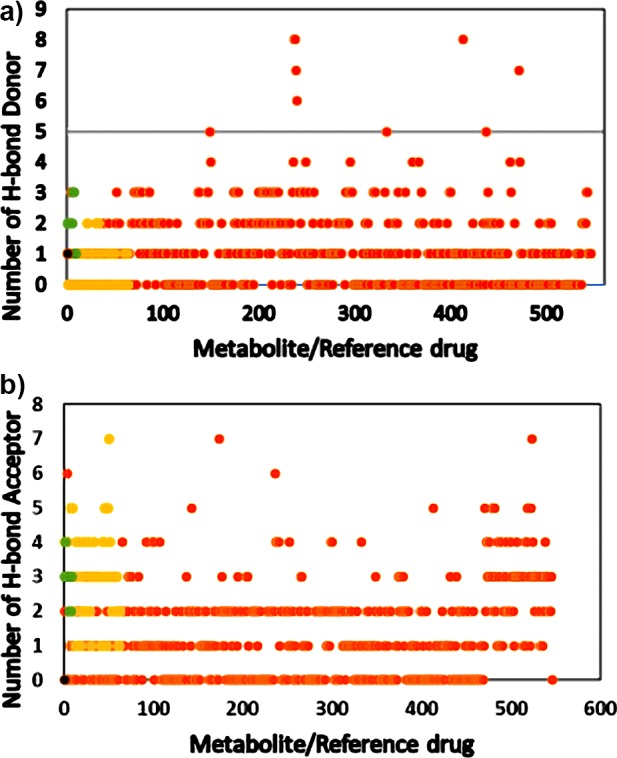
(a) H-bond donors of metabolites from C. sativa, its smoke, and other phytocannabinoids. Orange circle solid represents metabolite, yellow circle solid represents psychoactive reference compound, green circle solid represents endocannabinoid, black circle solid represents THC, and black and red lines represent Lipinski’s and the CNS cutoff, respectively. (b) H-bond acceptor of metabolites from C. sativa, its smoke, and other phytocannabinoids. (Orange circle solid) metabolite, (yellow circle solid) psychoactive reference compound, (green circle solid) endocannabinoid, (black circle solid) THC, black and red lines represent Lipinski’s and the CNS cutoff, respectively.
It is clear that CNS drugs and Schedule I−IV substances interact with their target receptors through hydrophobic interaction and less dependent on the H-bond network. THC showed only one H-bond donor and CBD had three H-bond donors. The CDK molecular descriptor tool calculates H-bond acceptors for the molecule when an oxygen or a nitrogen in the molecule has formal charge equivalent to or less than zero (formal charge ≤0). Figure 4b shows that the majority of cannabinoids, endocannabinoids, and the Schedule I−IV substances showed four H-bond acceptors.
Effect of the H-bond acceptor on drug efficacy is less pronounced from Lipinski’s point of view, and Figure 4b shows that Schedule I−IV substances having the H-bond acceptor value between 5 and 7 are able to penetrate the blood–brain barrier (BBB) and exhibit excellent psychoactive activity.
The number of rotatable bonds affects the molecular conformational freedom, and a molecule having less number of rotatable bonds has structural rigidity. It is important to freeze the bioactive conformation, and it is achieved by reducing the number of rotatable bonds.41Figure 5 shows the number of rotatable bonds for metabolites from cannabis, endocannabinoids, and the Schedule I−IV substances.
Figure 5.
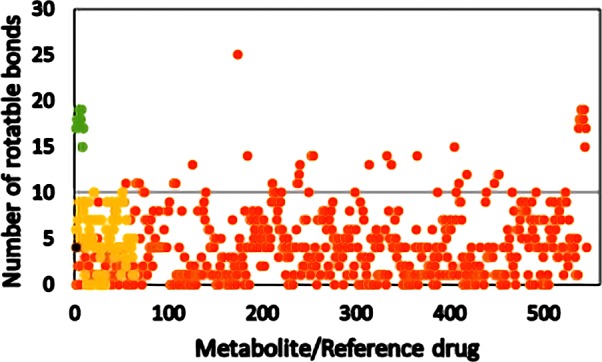
Number of rotatable bonds of metabolites from C. sativa, its smoke, and other phytocannabinoids. Orange circle solid represents metabolite, yellow circle solid represents psychoactive reference compound, green circle solid represents endocannabinoid, black circle solid represents THC, and black and red lines represent Lipinski’s and the CNS cutoff, respectively.
Compliance with Lipinski’s rule of five with a number of rotatable bonds (<10) is high for most cannabis metabolites and the Schedule I−IV substances. All of the endocannabinoids violate Lipinski’s rule with regard to the number of rotatable bonds being in the range of 15–19. It is to be noted that endocannabinoids are lipid moieties having a high number of freely rotatable bonds, they are synthesized and metabolized in the brain, and moreover they do not have to cross the blood–brain barrier.
Machine learning methods are recent tools in identifying the receptor targets based on structure- and ligand-dependent approaches in the drug discovery process.42 In this study, four classifiers, namely, multilayer perceptron, naïve-Bayes, support vector machine, and k-Nearest Neighbor were used to identify opiate- and THC/endocannabinoid-like molecules. Initially, the results of cross-validation performed on the reference candidates consisting of Schedule I−IV substances, THC endocannabinoids agonists, and negative controls provided the results shown in Table 1.
Table 1. Cross-Validation Results on the Reference Candidates for Four Classifiers: Multilayer Perceptron, naïve-Bayes, Support Vector Machine, and k-Nearest Neighbora.
| data type | classifier | TP rate | FP rate | precision | recall | F-measure | MCC | ROC area | PRC area | class |
|---|---|---|---|---|---|---|---|---|---|---|
| CDK | MLP | 0.973 | 0.056 | 0.986 | 0.973 | 0.980 | 0.899 | 0.994 | 0.976 | drug |
| CDK | MLP | 0.944 | 0.027 | 0.895 | 0.944 | 0.919 | 0.899 | 0.994 | 0.976 | no drug |
| CDK | NB | 0.946 | 0.111 | 0.972 | 0.946 | 0.959 | 0.803 | 0.950 | 0.980 | drug |
| CDK | NB | 0.889 | 0.054 | 0.800 | 0.889 | 0.842 | 0.803 | 0.971 | 0.889 | no drug |
| CDK | SVM | 0.986 | 0.056 | 0.986 | 0.986 | 0.986 | 0.931 | 0.965 | 0.984 | drug |
| CDK | SVM | 0.944 | 0.014 | 0.944 | 0.944 | 0.944 | 0.931 | 0.965 | 0.903 | no drug |
| CDK | kNN | 0.986 | 0.000 | 1.000 | 0.986 | 0.993 | 0.967 | 0.992 | 0.997 | drug |
| CDK | kNN | 1.000 | 0.014 | 0.947 | 1.000 | 0.973 | 0.967 | 0.992 | 0.936 | no drug |
| CDK–ADME | MLP | 0.986 | 0.000 | 1.000 | 0.986 | 0.993 | 0.967 | 0.999 | 1.000 | drug |
| CDK–ADME | MLP | 1.000 | 0.014 | 0.947 | 1.000 | 0.973 | 0.967 | 0.999 | 0.997 | no drug |
| CDK–ADME | NB | 0.986 | 0.222 | 0.948 | 0.986 | 0.967 | 0.821 | 0.909 | 0.959 | drug |
| CDK–ADME | NB | 0.778 | 0.014 | 0.933 | 0.778 | 0.848 | 0.921 | 0.983 | 0.913 | no drug |
| CDK–ADME | SVM | 0.986 | 0.000 | 1.000 | 0.986 | 0.993 | 0.967 | 0.993 | 0.997 | drug |
| CDK–ADME | SVM | 1.000 | 0.014 | 0.947 | 1.000 | 0.973 | 0.967 | 0.993 | 0.947 | no drug |
| CDK–ADME | kNN | 0.986 | 0.000 | 1.000 | 0.986 | 0.993 | 0.967 | 0.992 | 0.997 | drug |
| CDK–ADME | kNN | 1.000 | 0.014 | 0.947 | 1.000 | 0.973 | 0.967 | 0.992 | 0.936 | no drug |
| ADME | MLP | 0.986 | 0.056 | 0.986 | 0.986 | 0.986 | 0.931 | 0.999 | 1.000 | drug |
| ADME | MLP | 0.944 | 0.014 | 0.944 | 0.944 | 0.944 | 0.931 | 0.999 | 0.997 | no drug |
| ADME | NB | 0.986 | 0.222 | 0.948 | 0.986 | 0.967 | 0.821 | 0.887 | 0.948 | drug |
| ADME | NB | 0.778 | 0.014 | 0.933 | 0.778 | 0.848 | 0.821 | 0.981 | 0.941 | no drug |
| ADME | SVM | 0.986 | 0.056 | 0.986 | 0.986 | 0.986 | 0.931 | 0.965 | 0.984 | drug |
| ADME | SVM | 0.944 | 0.014 | 0.944 | 0.944 | 0.944 | 0.931 | 0.965 | 0.903 | no drug |
| ADME | kNN | 0.986 | 0.056 | 0.986 | 0.986 | 0.986 | 0.931 | 0.960 | 0.982 | drug |
| ADME | kNN | 0.944 | 0.014 | 0.944 | 0.944 | 0.944 | 0.931 | 0.960 | 0.892 | no drug |
TP: true positive; FP: false positive; MCC: Matthews correlation coefficient; ROC: receiver operating characteristic; PRC: precision recall.
