Abstract
Significant increases in rice yield and stress resistance are constant demands for breeders. However, high yield and high stress resistance are often antagonistic to each other. Here, we report several new rice mutants with high yield and excellent cold tolerance that were generated by simultaneously editing three genes, OsPIN5b (a panicle length gene), GS3 (a grain size gene) and OsMYB30 (a cold tolerance gene) with the CRISPR–Cas9 (clustered regularly interspaced short palindromic repeats-associated protein 9) system. We edited two target sites of each gene with high efficiency: 53% for OsPIN5b-site1, 42% for OsPIN5b-site2, 66% for GS3-site1, 63% for GS3-site2, 63% for OsMYB30-site1, and 58% for OsMYB30-site2. Consequently, the ospin5b mutants, the gs3 mutants, and the osmyb30 mutants exhibited increased panicle length, enlarged grain size and increased cold tolerance, respectively. Then nine transgenic lines of the ospin5b/gs3, six lines of ospin5b/osmyb30 and six lines of gs3/osmyb30 were also acquired, and their yield related traits and cold tolerance corresponded to the genes being edited. Additionally, we obtained eight ospin5b/gs3/osmyb30 triple mutants by editing all three genes simultaneously. Aside from the ospin5b/gs3/osmyb30-4 and ospin5b/gs3/osmyb30-25 mutants, the remaining six mutants had off-target events at the putative off-target site of OsMYB30-site1. The results also showed that the T2 generations of these two mutants exhibited higher yield and better cold tolerance compared with the wild type. Together, these results demonstrated that new and excellent rice varieties with improved yield and abiotic stress resistance can be generated through gene editing techniques and may be applied to rice breeding. Furthermore, our study proved that the comprehensive agronomic traits of rice can be improved with the CRISPR–Cas9 system.
Keywords: OsPIN5b, GS3, OsMYB30, cold tolerance, panicle length, grain length, CRISPR/Cas9
Introduction
Rice is one of the most important cereal crops in the world and feeds more than half of the world’s population (Khush, 1999). However, a shortage of rice has been an urgent threat for breeders as the human population has continued expanding for several decades (Wang and Li, 2008; Xing and Zhang, 2010). Rapid changes of climate have further restricted the rice yield. Therefore, it is an urgent matter for breeders to adopt advanced breeding methods that can not only improve the rice yield but also make rice more environmentally adaptable.
Precise gene engineering technologies, such as the transcription activator-like effector nucleases (TALENs) (Boch and Bonas, 2010), zinc finger nucleases (ZFNs) (Kim et al., 1996) and CRISPR–Cas9 (Jinek et al., 2012), have become fashionable strategies for developing new elite plant varieties to alleviate food insecurity owing to their high efficiency, speed and accuracy (Li et al., 2016; Li et al., 2017; Shen et al., 2018). Among those fashionable strategies, the CRISPR–Cas9 system is the most popular method to solve the outstanding issues currently because of its well-researched and constant improvement (Cong et al., 2013; Li et al., 2013; Mali et al., 2013).
In the past several decades, traditional hybrid breeders have been troubled by the cultivation of hybrid crops for the reason that their progeny did not have stable maintenance of their beneficial phenotypes owing to genetic segregation. Recently, researchers have made it possible to retain parental heterozygosity by the subsequent generations clonally through seeds using the CRISPR–Cas9 system. In their studies, clonal progeny was obtained through seeds when the edited BBM1, BBM2 and BBM3 genes substituting mitosis for meiosis (MiMe) and the expression of BBM1 are combined in the egg cell (Khanday et al., 2019). Researchers also edited four genes, REC8, PAIR1, OSD1 and MTL, in hybrid rice for clonal reproduction of F1 rice hybrids through seeds (Wang et al., 2019). Therefore, the extensive usage of this system provides new scientific guidance for adopting advanced techniques to develop elite rice varieties.