The performance of four classifiers for three types of data was assessed based on their accuracy, sensitivity, specificity, ROC area, and F-measure values. Accuracy quantifies the efficiency of each classifier to predict the true values. The Matthews correlation coefficient (MCC) allows one to judge the performance of a given classifier. MCC ranges from −1 to +1, where −1 indicates that it is a wrong binary classifier and +1 is the indication of the correct classifier. High positive values of ROC and PRC indicate that the classifier shows high performance for a given data set. Accuracy ranged from 85 to 99% for different classifiers with three data types. Sensitivity and specificity quantify the proportion of true positives and negatives, respectively. Both sensitivity and specificity ranged from 77 to 100%, where 77% was shown by the NB classifier with CDK–ADMET hybrid data type.
Diagnosis of Data Overfitting and Subsequent Data Analysis
Accuracy results in the training, cross-validation, and test for three data types (CDK, CDK–ADMET, and ADMET) using four classifiers on three unique datasets on reference candidates (training, validation, and test sets) are given in Table 2a–c.
Table 2. Accuracy Results on the Reference Candidates for Training, Cross-validation, and Test Results of Three Data Types using Four Classifiers: (a) CDK, (b) CDK–ADMET, and (c) ADMET.
| (a)
CDK | |||
|---|---|---|---|
| classifier | training | validation | test |
| NB | 98.64 | 72.72 | 57.14 |
| MLP | 100 | 90.90 | 85.71 |
| SMV | 98.64 | 81.81 | 71.42 |
| kNN | 100 | 81.81 | 57.14 |
| (b)
CDKADMET | |||
|---|---|---|---|
| classifier | training | validation | test |
| NB | 100 | 93.93 | 100 |
| MLP | 100 | 100 | 73.07 |
| SMV | 100 | 100 | 85.71 |
| kNN | 100 | 90.90 | 100 |
| (c)
ADMET | |||
|---|---|---|---|
| classifier | training | validation | test |
| NB | 98.64 | 90.90 | 71.42 |
| MLP | 100 | 100 | 100 |
| SMV | 100 | 100 | 85.71 |
| kNN | 100 | 100 | 85.71 |
Overfitting of data is diagnosed on the fact that whenever there is overfitting, the accuracy drastically reduces from training to test sets. Our results showed that for CDK data type, accuracy was significantly reduced for NB and kNN classifiers from training to test sets. In the case of CDK–NB combination, the training set was observed to be 98.64% and it was reduced to 57% for the test set. Similarly, in the case of CDK–kNN combination, the accuracy of 100% observed for the training set was reduced to 57% for the test set. It can be concluded that NB and kNN classifiers overfit the data for CDK data type and hence these two classifiers were not included for analysis involving CDK data type.
The list of candidates classified as a drug by all four classifiers from three data types is given in Supplement 1. The four classifiers eliminated 138 candidates from the original database based on the comparison of predicted physiochemical and clinical descriptors of Schedule I−IV drugs, endocannabinoid agonists, and negative controls. In the remaining 330 candidates, about 41% of the candidates in the list belong to volatile metabolites, and these volatile compounds gain importance, as smoking is the most common route of consumption of cannabis products. Smoking is the quickest way to have a rapid onset of psychoactive effect due to the high bioavailability of THC in the systemic circulation. Interestingly, three alkanes, namely, 3-methyl heptane, 4-methyl decane, and nonane were also found in the volatile mixture of cannabis smoke. Alkanes possess anesthetic effect, and the anesthetic potency decreases with increasing chain; moreover, no anesthetic effect is observed from n-undecane (C-11).43
As a next step of filtering the psychoactive cannabinoid candidates, nonmetric multidimensional scaling was performed on 330 molecular candidates obtained from machine learning methods. Multidimensional scaling (MDS) is a valuable tool to obtain a degree of similarity among the members of a group in a given data set.44−46 MDS has two methods, that is, metric and nonmetric MDS. Metric MDS measures similarity-based interpoint distances between the members of the group and nonmetric MDS operates by the relative ordering of similarity in an ensemble of data points. Nonmetric MDS has the advantage over metric MDS as the magnitude of similarity of input data points is unreliable or sometimes difficult to measure with high accuracy. MDS has been applied to analyze the patterns of gene expression47 and evolutionary pathways of GPCR.48 Nonmetric MDS was performed on 330 candidates using physiochemical molecular descriptors and later using ADMET descriptors. In the NMDS map, the outliers were excluded and the remaining candidates were retained for further analysis. NMDS on physiochemical descriptors resulted in 165 candidates (Supplement 2), and the subsequent NMDS using clinical descriptors provided 112 candidates (Supplement 1). NMDS maps for CDK and ADMET are shown in Figures 6 and 7.
Figure 7.
Nonmetric MDS map for ADMET data type for 1–15 metabolites in 165 dataset generated by NM-MDS on the CDK dataset. C, P, E, and T denote metabolite, opioid reference drug, endocannabinoid, and THC, respectively.
Structure–Activity Relationship of Cannabinoids
A common scaffold of THC was compared by three-dimensional (3D) alignment with every candidate in the group of 112 members from NM-MDS analysis, and the score obtained for 3D alignment was listed from the highest to lowest value. The minimum cutoff value of 0.5 was fixed for 3D alignment, as the cannabidiol having less psychoactive character compared to THC has a homology score of 0.558, and hence it is taken as the baseline for assessing the structure–activity relationship. The candidates having 3D homology till 0.5 were selected for study on the structure–activity relationship and scaffold homology of each metabolite with THC, and changes in pharmacophores of each metabolite in comparison to THC and their corresponding chemical structures are given in Tables 3A and 3B. In addition, 3D-aligned structures of each metabolite with THC are provided in Supplement 4.
Table 3A. Changes in Pharmacophores of the THC Scaffold Leading to Various Cannabinoid Metabolites Found in C. sativa.
| sl. no. | cannabinoid | scaffold homology To THC | 1-methyl cyclohexene | benzopyran | phenolic group | C-3-side chain | any other cause |
|---|---|---|---|---|---|---|---|
| 1 | 8a-hydroxy-delta-9-tetrahydrocannabinol | 0.95 | OH at 8th position | ||||
| 2 | delta-9-tetrahydrocannabinol-C-4 | 0.945 | one CH2 less to THC | ||||
| 3 | delta-8-tetrahydrocannabinolic acid | 0.907 | COOH at position 2 | low BBB | |||
| 4 | 9β,10β-epoxyhexahydrocannabinol | 0.859 | loss of unsaturation at position 9 and converted to the epoxide ring | ||||
| 5 | 9α,10α-tetrahydrocannabinol epoxide | 0.859 | loss of unsaturation at position 9 and converted to the epoxide ring | ||||
| 6 | 7,8-dihydrocannabinol | 0.825 | additional double bond at position 7 | ||||
| 7 | 9,10-anhydrocannabitriol | 0.825 | epoxide ring at position 9 and a double bond at position 10a | ||||
| 8 | 10-hydroxy-9-oxo-delta-8-tetrahydrocannabinol | 0.802 | double bond at position 8 and carbonyl at position 9 and OH group at position 10 | ||||
| 9 | 10-oxo-delta-6a-tetrahydrocannabinol (OTHC) | 0.744 | carbonyl at position 10 | double bond at position 6a | |||
| 10 | cannabichromanone (CBCF) | 0.73 | ring opens | carbonyl at position 10a | |||
| 11 | delta-8-tetrahydrocannabinol (Δ8-THC) | 0.722 | double bond at position 8 instead of 9 | ||||
| 12 | isocannabitrol | 0.709 | OH group at positions 8, 9 and ethylene group at position 10 | ||||
| 13 | 10α-hydroxy-delta-9,11-hexahydrocannabinol | 0.706 | ethylene group at position 9 and OH group at position 8 | ||||
| 14 | compound-3 | 0.692 | ring opens, OH group at position 7 and a double bond at position 9 oxidized to aldehyde | ||||
| 15 | delta-9-tetrahydrocannabinolic acid B (THCA-B) | 0.681 | COOH group at position 4 | low BBB | |||
| 16 | delta-9-cis-tetrahydrocannabivarin | 0.678 | cis isomer at position 9 | propyl group instead of pentyl | |||
| 17 | compound-2 | 0.673 | ring opens, double bond at C-9–C-10 oxidized to ketone and aldehyde | ||||
| 18 | cannabichromaonone D | 0.654 | ring fused with both phenolic and pyran rings | ||||
| 19 | compound-4 | 0.654 | ring opens and fused with a phenolic group | keto group at position 10a | |||
| 20 | delta-9-nor-tetrahydrocannabinol | 0.647 | ethylene group at position C-8 | butyl group at instead of pentyl | |||
| 21 | 2-formyl-delta-9-tetrahydrocannabinol | 0.645 | ethylene group at position C-8 | formyl group at position 2 | |||
| 22 | cannabicyclolic acid (CBLA) | 0.639 | ring converted to fused cyclopentyl and cyclobutyl groups | COOH group at position 2 | low BBB | ||
| 23 | 8-oxo-delta-9-tetrahydrocannabinol | 0.638 | oxo group at position 8 | ||||
| 24 | hexahydrocannabinol | 0.636 | double bond at C-9 is reduced | ||||
| 25 | bis-nor-cannabitriol | 0.632 | double bond at C-9 reduced, OH group at C-9 added, vinyl alcohol is added to position C-10 | ||||
| 26 | cannabinol (CBN) | 0.63 | converted to benzene ring | ||||
| 27 | 6a-R-cannabichromanone B | 0.627 | ring opens | keto group at position 10a and methyl 3-hydroxy propenyl ketone is added to position 6a | low BBB | ||
| 28 | 7-hydroxy cannabinol | 0.618 | ring converted to hydroxy benzene | ||||
| 29 | 7-hydroxy cannabichromane | 0.616 | ring opens | 2-methyl-2-pentene attached to position 6 | |||
| 30 | delta-7-trans-isotetrahydrocannabinol | 0.614 | ring opens | ring opens | |||
| 31 | 10-ethoxy-9-hydroxy-delta-6a-tetrahydrocannabinol | 0.608 | ethoxy group at position 10 and OH group at position 9 | ||||
| 32 | delta-9-tetrahydrocannabinolic acid A (THCA-A) | 0.608 | COOH at position 2 | low BBB | |||
| 33 | cannabinol methyl ether (CBNM) | 0.592 | same as cannabinol | ||||
| 34 | delta-9-nor-tetrahydrocannbinolic acid | 0.592 | ethylene group at position 8 | COOH at position 2 | butyl group at instead of pentyl | ||
| 35 | delta-9-tetrahydrocannabiorcol (THC-C1) | 0.587 | methyl group instead of pentyl | ||||
| 36 | anhydrocannabimovone | 0.586 | pyran is replaced by tetrahydrofuran | ||||
| 37 | 10-O-ethyl bis-nor cannabitriol | 0.584 | double bond at C-9 reduced, OH group at C-9 added, ethyl vinyl ether at position 10 | ||||
| 38 | bis-nor-cannabichromanone | 0.574 | ring opens | propyl instead of pentyl group | |||
| 39 | cannabielsoin (CBE) | 0.554 | pyran is replaced by tetrahydrofuran | ||||
| 40 | delta-9-tetrahydrocannabinolic acid-C-4 (THCA-C-4) | 0.554 | COOH at position 2 | butyl instead of pentyl group | |||
| 41 | cannabicyclovarin (CBLV) | 0.547 | ring converted to fused cyclopentyl and cyclobutyl groups | propyl instead of pentyl group | |||
| 42 | delta-9-tetrahydrocannabiorcolic acid (THCA-C1) | 0.547 | COOH at position 2 | methyl group instead of pentyl | |||
| 43 | O-propyl-cannabidiol | 0.547 | ring opens, propyl group at OH group | ||||
| 44 | cannabidiol (CBD) | 0.539 | ring opens | ||||
| 45 | 3-hydroxy-delta-4,5-cannabichromene | 0.536 | ring opens | methyl at position 7 is substituted by 3-hydroxy-pentene group | |||
| 46 | 3,4,5,6-tetrahydro-7-hydroxy-a,a-2-trimethyl-9-n-propyl-2,6-methano-2H-1-benzoxocin-5-methanol | 0.535 | ring opens | cyclopentyl group attached to ortho and OH groups | |||
| 47 | cannabicoumaronone | 0.53 | 4,5-dehydrofuran group fused with benzopyran and phenolic groups | ||||
| 48 | delta-7-cis-isotetrahydrocannabivarin | 0.528 | ring opens | cyclopentyl group attached to ortho and OH groups | propyl instead of pentyl group | ||
| 49 | cannabidivarin (CBDV) | 0.522 | ring opens | propyl instead of pentyl group | |||
| 50 | cannabinodivarin (CBVD) | 0.522 | replaced by benzene ring | ring opens | propyl instead of pentyl group | ||
| 51 | cannabiglendol | 0.514 | replaced by tetrahydrofuran group | propyl instead of pentyl group | |||
| 52 | cannabidiol monomethyl ether (CBDM) | 0.508 | ring opens, methyl group at OH group | ||||
| 53 | 4-acetoxy cannabichromene | 0.501 | ring opens | methyl at position 7 is substituted by 2-methyl-pentene group | acetoxy group at position 4 | ||
| 54 | 7-R-cannabicoumarononic acid | 0.5 | ring opens | 4,5-dehydrofuran group fused with benzopyran and phenolic groups | COOH at position 2 |
Table 3B. Chemical Structures of 54 Metabolites Having High 3D Homology with THC.
The metabolite having the highest scaffold homology (0.95) is 8a-hydroxy-delta-9-tetrahydrocannabinol. As shown in Supplement 4-1, it has an additional hydroxy moiety at 8th position in the cyclohexene ring and it makes the molecule more hydrophilic than THC. THC is highly lipophilic and the hydrophobic regions such as cyclohexene ring, C-5 chain on the phenolic moiety, and pyran ring interact with membrane lipid hydrophobic acyl chains and hydrophobic amino acids in transmembrane helices VI and VII of GPCR as reported in the literature.49 Introducing an OH group at 8th position on the cyclohexene ring reduces the partition distribution into the membrane, and it causes unfavorable energy of the OH group interacting with hydrophobic membrane lipids and the amino acids in the receptor. Liposome-mediated delivery and masking the OH group by alkylating with a methyl or an ethyl group may increase the log P value, thereby increasing membrane solubility and favorable interaction energy with the receptor. Delta-9-tetrahydrocannabinol-C-4 (Supplement 4-2) has homology score of 0.945 and C-5 chain at position 3 of the phenol group is reduced to C-4 and any decrease in hydrophobic chain length reduces the affinity with the receptor and potency, as the hydrophobic interaction of C-5 chain with membrane lipids and receptor is vital for receptor activation. Delta-9-cis-tetrahydrocannabivarin (Supplement 4-16), apart from cis configurational change at 9th position in cyclohexane ring, has a propyl group instead of pentyl at C-3 position of the phenolic group, which reduces its affinity for the receptor. Delta-9-tetrahydrocannabiorcol (THC-C1) (Supplement 4-35) has a methyl group instead of pentyl at the C-3 position of the phenolic group, and it reduces the affinity for the CB1 receptor significantly.
Epoxide ring derivatives such as 9β,10β-epoxyhexahydrocannabinol (Supplement 4-4), 9α,10α-tetrahydrocannabinol epoxide (Supplement 4-5), and 9,10-anhydrocannabitriol (Supplement 4-7) have epoxide rings at positions 8–9 in the cyclohexene ring, this epoxide ring is highly susceptible to hydrolysis under acidic conditions existing in the stomach, and the end product 8,9-dihydroxy THC is more polar than THC and hence it may have lower capacity to cross the blood–brain barrier and partition into the membranes is reduced similar to 8-hydroxy THC.
The metabolite 7,8-dihydrocannabinol (Supplement 4-6) has an additional double bond at 10–6a position in the cyclohexene ring and makes it more reactive to undergo metabolism in the liver. Otherwise, it may be expected to have bioactivity equivalent to THC. In 10-hydroxy-9-oxo-delta-8-tetrahydrocannabinol (Supplement 4-8) and 10-oxo-delta-6a-tetrahydrocannabinol (Supplement 4-9) (OTHC), additional OH and carbonyl groups at positions 10a and 9 in the cyclohexene ring make the overall molecule more polar having one more H-bond donor and H-bond acceptor. The extra OH and carbonyl groups may result in dimerization by head-to-tail mode of interaction. An increase in polarity reduces the partition distribution into the membrane to interact with GPCR.
Delta-8-THC (Supplement 4-11) is an isomer of THC, which differs only in the position of double bond in the cyclohexene ring. Delta-8-THC is less psychoactive than delta-9-THC, and it may be due to low solubility of delta-8-THC in water.50 Isocannbitrol (Supplement 4-12) having two additional OH groups at 8, 9 positions in the cyclohexene ring makes it highly hydrophilic, and similar to 8-hydroxy-THC, isocannbitrol may have lower partition distribution into the membranes for receptor activation. Similarly, 10-hydroxy-delta-9,11-hexahydrocannabinol (Supplement 4-13) has an extra OH group in the cyclohexene ring and is more polar than delta-9-THC.