Rice yield is related to many agricultural traits: panicle number/plant, grain number/panicle, thousand grain weight and so on. Many important agricultural trait-related genes provide valuable resources for the application of bioengineering methods to improve the quality of crops. For example, OsPIN5b, an endoplasmic reticulum (ER) localized protein, has been inferred to be an auxin carrier and has important functions in auxin balance and transport. The expression of OsYUC1, an IAA related gene, is significantly increased in the OsPIN5b overexpression plants. Decreased expression of OsPIN5b leads to pleiotropic effects characterized as longer panicles, higher tiller number and higher rice yield compared with the wild type (Lu et al., 2015). GS3, which is widely used in the study of rice yield improvement, is comprised of four different functional domains that participate in the grain size regulatory network: an organ size regulation (OSR) domain, a transmembrane domain, a tumor necrosis factor receptor/nerve growth factor receptor (TNFR/NGFR) family cysteine-rich domain and a von Willebrand factor type C (VWFC) domain. Functional loss of the OSR domain leads to long grains, while changing the structure of the TNFR/NGFR and VWFC domains produces the opposite effect (Mao et al., 2010).
Furthermore, rice yield can be affected by many environmental factors. For instance, the rice growth and yield can be influenced by cold stress, which is one of the important environmental factors. To date, several cold stress-related genes have been cloned, including COLD1, OsSRFP1, and SGD1 (Fang et al., 2015; Ma et al., 2015b; Wang et al., 2017). Another cold-responsive R2R3-type MYB gene OsMYB30, can bind to the promoters of β-amylase (BMY) genes as a transcription factor and has a negative influence in rice cold tolerance. Under cold stress, OsMYB30 and OsJAZ9 function as a complex to suppress the expression of BMY genes, thus affecting the starch degradation and maltose accumulation, which may contribute to an increase in cold sensitivity (Lv et al., 2017). Because of their effects on rice yield, cold tolerance related genes should be given more attention for their application in coping with cold disasters and in the expansion of rice cultivation areas into the subtropical zone.
Here, we report several novel rice mutants produced by editing OsPIN5b, GS3 and OsMYB30 with the CRISPR–Cas9 system in Nipponbare, which is a typical japonica rice with its whole genome sequence available and is widely used as a donor in rice breeding (International Rice Genome Sequencing, 2005). Our study revealed that the ospin5b mutants, the gs3 mutants and the osmyb30 mutants exhibited increased panicle length, enlarged grain size and enhanced cold tolerance, respectively. When two of the three genes were edited, the transgenic plants had the corresponding phenotypes, proving that we could edit three genes simultaneously to obtain the new elite varieties. The results then showed that the two triple mutants, ospin5b/gs3/osmyb30-4 and ospin5b/gs3/osmyb30-25, in which all three genes were edited simultaneously without off-target events, exhibited higher yield and better cold tolerance compared to the wild type. In summary, we provided an example of how to improve the comprehensive agricultural traits of rice through use of the CRISPR–Cas9 system, which may offer new possibilities for rice breeding.
Materials and Methods
Plant Materials
Nipponbare was used for rice transformation in this research. These materials under natural field conditions were grown in the Wuhan experimental fields of Wuhan University to investigate the yield related traits. The seedlings used for cold stress treatments were grown in a greenhouse or a chamber at 4°C.
Vector Construction
All of the vectors were constructed as previously described (Ma et al., 2015a; Ma and Liu, 2016). First, the target adaptors were cloned into the pYLgRNA-OsU6a vector by using T4 DNA ligase. Then, the ligated products as a template were amplified using U-F and gR-R primers ( Table S2 ). Next, secondary PCR was employed to ligate the six cassettes. Finally, BsaI and T4 DNA ligase were used to clone the cassettes into the pYLCRISPR/Cas9Pubi-H vector.
Off-Target Detection
The potential off-target sites were predicted on the website http://crispr.hzau.edu.cn/CRISPR/. The primers ( Table S2 ), designed from Primer premier 5, were used for amplifying the off-target sequences. Then, the PCR products were sequenced for the detection of off-target events.
Measurements of Rice Traits
Plant height, panicle number/plant, grain number/panicle, pollen fertility, seed-setting rate and heading date were measured with three random T2 generation plants. Ten randomly grains from each plant were selected to measure the grain length and three T2 generation plants repeats. Grain weight was calculated with 1,000 grains and three repeats.
Cold Stress Treatments
The cold stress treatments were performed as previous studies (Lv et al., 2017). The wild type and transgenic lines were grown in a greenhouse with normal environment until three-leaf stage, and then the all seedlings were transferred into a chamber at 4°C. After 5 to 10 days, the seedlings were transferred back to the normal environment to record their survival rates.