Delta-9-nor-tetrahydrocannabinol (Supplement 4-20) has an ethylene group in the cyclohexene group and the alkyl chain at C-3 of the phenolic group is reduced to butyl instead of pentyl group. Dramatic reduction in the scaffold homology (0.647) with the introduction of the ethylene group at position 8 of the cyclohexene group generates steric hindrance, and reduction in chain length at the C-3 position of the phenolic group decreases the affinity for the receptor. In 2-formyl-delta-9-tetrahydrocannabinol (Supplement 4-21), the formyl group at position 2 in the phenolic group may form intra or intermolecular H-bonding with OH group and may lead to dimerization. As the phenolic OH group may be engaged intra- or intermolecularly in 2-formyl-delta-9-tetrahydrocannabinol, its involvement with receptor activation may be affected. In addition, the ethylene group at position 8 of the cyclohexene ring may generate steric effects similar to delta-9-nor-tetrahydrocannabinol. With 8-oxo-delta-9-tetrahydrocannabinol (Supplement 4-23), the introduction of the oxo group increases the polarity and hence it reduces the partition distribution into the membrane to interact with the receptor.
Hexahydrocannabinol (Supplement 4-24) has a reduced cyclohexane ring instead of a cyclohexene ring, which alters the orientation of this ring inside the membrane on receptor interaction due to conformational flexibility.
In cannabinol (Supplement 4-26), 7-hydroxy cannabinol (Supplement 4-28), and cannabinol methyl ether (Supplement 4-33), replacement of the nonplanar cyclohexene ring with a benzene ring abolishes the bioactivity, but its affinity for receptor increases, leading to the antagonistic mode of interaction with GPCR. The aromatic ring in cannabinol has π-electron cloud, which may interact with positively charged amino acid Lys or Arg due to cation−π interaction, resulting in deviation from bioactive conformation. Cation−π binding in cannabinol and 7-hydroxy cannabinol renders a stronger affinity for the receptor.
Bis-nor-cannabitrol (Supplement 4-25) and 10-ethoxy-9-hydroxy-delta-6a-tetrahydrocannabinol (Supplement 4-31) have hydroxyl groups at positions 9 and 10 in the cyclohexene ring, which makes it highly hydrophilic and hence its partition distribution into the membrane reduces significantly. 10-O-Ethyl bis-nor cannabitriol (Supplement 4-37) has a hydroxyl group at position 9 in the cyclohexyl ring, which makes it more hydrophilic, and reducing the chain length from a pentyl to a propyl group at the C-3 position of the phenolic group leads to low partition distribution into the membrane and reduces the affinity for the CB1 receptor.
In the case of cannabicycloic acid (Supplement 4-22), delta-8-tetrahydrocannabinolic acid (Supplement 4-3), THCA-A (Supplement 4-32), THCA-B (Supplement 4-15), delta-9-tetrahydrocannabinolic acid-C-4 (THCA-C-4) (Supplement 4-40), delta-9-tetrahydrocannabiorcolic acid (THCA-C1) (Supplement 4-42), and delta-9-nor-tetrahydrocannbinolic acid (Supplement 4-34), the COOH group at various positions of the phenolic group results in negatively charged species under physiological conditions. Generally, charged species have a lower blood–brain barrier,51 and our predicted ADMET values are in good agreement with the introduction of the carboxylic group in the phenolic group.
The five metabolites from cannabis such as compound-2 (Supplement 4-17), compound-3 (Supplement 4-14), 6a-R-cannabichromanone B (Supplement 4-27), 7-hydroxy cannabichromane (Supplement 4-29), and bis-nor-cannabichromanone (Supplement 4-38) lack the cyclohexene ring and substituted ketones occupy position 3 in the benzopyran ring. As per the structure–activity relationship rules of cannabinoids, a nonplanar cyclohexene ring is important but not essential for bioactivity. When the nonplanar cyclohexene ring is replaced by a bulky substituent attached to positions 3 and 4, still the molecules can show bioactivity. However, in the above five metabolites, no bulky substitution is attached to occupy positions 3 and 4 in the benzopyran ring and hence all of these five metabolites may not show bioactivity as THC.
Cannabichromanone (Supplement 4-10) has a fused tetrahydrofuran ring connecting phenolic OH and the benzopyran ring. It replaces the cyclohexene ring, and the 2-butanone group is attached to the C-3 position of the benzopyran ring. It is more nonpolar than THC, and the missing OH moiety at the phenolic group may reduce the favorable interaction energy, thereby reducing the affinity for the CB1 receptor.
In the case of delta-7-trans-isotetrahydrocannabinol (Supplement 4-30), the benzopyran ring is broken and as per SAR rules, the loss of benzopyran ring results in the absence of any bioactivity. Cannabichromanone D (Supplement 4-18) has a ring fused at position 3 of the benzopyran ring and the OH group of the phenolic group. It also has a carbonyl group at position 4 of the benzopyran ring. SAR rules of cannabinoids state that even with substitution of the hydroxyl group of phenolic pharmacophore, the molecule retains bioactivity. Hence, cannabichromanone D may show bioactivity, as three out of four pharmacophores may be enough to elicit receptor activation.
In all three metabolites, anhydrocannabimovone (Supplement 4-36), cannabielsoin (Supplement 4-39) (CBE), and cannabiglendol (Supplement 4-51), the 6-membered benzopyran ring is replaced by a 5-membered tetrahydrofuran ring. As the benzopyran ring is essential for bioactivity, its replacement may result in loss of bioactivity.
Delta-7-cis-isotetrahydrocannabivarin (Supplement 4-48) has bulky fused cyclohexyl and cyclopropyl groups that are attached to phenolic OH and C-6 position. The missing pharmacophore, benzopyran group, and propyl group at C-3 of the phenolic group instead of the pentyl group that reduces the affinity for the receptor and leads to complete loss of bioactivity.
In O-propyl-cannabidiol (Supplement 4-43), cannabidiol (CBD) (Supplement 4-44), and cannabidiol monomethyl ether (CBDM) (Supplement 4-52), the important pharmacophore benzopyran ring is opened, and it results in loss of bioactivity.
In the 3,4,5,6-tetrahydro-7-hydroxy-a,a-2-trimethyl-9-n-propyl-2,6-methano-2H-1-benzoxocin-5-methanol (Supplement 4-46), the absence of an important pharmacophore benzopyran ring results in loss of bioactivity. In addition, the pentyl group at the C-3 position of the phenolic group is replaced by a propyl group, and it reduces the affinity for the CB1 receptor.
Cannabicyclovarin (CBLV) (Supplement 4-41) has bulky fused cyclopentyl and cyclobutyl groups on the benzopyran ring, it is well tolerated for replacement of the cyclohexene group, and it has a propyl group at C-3 of the phenolic ring, which reduces the affinity for the CB1 receptor.
In 3-hydroxy-delta-4,5-cannabichromene (Supplement 4-45) and 4-acetoxy cannabichromene (Supplement 4-53), the basic cannabichromene has a missing cyclohexene ring, and the absence of bulky groups at 3, 4 positions in the benzopyran group makes it biologically inactive. Moreover, in the case of 4-acetoxy cannabichromene, it can undergo hydrolysis in the stomach giving rise to a hydrophilic 4-hydroxy phenolic group derivative. The hydroxyl group in 3-hydroxy-delta-4,5-cannabichromene and post-metabolic 4-acetoxy cannabichromene may have reduced partition into the membrane for receptor activation.
In cannabidivarin (Supplement 4-50) and cannabinodivarin (Supplement 4-49), the benzopyran ring is opened and it is similar to cannabidiol. The difference between cannabidivarin and cannabidiol is that the pentyl group is present at C-3 of the phenolic group in cannabidiol, and it is replaced by a propyl group in cannabidivarin. Hence, cannabidivarin is expected to have less affinity for the CB1 receptor than cannabidiol. Cannabidivarin resembles cannabinol, in which both have benzene rings instead of the cyclohexene ring. They differ with respect to the benzopyran and pentyl groups at C-3 of phenolic pharmacophore.
7-R-Cannabicoumarononic acid (Supplement 4-54) has an additional COOH group at position 2 in the phenolic group and the benzopyran ring is replaced by a 5-membered tetrahydrofuran ring. The introduction of the COOH group at position 2 of the phenolic group decreases the BBB, and the replacement of benzopyran by a THF ring results in complete loss of bioactivity.
Metabolites Detected in the Cannabis Smoke Activating Other Receptors
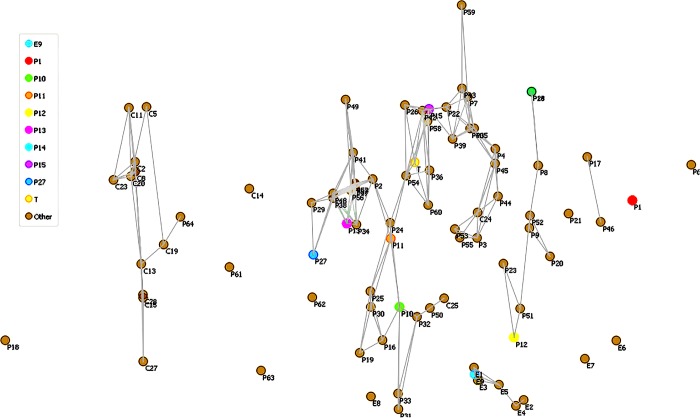
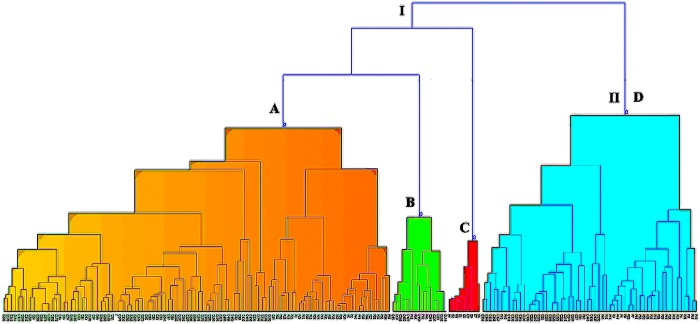
To find the metabolites in cannabis smoke having the possibility of activating other receptors in the brain, a hierarchical cluster analysis was performed using the positive reference drugs and 112 metabolites obtained from MDS analysis. The resulting dendrogram is presented in Figure 8.
Figure 8.
Dendrogram from hierarchical cluster analysis (HCA) showing 112 metabolites obtained from nonmetric MDS analysis on the ADMET descriptor of 165 metabolites. (A) Region highly populated with metabolites having THC-like scaffold. (B) Region with metabolites having no THC-like scaffold. (C) Endocannabinoids. (D) Region having metabolites with less populated THC-like scaffold.
The dendrogram shows that there are two major clusters I and II. Cluster I has 3 subclusters A–C and II has subcluster D. Subcluster C has all of the endocannabinoids, and they do not cluster with any reference drugs. The reference drugs clustered with metabolites are shown in Table 4.
Table 4. Metabolites from C. sativa, Smoke, and Other Cannabinoids Directly Connected with the Reference Drug in the Dendrogram Obtained from HCA.
| drug-metabolite cluster in the dendrogram | drug (P) | metabolite (C) |
|---|---|---|
| P18-C19 | ethanol | 2-chloroacetophenone |
| P18-C45 | ethanol | 3-methyl acetophenone |
| P18-C268 | ethanol | cuminaldehyde |
| P18-C427 | ethanol | propofol |
| P18-C455 | ethanol | thymol |
| P10, P47-C232, C436, C437, C441 | cocaine (P10), proheptazine (P47) | cannabioxepane (C232), radulanin A (C436), radulanin H (C437), radulanin L (C441) |
| P60-C84, C193 | lysergide | 7-R-cannabicoumarononic acid (C84), cannabicoumaronone (C193) |
| P61, P63-C246 | midomafetamine (P61), mescaline (P63) | carbofuran |
| P64 | nicotine | tetrahydrozoline |
| P42, P43-C418 | phenazocine (P42), phenomorphan (P43) | perrottetinenic acid |
| P62-C51, C57 | methaqualone | 4,5-dihydroxy-2,3,6-trimethoxy-9,10-dihydrophenanthrene (C51), 4-hydroxy-2,3,6,7-tetramethoxy-9,10-dihydrophenanthrene (C57) |
Subcluster A showed that only one reference drug (P60) clustered with cannabicoumaronone (C193) and 7-R-cannabicoumarononic acid (C84). In the subcluster B, ethanol (P18), the most abused addictive substance matches its physiochemical descriptors with five metabolites, propofol (C427), thymol (C455), 3-methyl acetophenone (C45), 2-chloroacetophenone (C19), and cuminaldehyde (C268). Ethanol exerts its neurophysiological effects through γ amino butyric acid (GABA) and glycine receptors.52 Propofol is an established anesthetic used for surgical applications.53 Propofol, a well-known anesthetic used for regular surgeries, interacts with the GABA receptor.54 Carbofuran is the carbamate insecticide used to control pests that infect C. sativa, and it is shown to bind melatonin and serotonin receptors, which affect biorthyms55 and metabolic and psychological processes,56 respectively. Thymol, a natural monoterpene, is isolated from Thymus vulgaris. Thymol, a monoterpene compound, is found to possess neuroprotective properties.57 It is used as an antimicrobial agent58 Thymol has been reported to bind with GABA receptors on its allosteric site.59 In the case of acetophenone derivatives, the basic acetophenone scaffold itself is reported to have slight narcotic properties on smelling its vapors.60 Both 2-chloroacetophenone and 3-methyl acetophenone have very high predicted blood–brain barrier values of 0.995 and 0.981, respectively. Combining both high BBB and basic scaffold acetophenone with narcotic properties, both the derivatives may have protein targets in the brain and exhibit psychoactive effects. Cuminaldehyde has a BBB of 0.951, and its parent compound cumene has a narcotic property and is aproven CNS depressant and hence the aldehyde derivative of cumene may bind to the GABA receptor and exhibit psychoactive property. It is very interesting to note that the correct selection of physiochemical and ADMET descriptors has enabled us to predict the metabolites from smoke activating GABA/glycine receptors by comparison with ethanol having no structural homology with the metabolites.
Tetrahydrozoline is a nasal solution and highly toxic causing sedation and muscle weakness on ingestion.61 Tetrahydrofuran interacts with α-1-adrenoreceptors,62 whereas its reference counterpart nicotine interacts with the nicotine receptor. Similarly, carbofuran, an insecticide used during horticulture of the cannabis plant, contaminates the product and is reported to interact with the melatonin receptor,63 whereas its counterpart reference drugs mescaline and midomafetamine interact with opioid and serotonin receptors, respectively. Remaining metabolites are cannabinoids, their scaffold homology with their opioid reference drug counterparts is much smaller, and their potential of interaction with opioid receptors is low to consider in this study.
In the identification of psychoactive metabolites from C. sativa, its smoke, and other phytocannabinoids, apart from SAR, there are four control points, which clearly have deciding factors with the exhibition of psychoactive character for metabolites. The first control point is the plasma protein binding (PPB), which controls the quantity of free available metabolite in serum for biological action. The second control point resides in the liver and the extent of metabolic changes it undergoes in the liver. The third control point is at the entry of the blood–brain barrier and finally its partition coefficient into the membrane to interact with the receptor. In the case of oral administration of cannabis products, the second control point is more important than for the smoking route of inhalation. Some of the metabolites may preserve the THC scaffold, but the functional group modification may affect the four control points depending on its polarity. More polar molecules would have less PPB, high metabolic activity, less BBB, and lower partition distribution into the membrane. On crossing overall four control points, the molecule must have functional groups giving rise to favorable interaction energy with the receptor for optimal binding and eliciting functional response. The scaffold homology factor calculated for 54 metabolites may directly correlate with the strength of the CB1 receptor binding and degree of psychoactive character. Orally administered cannabis has less psychoactive effects than the smoking route as the oral route64 produces metabolites by the action of liver enzymes such as CYP450 and resulting in modification of the scaffold leading to less psychoactive effects. Preservation of the cannabinoid scaffold has a much higher probability with the smoking route than the oral route of administration. Cannabinol and delta-8-THC are classified as psychoactive,65 and cannabidiol is considered nonpsychoactive in both oral and intravenous administrative routes. This observation correlates well with scaffold homology, as delta-8-THC (0.722) and cannabinol (0.63) have higher scaffold homology than cannabidiol (0.539). The relative proportion of each metabolite is an important factor in exerting the psychoactive character, as THC is the only metabolite present in the highest proportion in C. sativa.65 It will be interesting to test this cannabinoid scaffold homology factor in ranking the psychoactive character with more in vitro and in vivo experiments with various proportions or pure substances of each metabolite. If the cannabinoid scaffold theory is true, the metabolic engineering on cannabis plant can produce the product rich in metabolites with more medicinal benefits and less harmful effects of THC in the future.
Conclusions
Machine learning tools combined with multivariate methods applied to physiochemical and ADMET descriptors of metabolites of C. sativa, its smoke, and other phytocannabinoids lead to the following conclusions. First, computational tools identified psychoactive metabolites by comparing with agonists of various receptors such as cannabinoid, opioid, GABA/glycine. Second, this study showed that scaffold homology of cannabinoids may correlate well with the degree of psychoactive character. Third, some of the metabolites present in the smoke such as propofol and thymol have the possibility of interacting with GABA and glycine receptors. More in vitro and in vivo studies are required to understand the structural, physiochemical, and clinical parameters controlling the psychoactive character of cannabinoids present in C. sativa. Better understanding will lead to C. sativa plants with a new set of metabolite matrix having beneficial medicinal properties in the future.