RNA Extraction and qRT-PCR
Total RNA of the wild type and transgenic lines were extracted with TRIzol reagent according to the product manual. Then, the extracted RNA was treated and purified with chloroform and DNase I, and the purified RNA was used to synthesize cDNA using M-MLV (Invitrogen). Quantitative reverse transcription–PCR (qRT-PCR) was performed with the primers ( Table S2 ) using the Lightcycler 480 (Roche), taking the actin gene as the control.
Selection of Target Sequences
Target sequences were predicted on the website http://crispr.hzau.edu.cn/CRISPR/ and the secondary structure between the target sequence and sgRNA were predicted on the website http://unafold.rna.albany.edu/?q=mfold/RNA-Folding-Form2.3. We designed two sgRNAs for OsPIN5b (Os08g0529000), two for GS3 (Os03g0407400), and two for OsMYB30 (Os02g0624300) with the primers ( Table S2 ).
Rice Transformation
The CRISPR/Cas9 vector was transformed into A. tumefaciens strain EHA105 by electroporation. Transformation of rice was performed using a method described previously (Hiei et al., 1994). The genomic DNA was extracted from the T0 generation and the target sites were amplified by PCR ( Figure S1 ) with the primers ( Table S2 ). The PCR products were sequenced for the detection of mutations.
Homology Modeling
The relative protein structures were modeled by SWISS-MODEL. Amino acid sequences of the wild type and mutants were imported to the predict website for modeling the optimum protein structures ( Figure S2 ). According to the GMQE (Global Model Quality Estimation) values and the QMEAN4 scores, we selected 5yzg.1.L and 5a1s.1.B as optimum templates for OsPIN5b and OsMYB30, respectively. The predicted protein structures were analyzed by Swiss-PdbViewer software.
Results
The CRISPR–Cas9 System Had High Efficiency in Editing Three Genes in Rice
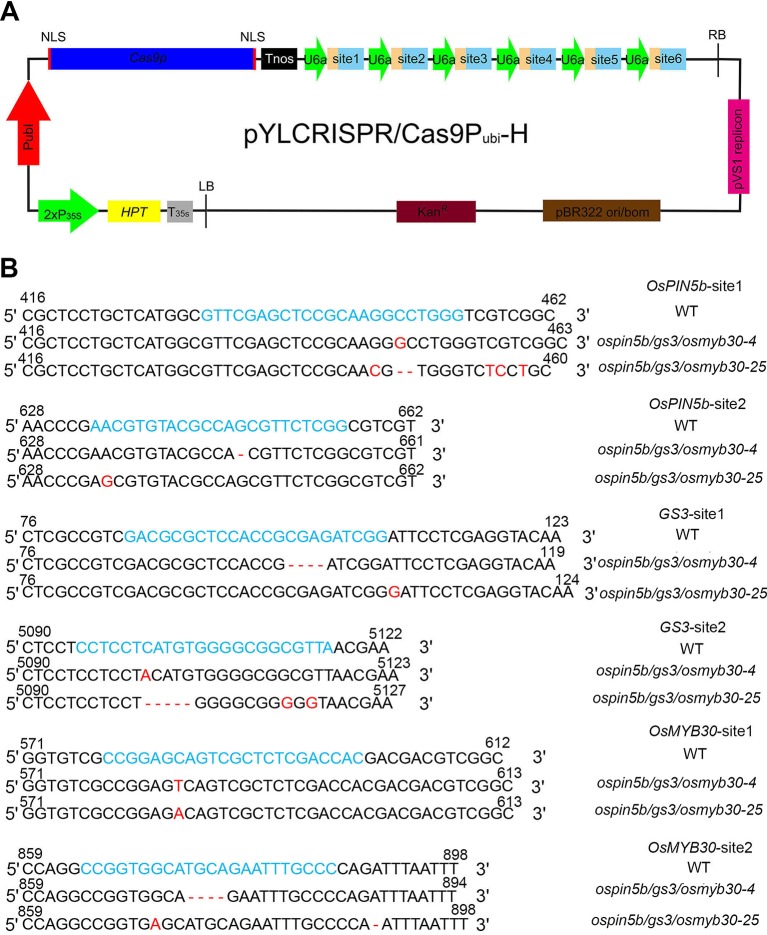
To obtain rice mutants with both high yield and good cold tolerance, we selected these three genes to edit: the panicle length gene OsPIN5b, the grain size gene GS3 and the cold tolerance gene OsMYB30. Two target sites of each gene were selected: OsPIN5b-site1 and OsPIN5b-site2, GS3-site1 and GS3-site2, and OsMYB30-site1 and OsMYB30-site2 ( Figure S1 ). We used Golden Gate assembly to clone six sgRNA cassettes into the CRISPR/Cas9 vector; the structure of the CRISPR/Cas9 vector was shown in Figure 1A . By examination of the PCR products of the target sites, we finally obtained a total of thirty-eight independent transgenic lines. The results showed that the CRISPR/Cas9 system had a high editing efficiency in the T0 generation: 53% for OsPIN5b-site1, 42% for OsPIN5b-site2, 66% for GS3-site1, 63% for GS3-site2, 63% for OsMYB30-site1, and 58% for OsMYB30-site2 ( Table 1 ). A high frequency of homozygous or biallelic mutations were also found after analyzing the results with the Degenerate Sequence Decoding (DSD) website http://dsdecode.scgene.com/ (Liu et al., 2015). Finally, we identified eight triple mutants (ospin5b/gs3/osmyb30) in which all three genes were edited with high efficiency (21%) from these thirty-eight independent transgenic lines ( Table 1 ). Thus, these studies demonstrated that the CRISPR/Cas9 system has high editing efficiency in manipulating multiple agronomic trait-related genes simultaneously.