Experimental Section
Database
The database containing 468 metabolites was generated from peer-reviewed articles on chemical constituents of C. sativa. It is comprises phytocannabinoids66−70 and noncannabinoids71 from C. sativa, phytocannabinoids from higher plants, liverworts, and fungi,72 and volatile compounds derived from C. sativa.73 Positive controls (74) were Schedule I−IV substances collected from the Yellow list of narcotic drugs under International control,74 the endocannabinoid agonists acting on CB1 and CB2 receptors.75 Eighteen negative controls were the small molecule metabolites found in the human serum having no psychoactive property. THC was added to the positive control list, as it is a psychoactive drug representing the cannabinoid category.
Physiochemical and ADMET Descriptors
For data mining and multivariate analysis, 32 physiochemical descriptors were calculated using the CDK molecular descriptor calculator.76 Twenty nine ADMET properties were predicted using the online platform from ADMETlab based on the comprehensive database of about 300 000 compounds.77 All of the physiochemical (Supplement 1) and ADMET descriptors (Supplement 2) of the entire database are provided in Supplements 1 and 2. Chemical structures of all of the 468 metabolites are provided in Supplement 3.
Computational Strategy
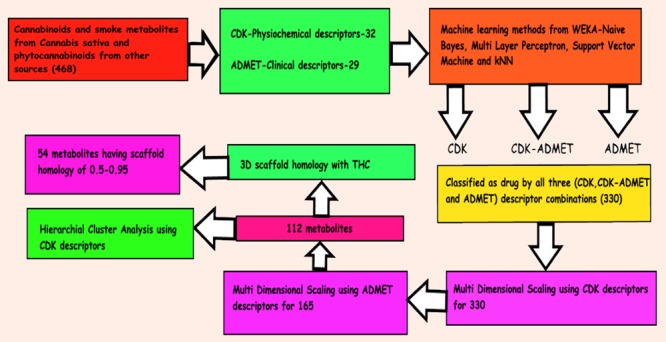
The computational strategy for classification and prediction is outlined in the TOC Figure. Initially, classification was performed by using four classifiers, namely, naïve-Bayes, multilayer perceptron, support vector machine, and k-nearest neighbor. Open-source application, Weka 3.8.3 from The University of Waikato, New Zealand, was used for this purpose.78 All of the members of the database were classified as drug (psychoactive) or no drug (not psychoactive) by all four classifiers using CDK (purely physiochemical), hybrid CDK–ADMET, and ADMET (purely predicted clinical descriptors) descriptors separately. Cross-validation (10-fold) was performed using all four classifiers on the training set having positive and negative controls under the same testing conditions for test candidates. The test candidate was classified as a “drug” when all four classifiers predict the test candidate as a drug for all three descriptor combinations (CDK, CDK–ADMET, and ADMET).
Detection of Overfitting of Data
To diagnose the overfitting of data, the training data was divided into 80% training set, 10% validation set, and 10% test set. All of the training, validation, and test sets had unique positive and negative controls, and each set had 6 unique negative controls. Training, validation, and test sets had 74, 11, and 7 unique candidates, respectively. The test set was designed to be tough having six negative controls and THC as positive control where it is only the psychoactive substance in the C. sativa. Training, validation, and test were performed for CDK, CDK–ADMET, and ADMET data types for all four (NB, MLP, SVM, and kNN) classifiers.
Nonmetric Multidimensional Scaling (NM-MDS)
Nonmetric MDS was executed for each set of 15 metabolites along with 74 reference drugs using Orange software (version 3.15.0) developed by Bioinformatics Lab at the University of Ljubljana, Slovenia, in collaboration with the open-source community.79
Scaffold Homology Analysis
Scaffold homology analysis was performed using the Marvin sketch 18.20 module from ChemAxon software solutions. The common scaffold of structures free of explicit hydrogen was compared with THC by 3D alignment option in the accurate mode, and the resulting pictures were extracted in PNG format.
Cluster Analysis
The final list of 112 predicted drug candidates was made to go through cluster analysis using CDK descriptor combinations. From the dendrograms, the predicted drug candidates clustering with positive controls were analyzed for possible respective receptor activation.
Acknowledgments
I dedicate this article to the late Prof. C. Ramakrishnan, who was an excellent teacher and researcher at the Molecular Biophysics Unit, Indian Institute of Science, Bangalore, India. I sincerely thank NACPT Director, Rathi Param, for her timely support and gratefully acknowledge the Ministry of Advanced Education and Skills Development for financial support during the preparation of this manuscript.
Glossary
Abbreviations
- GABA
γ amino butyric acid
- ADMET
absorption, distribution, metabolism, excretion, toxicity
- SAR
structure–activity relationship
- THC
tetra hydro cannabinol
- UNDOC
United Nations Office on Drugs and Crime
- CNS
central nervous system
- TPSA
total polar surface area
- BBB
blood–brain barrier
- GPCR
G-protein coupled receptor
- CB1
cannabinoid 1
- HCA
hierarchical cluster analysis
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b02663.
Financial support rendered by the Ministry of Advanced Education and Skills Development is gratefully acknowledged.
The author declares no competing financial interest.
Supplementary Material
References
- Drugs and Age, World Drug Report. United Nations Publication, Sales No. E.18.XI.9, 2018.
- Analysis of Drug Markets: Opiates, Cocaine, Cannabis, Synthetic Drugs, World Drug Report. United Nations Publication, Sales No. E.18.XI.9, 2018.
- National Academies of Sciences, Engineering, and Medicine . Therapeutic Effects of Cannabis and Cannabinoids. In The Health Effects of Cannabis and Cannabinoids: The Current State of Evidence and Recommendations for Research; The National Academies Press: Washington, DC, 2017; pp 85–140. [PubMed] [Google Scholar]
- Allison K. Weighing the Benefits and Risks of Medical Marijuana Use: A Brief Review. Pharmacy 2018, 6, 128. 10.3390/pharmacy6040128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federica P.; Vittoria B.; Virginia B.; Marco B.; Stefania B.; Lorenzo C. Cannabis sativa L. and Nonpsychoactive Cannabinoids: Their Chemistry and Role against Oxidative Stress, Inflammation, and Cancer. BioMed Res. Int. 2018, 1691428 10.1155/2018/1691428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouhamed Y.; Vishnyakov A.; Qorri B.; Sambi M.; Frank S. S.; Nowierski C.; Lamba A.; Bhatti U.; Szewczuk M. R. Therapeutic potential of medicinal marijuana: an educational primer for health care professionals. Drug, Healthcare Patient Saf. 2018, 10, 45–66. 10.2147/DHPS.S158592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegason P. D.; Jacob M. V.; Sarah S. S. The Role of Cannabis within an Emerging Perspective Schizophrenia. Medicines 2018, 5, 86. 10.3390/medicines5030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidra Z.; Deepak K.; Muhammad T. K.; Pirthvi R. G.; Kiran F. N. U. Epilepsy and Cannabis: A Literature Review. Cureus 2018, 10, e3278 10.7759/cureus.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernanda F. P.; Alvaro C. L.; Jaime E. C. H.; Jose A. C.; Regina H. S.; Vanessa C. A. Cannabidiol as a promising strategy to treat and prevent movement disorders?. Front. Pharmacol. 2018, 9, 482. 10.3389/fphar.2018.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexanabinol in Patients With Brain Cancer - Full Text View - ClinicalTrials.gov [Internet]. [cited 2017 Feb 10]. https://clinicaltrials.gov/ct2/show/NCT01654497?term=cannabinoid+AND+cancer&rank=33.
- Molly B.; Zenab T. K.; Mohammad B. K.; Maish K.; Ayobami W.; Bhagelu R. A.; Ali S. A.; David C. H.; Md N. H.; Babak B.; Krishnan M. D.; Kumar V. Selective activation of cannabinoid receptor-2 reduces neuroinflammation after traumatic brain injury via alternative macrophage polarization. Brain, Behav., Immun. 2018, 68, 224–237. 10.1016/j.bbi.2017.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman P.; Gilaad G. K.; Keith A. S.; Cynthia H. S. Insights into the role of cannabis in the management of inflammatory bowel disease. Ther. Adv. Gastroenterol. 2019, 12, 1756284819870977 10.1177/1756284819870977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudroff T.; Sosnoff J. Cannabidiol to Improve Mobility in People with Multiple Sclerosis. Front. Neurol. 2018, 9, 183. 10.3389/fneur.2018.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherelle L. C.; Christopher W. V. Plant-Based Cannabinoids for the Treatment of Chronic Neuropathic Pain. Medicines 2018, 5, 67. 10.3390/medicines5030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith A. S.; Nissar A. D.; Linda A. P. Regulation of nausea and vomiting by cannabinoids and the endocannabinoid system. Eur. J. Pharmacol. 2014, 722, 134–146. 10.1016/j.ejphar.2013.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leos L.; Jan J.; Jiri S.; Monika P.; Regina D. Medical cannabis in the treatment of cancer pain and spastic conditions and options of drug delivery in clinical practice. Biomed. Pap. 2018, 162, 18–25. 10.5507/bp.2018.007. [DOI] [PubMed] [Google Scholar]
- Hillig K. Genetic evidence for speciation in Cannabis (Cannabaceae). Genet. Resour. Crop Evol. 2005, 52, 161–180. 10.1007/s10722-003-4452-y. [DOI] [Google Scholar]
- Small E.; Beckstead H. D. Common cannabinoid phenotypes in 350 stocks of Cannabis. Lloydia 1973, 36, 144–165. [PubMed] [Google Scholar]
- Zerrin A. Cannabis, a complex plant: different compounds and different effects on individuals. Ther. Adv. Psychopharmacol. 2012, 2, 241–254. 10.1177/2045125312457586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brighenti V.; Pellati F.; Steinbach M.; Maran D.; Benvenuti S. Development of a new extraction technique and HPLC method for the analysis of non-psychoactive cannabinoids in fibre-type Cannabis sativa L. (hemp). J. Pharm. Biomed. Anal. 2017, 143, 228–236. 10.1016/j.jpba.2017.05.049. [DOI] [PubMed] [Google Scholar]
- Andre C. M.; Hausman J. F.; Guerriero G. Cannabis sativa: The plant of the thousand and one molecule. Front. Plant Sci. 2016, 7, 1–17. 10.3389/fpls.2016.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devane W. A.; Dysarz F. A.; Johnson M. R.; Melvin L. S.; Howlett A. C. Determination and characterization of a cannabinoid receptor in rat brain. Mol. Pharmacol. 1988, 34, 605–613. [PubMed] [Google Scholar]
- Shenglong Z.; Ujendra K. Cannabinoid Receptors and the Endocannabinoid System: Signaling and Function in the Central Nervous System. Int. J. Mol. Sci. 2018, 19, 833. 10.3390/ijms19030833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryberg E.; Vu H. K.; Larsson N.; Groblewski T.; Hjorth S.; Elebring T.; Sjorgren S.; Greasley P. J. Identification and characterisation of a novel splice variant of the human CB1receptor. FEBS Lett. 2005, 579, 259–264. 10.1016/j.febslet.2004.11.085. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mariscal I.; Krzysik-Walker S. M.; Doyle M. E.; Liu Q. R.; Cimbro R.; Calvo S. S. C.; Ghosh S.; Ciesla L.; Moaddel R.; Carlson O. D.; Rafal P. W.; O’Connell J. F.; Josephine M. E. Human CB1 receptor isoforms, present in hepatocytes and -cells, are involved in regulating metabolism. Sci. Rep. 2016, 6, 33302 10.1038/srep33302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing-Rong L.; Chun-Hung P.; Akitoyo H.; Chuan-Yun L.; Zheng-Xiong X.; Alvaro L.-B.; Maria-Paz V.; Hiroki I.; Tadao A.; Emmanuel S. O.; George R. U. Species differences in cannabinoid receptor 2 (CNR2 gene): identification of novel human and rodent CB2 isoforms, differential tissue expression, and regulation by cannabinoid receptor ligands. Genes, Brain Behav. 2009, 8, 519–530. 10.1111/j.1601-183X.2009.00498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. Y.; Bi G. H.; Li X.; Li J.; Qu H.; Zhang S. J.; Li C. Y.; Onaivi E. S.; Gardner E. L.; Xi Z. X.; Liu Q.-R. Species differences in cannabinoid receptor 2 and receptor responses to cocaine self-administration in mice and rats. Neuropsychopharmacology 2015, 40, 1037–1051. 10.1038/npp.2014.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z.; Ting X.; Saifei L.; Fulai Z.; Ming-Wei W. Application of chemical biology in target identification and drug discovery. Arch. Pharmacal Res. 2015, 38, 1642–1650. 10.1007/s12272-015-0643-2. [DOI] [PubMed] [Google Scholar]
- Tiejun C.; Qingliang L.; Zhigang Z.; Yanli W.; Stephen H. Structure-Based Virtual Screening for Drug Discovery: a Problem-Centric Review. AAPS J. 2012, 14, 133–141. 10.1208/s12248-012-9322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A. C.; David L. P.; Thomas D. M. Drug–target residence time and its implications for lead optimization. Nat. Rev. Drug Discovery 2006, 5, 730–739. 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- Feixiong C.; Weihua L.; Guixia L.; Yun T. In Silico ADMET Prediction: Recent Advances, Current Challenges and Future Trends. Curr. Top. Med. Chem. 2013, 13, 1273–1289. 10.2174/15680266113139990033. [DOI] [PubMed] [Google Scholar]
- Jagannathan R. Characterization of Drug-like Chemical Space for Cytotoxic Marine Metabolites Using Multivariate Methods. ACS Omega 2019, 4, 5402–5411. 10.1021/acsomega.8b01764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas A. M. Improving Drug Design: An Update on Recent Applications of Efficiency Metrics, Strategies for Replacing Problematic Elements, and Compounds in Nontraditional Drug Space. Chem. Res. Toxicol. 2016, 29, 564–616. 10.1021/acs.chemrestox.6b00043. [DOI] [PubMed] [Google Scholar]
- Ottl J.; Leder L.; Schaefer J. V.; Dumelin C. E. Encoded Library Technologies as Integrated Lead Finding Platforms for Drug Discovery. Molecules 2019, 24, 1629 10.3390/molecules24081629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-Chen L.; Stefano E. R.; Wen T.; Russ B. A. Machine learning in chemoinformatics and drug discovery. Drug Discovery Today 2018, 23, 1538–1546. 10.1016/j.drudis.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C. A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discovery Today: Technol. 2004, 1, 337–341. 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Pajouhesh H.; George R. L. Medicinal Chemical Properties of Successful Central Nervous System Drugs. Neurotherapeutics 2005, 2, 541–553. 10.1602/neurorx.2.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm K.; Stenberg P.; Luthman K.; Artursson P. Polar molecular surface properties predict the intestinal absorption of drugs in humans. Pharm. Res. 1997, 14, 568–571. 10.1023/A:1012188625088. [DOI] [PubMed] [Google Scholar]
- Kelder J.; Grootenhuis P. D. J.; Bayada D. M.; Delbressine L. P. C.; Ploemen J. P. Polar molecular surface as a dominating determinant for oral absorption and brain penetration of drugs. Pharm. Res. 1999, 16, 1514–1519. 10.1023/A:1015040217741. [DOI] [PubMed] [Google Scholar]
- Veber D. F.; Johnson S. R.; Cheng H.-Y.; Smith B. R.; Ward K. W.; Kopple K. D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Lopez-Tapia F.; Lou Y.; Brotherton-Pleiss C.; Kuglstatter A.; So S. S.; Kondru R. A. Potent seven-membered cyclic BTK (Bruton’s tyrosine Kinase) chiral inhibitor conceived by structure-based drug design to lock its bioactive conformation. Bioorg. Med. Chem. Lett. 2019, 29, 1074–1078. 10.1016/j.bmcl.2019.03.001. [DOI] [PubMed] [Google Scholar]
- Heck G. S.; Pintro V. O.; Pereira R. R.; de Ávila M. B.; Levin N. M. B.; de Azevedo W. F. Supervised Machine Learning Methods Applied to Predict Ligand- Binding Affinity. Curr. Med. Chem. 2017, 24, 2459–2470. 10.2174/0929867324666170623092503. [DOI] [PubMed] [Google Scholar]
- Fatma T.The correlation between critical anaesthetic dose and melting temperatures in synthetic membranes. Thesis, University of Copenhagen, 2015. [Google Scholar]
- Michael C. H.; Megan H. P.; Stephen D. G. Multidimensional scaling. Rev. Cogn. Sci. 2013, 4, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springall A. A review of multidimensional scaling. J. Appl. Stat. 1978, 5, 146–192. 10.1080/768370750. [DOI] [Google Scholar]