Figure 1.
The ospin5b/gs3/osmyb30-4 and ospin5b/gs3/osmyb30-25 were precisely edited with CRISPR/Cas9 system. (A) Schematic diagram of the CRISPR/Cas9 vector. The pYLCRISPR/Cas9 binary vector was based on the pCAMBIA1300 backbone which contained the Kanamycin resistance gene. HPT encoded hygromycin B phosphotransferase, which could be driven by the cauliflower mosaic virus 35S promoter (P35S). The Cas9 was driven by the maize ubiquitin promoter (Pubi) and used to edit target sites. Tnos was the terminator of nopaline synthase gene which was chose to terminate the expression of Cas9 gene. U6a, small nuclear RNA promoters, was employed to facilitate the expression of multiple sgRNA cassettes. The six sgRNA cassettes also were inserted behind U6a. (B) Six target sequences alignment in ospin5b/gs3/osmyb30-4 and ospin5b/gs3/osmyb30-25. Blue represents the target sequences. Red represents the changing base.
Table 1.
The editing efficiency of CRISPR/Cas9 system in rice.
| Target Site | No. of Independent Plants | No. of plants with mutation | Mutation rate (%) | ||
|---|---|---|---|---|---|
| OsPIN5b-Site1 | 38 | 20 | 53 | ||
| OsPIN5b –Site2 | 38 | 16 | 42 | ||
| GS3-Site1 | 38 | 25 | 66 | ||
| GS3-Site2 | 38 | 24 | 63 | ||
| OsMYB30-Site 1 | 38 | 24 | 63 | ||
| OsMYB30-Site 2 | 38 | 22 | 58 | ||
| Target gene | No. of independent plants | No. of plants with mutation | Mutation rate (%) | ||
| OsPIN5b | 24 | 38 | 63 | ||
| GS3 | 25 | 38 | 66 | ||
| OsMYB30 | 26 | 38 | 68 | ||
| OsPIN5b/GS3/OsMYB30 | 8 | 38 | 21 | ||
OsMYB30-site1 Had Off-Target Events
Previous studies have demonstrated that off-target events are unavoidable when using the CRISPR/Cas9 system (Lawrenson et al., 2015). We wondered if there might have been off-target events that may disturb the comprehensive phenotypes in the eight ospin5b/gs3/osmyb30 mutants. First, we selected two putative off-target sites for each target on the website http://crispr.hzau.edu.cn/CRISPR/. By amplifying the putative off-target sites of the eight triple-mutant lines, we found that only two of them, ospin5b/gs3/osmyb30-4 and ospin5b/gs3/osmyb30-25, did not experience off-target events and that the remaining six had off-target events at the first putative off-target sites of OsMYB30-site1 ( Figure 1B , Table 2 ). It was unexpected that the off-target rate of the first putative off-target site of OsMYB30-site1 was higher than that of the other putative off-target sites. Previous studies have put forward that mismatches between the seed sequence (12bp approach to the PAM) and the target sequence are important for decreasing the off-target frequency (Tsai et al., 2015). Sequence alignment of the twelve putative off-target sites showed that the most frequent off-target site had a higher similarity to its target site in both the seed sequence and target sequence compared with the other putative off-target sites. These results suggest that we should select target sites with low sequence similarity to other genes in the seed sequence and carefully target sequences to decrease the frequency of off-target events.