- Jan de L.; Willem H.. Theory of Multidimensional Scaling. In Handbook of Statistics, 2nd ed.; Krishnaiah P. R.; Kanal L. N., Eds.; North-Holland Publishing Company, 1982; pp 285–316. [Google Scholar]
- Taguchi Y.-h.; Oono Y. Relational patterns of gene expression via non-metric multidimensional scaling analysis. Bioinformatics 2005, 21, 730–740. 10.1093/bioinformatics/bti067. [DOI] [PubMed] [Google Scholar]
- Julien P.; Hervé A.; Matthieu M.; David T.; Marie C. Multidimensional Scaling Reveals the Main Evolutionary Pathways of Class A G-Protein-Coupled Receptors. PLoS One 2011, 6, e19094 10.1371/journal.pone.0019094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H.; Kiran V.; Mengchen P.; Lu Q.; Gye W.-H.; Yiran W.; Suwen Z.; Wenqing S.; Shanshan L.; Anisha K.; Robert B. L.; Edward L. S.; Jo-Hao H.; Nikolai Z.; Han Z.; Irina K.; Beili W.; Qiang Z.; Michael A. H.; Laura M. B.; Alexandros M.; Raymond C. S.; Zhi-Jie L. Crystal Structure of the Human Cannabinoid Receptor CB1. Cell 2016, 167, 750–762. 10.1016/j.cell.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketan H.; Waseem G.; Mahmoud A. E.; Michael A. R.; Soumyajit M. Enhanced Solubility, Stability, and Transcorneal Permeability of Delta-8-Tetrahydrocannabinol in the Presence of Cyclodextrins. AAPS PharmSciTech 2011, 12, 723–731. 10.1208/s12249-011-9639-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerry F.; Roger M. B.. Bioavailability of Drugs to the Brain and the Blood-Brain Barrier; Monograph series; National Institute of Drug Abuse, 1992. [Google Scholar]
- Karina P. A.; Armando G. S.; David M. L. Alcohol and the Brain: Neuronal Molecular Targets, Synapses, and Circuits. Neuron 2017, 96, 1223–1238. 10.1016/j.neuron.2017.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinori K.; Masamitsu S.; Shinichi Y.; Toru I.; Hideaki H. The Experimental and Clinical Pharmacology of Propofol, an Anesthetic Agent with Neuroprotective Properties. CNS Neurosci. Ther. 2008, 14, 95–106. 10.1111/j.1527-3458.2008.00043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H.; Watanabe Y.; Isshiki A.; Uchino H. Neuroprotective properties of propofol and midazolam, but not pentobarbital, on neuronal damage induced by forebrain ischemia, based on the GABAA receptors. Acta Anaesthesiol. Scand. 1999, 43, 153–162. 10.1034/j.1399-6576.1999.430206.x. [DOI] [PubMed] [Google Scholar]
- Ramesh C. G. Carbofuran toxicity. J. Toxicol. Environ. Health, Part A 1994, 43, 383–418. 10.1080/15287399409531931. [DOI] [PubMed] [Google Scholar]
- Marina P.-G.; Margarita L. D.; Rajendram V. R. Carbamate Insecticides Target Human Melatonin Receptors. Chem. Res. Toxicol. 2017, 30, 574–582. 10.1021/acs.chemrestox.6b00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayate J.; Sheikh A.; MF Nagoor Meeran.; Suraiya A. A.; Shreesh O. Neuroprotective Effects of Thymol, a Dietary Monoterpene Against Dopaminergic Neurodegeneration in Rotenone-Induced Rat Model of Parkinson’s Disease. Int. J. Mol. Sci. 2019, 20, 1538. 10.3390/ijms20071538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpanen T. J.; Worthington T.; Hendry E. R.; Conway B. R.; Lambert P. A. Antimicrobial efficacy of chlorhexidine digluconate alone and in combination with eucalyptus oil, tea tree oil and thymol against planktonic and biofilm cultures of Staphylococcus epidermidis. J. Antimicrob. Chemother. 2008, 62, 1031–1036. 10.1093/jac/dkn325. [DOI] [PubMed] [Google Scholar]
- Priestley C. M.; Williamson E. M.; Wafford K.; Sattelle D. B. Thymol, a constituent of thyme essential oil, is a positive allosteric modulator of human GABAA receptors and a homo-oligomeric GABA receptor from Drosophila melanogaster. Br. J. Pharmacol. 2003, 140, 1363–1372. 10.1038/sj.bjp.0705542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services . Hazardous Substances Data Bank (HSDB, online database); National Toxicology Information Program, National Library of Medicine: Bethesda, MD, 1993. [Google Scholar]
- Henry A. S.; Jacqualine R.; Tama S. S. Drug facilitated sexual assault using an over-the-counter ocular solution containing tetrahydrozoline (Visine). Leg. Med. 2007, 9, 192–195. 10.1016/j.legalmed.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Lowry J. A.; Brown J. T. Significance of the imidazoline receptors in toxicology. Clin. Toxicol. 2014, 52, 454–469. 10.3109/15563650.2014.898770. [DOI] [PubMed] [Google Scholar]
- Marina P.-G.; Margarita L. D.; Rajendram V. R. Carbamate Insecticides Target Human Melatonin Receptors. Chem. Res. Toxicol. 2017, 30, 574–582. 10.1021/acs.chemrestox.6b00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimalakonda K. C.; Seely K. A.; Bratton S. M.; Brents L. K.; Moran C. L.; Endres G. W.; et al. Cytochrome P450-mediated oxidative metabolism of abused synthetic cannabinoids found in K2/Spice: identification of novel cannabinoid receptor ligands. Drug Metab. Dispos. 2012, 40, 2174–2184. 10.1124/dmd.112.047530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kögel C. C.; López-Pelayo H.; Balcells-Olivero M. M.; Colom J.; Gual A. Psychoactive constituents of cannabis and their clinical implications: a systematic review. Adicciones 2018, 30, 140–151. [DOI] [PubMed] [Google Scholar]
- Radwan M. M.; ElSohly M. A.; Slade D.; Ahmed S. A.; Khan I. A.; Ross S. A. Biologically Active Cannabinoids from High-Potency Cannabis sativa. J. Nat. Prod. 2009, 72, 906–911. 10.1021/np900067k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safwat A. A.; Samir A. R.; Desmond S.; Mohamed M. R.; Ikhlas A. K.; Mahmoud A. E. Structure determination and absolute configuration of cannabichromanone derivatives from high potency Cannabis sativa. Tetrahedron Lett. 2008, 49, 6050–6053. 10.1016/j.tetlet.2008.07.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed M. R.; Mahmoud A. E.; El-Alfy A. T.; Safwat A. A.; Desmond S.; Afeef S. H.; Susan P. M.; Lisa W.; Suzanne S.; Stephen J. C.; Samir A. R. Isolation and Pharmacological Evaluation of Minor Cannabinoids from High-Potency Cannabis sativa. J. Nat. Prod. 2015, 78, 1271–1276. 10.1021/acs.jnatprod.5b00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melissa M. L.; Yi Y.; Ewa W.; Hance A. C.; Lakshmi P. K. Chemical Profiling of Medical Cannabis Extracts. ACS Omega 2017, 2, 6091–6103. 10.1021/acsomega.7b00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf B.Chapter 2: Chemistry and Analysis of Phytocannabinoids and Other Cannabis Constituents. In Marijuana and the Cannabinoids; Springer, 2007. [Google Scholar]
- Mohamed M. R.; Mahmoud A. E.; Desmond S.; Safwat A. A.; Lisa W.; El-Alfy A. T.; Ikhlas A. K.; Samir A. R. Non-cannabinoid constituents from a high potency Cannabis sativa variety. Phytochemistry 2008, 69, 2627–2633. 10.1016/j.phytochem.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumír O. H.; Stefan M. M.; Eduardo M.; Orazio T.-S.; Giovanni A. Phytocannabinoids: a unified critical inventory. Nat. Prod. Rep. 2016, 33, 1357–1392. 10.1039/C6NP00074F. [DOI] [PubMed] [Google Scholar]
- Somchai R.; Jacek A. K. Characterizing the Smell of Marijuana by Odor Impact of Volatile Compounds: An Application of Simultaneous Chemical and Sensory Analysis. PLoS One 2015, 10, e0144160 10.1371/journal.pone.0144160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- List of narcotic drugs under International control (Yellow List) from International Narcotics Control Board, 57th edition; 2018.
- Kazuhito T.; Toru U.; Yasuo O.; Natsuo U. Endocannabinoids and related N-acylethanolamines: biological activities and metabolism. Inflammation Regener. 2018, 38, 28. 10.1186/s41232-018-0086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle N.; Guha R.; Willighagen E.; Adams S.; Alvarsson J.; Bradley J. C.; et al. Open Data, Open Source and Open Standards in chemistry: The Blue Obelisk five years on. J. Cheminf. 2011, 3, 37. 10.1186/1758-2946-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jie D.; Ning-Ning W.; Zhi-Jiang Y.; Lin Z.; Yan C.; Defang O.; Ai-Ping L.; Dong-Sheng C. ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminf. 2018, 10, 29. 10.1186/s13321-018-0283-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibe F.; Mark A. H.; Ian H. W.. The WEKA Workbench. Online Appendix for “Data Mining: Practical Machine Learning Tools and Techniques”, 4th ed.; Morgan Kaufmann, 2016. [Google Scholar]
- Demsar J.; Curk T.; Erjavec A.; Gorup C.; Hocevar T.; Milutinovic M.; Mozina M.; Polajnar M.; Toplak M.; Staric A.; Stajdohar M.; Umek L.; Zagar L.; Zbontar J.; Zitnik M.; Zupan B. An evaluation of machine learning methods for prominence detection in French. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.