Table 2.
The putative off-target event in the ospin5b gs3 osmyb30 triple mutant lines.
| Target Site | Putative off-target site | The sequence of putative off-target site | No. of plants | No. of plants with mutations | Mutation rate (%) |
|---|---|---|---|---|---|
| OsPIN5b-Site1 | Chr3:9566074–9566052 | CCTCGAGCTCTGCAAGGCTTTGG | 8 | 0 | 0 |
| Chr1:19390027–19390049 | TTTCGAGCTGCGCAAGGCGCGGG | 8 | 0 | 0 | |
| OsPIN5b-Site2 | Chr7: 8747238–8747260 | CTCGTGTGCGCCAACGTTCTCGG | 8 | 0 | 0 |
| Chr3:24251589–24251567 | AACGTGCTCGTCAGCGTCCTCGG | 8 | 0 | 0 | |
| GS3-Site 1 | Chr8: 2495427–2495449 | GACGCGCTCCACCGCGCGCTCGG | 8 | 0 | 0 |
| Chr7:14816697–14816719 | AACTCGCTCAACCGCGAGAGGGG | 8 | 0 | 0 | |
| GS3-Site 2 | Chr9: 1832101–1832123 | TCGCGCCACCCCACACGAGGTGG | 8 | 0 | 0 |
| Chr7: 27472889–27472911 | TAACCCCGCACCGCATGAGGCGG | 8 | 0 | 0 | |
| OsMYB30-Site 1 | Chr3:13108929–13108951 | GTGGCGGAGAGCGACTGCACGGG | 8 | 6 | 75 |
| Chr5: 1475321–1475343 | CTGCGCGAGAGCGACTGCGCCGG | 8 | 0 | 0 | |
| OsMYB30-Site 2 | Chr4: 24538627–24538649 | TGACAAATGCTGCATGCCATGGG | 8 | 0 | 0 |
| Chr2:28909447–28909469 | CGTCAAATTCTGCATGACAGTGG | 8 | 0 | 0 |
The green font represented the PAM motif (NGG); the red font represented the mismatch bases.
The Panicle Length Was Obviously Increased in the ospin5b Mutants
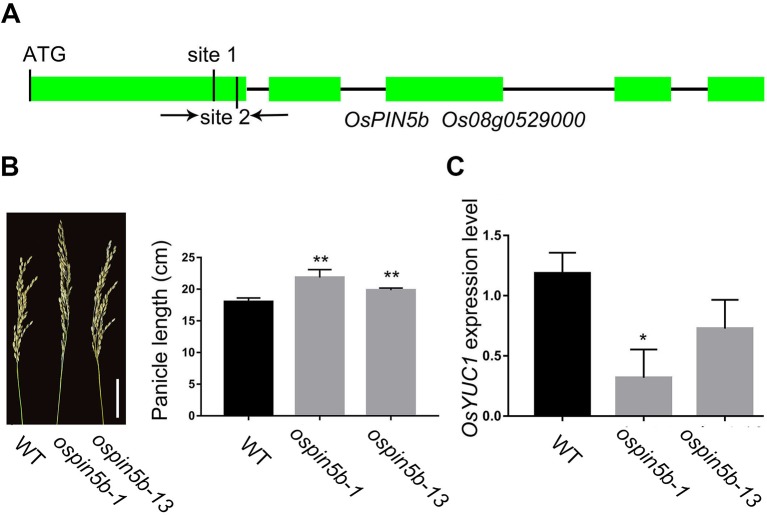
OsPIN5b, located in the endoplasmic reticulum (ER), is involved in the balance and transport of auxin in rice (Lu et al., 2015). The ospin5b knockdown mutant showed more tiller numbers and longer panicles compared with the wild type. In this study, we selected two sites located in the first exon to change the coding region of OsPIN5b with the CRISPR–Cas9 system ( Figure 2A ). Finally, we identified 24 mutant lines of OsPIN5b from 38 independent lines (63%) ( Table 1 ). T2 generation of the ospin5b mutants, ospin5b-1 and ospin5b-13, in which only OsPIN5b had been edited, were planted in the field for further phenotype analysis. The average panicle length of the wild type is 18.3 cm, while the ospin5b-1 and ospin5b-13 mutants achieved 22.6 cm and 19.8 cm, respectively ( Figure 2B ).
Figure 2.
CRISPR/Cas9-induced ospin5b mutants and the expression level of cell cycle-related genes. (A) Schematic representation of the OsPIN5b genomic region. The two target sites of OsPIN5b and PCR primers were marked in first exon. (B) The phenotype of panicles and data statistics of ospin5b-1, ospin5b-13 and WT, Bar = 5 cm. (C) The expression level of marker gene OsYUC1 in WT, ospin5b-1 and ospin5b-13. Data are means ± SD from three biological replicates. Student’s t test, **P < 0.001, *0.001 < P < 0.05.
Previous studies have proven that the expression level of many auxin biosynthesis and transport related genes in the ectopic expression OsPIN5b lines are different from that in the wild type. It is worth noting that the expression level of YUCCA genes and the content of IAA are affected in the ectopic expression lines. Therefore, we took OsYUCCA1 as a marker gene to measure the effect of the CRISPR–Cas9 system in vivo. As expected, the expression level of the marker gene was significantly decreased in the ospin5b-1 and ospin5b-13 transgenic lines compared with the wild type ( Figure 2C ). Together, these results revealed that the editing efficiency of the CRISPR–Cas9 system was high in the OsPIN5b target sites, and the panicle length of the ospin5b mutants was obviously increased compared with the wild type.
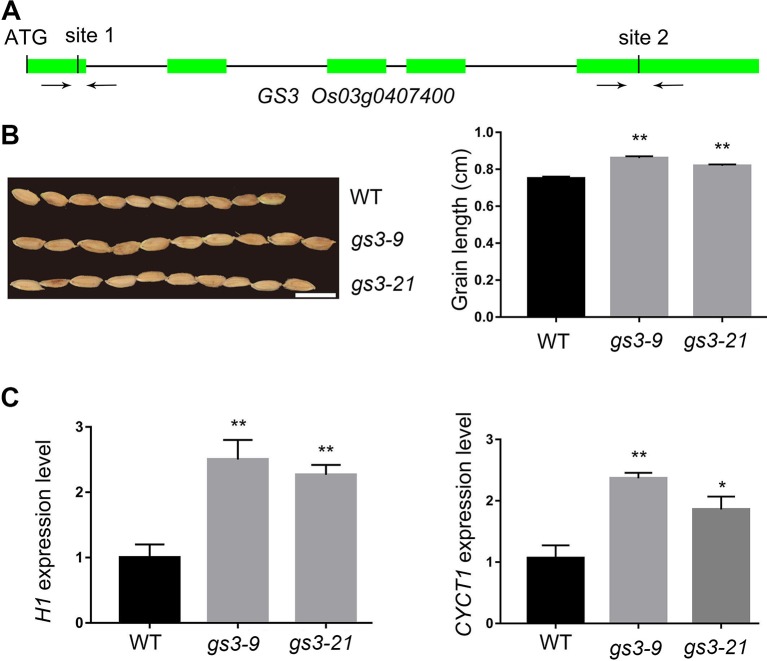
The Grain Size Was Strikingly Enlarged in the GS3 Edited Lines
The GS3 gene has been cloned in the Minghui 63 background. A substitution of C to A in the allele of GS3 resulted in a 178-amino-acid truncation and, thus, an altered grain size (Fan et al., 2006). In this study, aiming to interrupt the function of GS3, we chose two target sites from the website http://crispr.hzau.edu.cn/CRISPR/ to edit with the CRISPR–Cas9 system. The two sites are located in the first and fifth exons of GS3 ( Figure 3A ). Finally, we acquired 25 GS3 edited lines among the 38 independent transgenic lines (66%) ( Table 1 ). In the 25 edited lines, some of the mutations were homozygous, which might happen before the embryogenic cell division ( Table S1 ) (Zhang et al., 2014). T2 generation of the gs3 edited lines, gs3–9 and gs3–21, in which only GS3 had been edited, were planted for grain length characterization. The average grain length of the wild type was 7.5 mm, while it was 8.6 mm of gs3–9 and 8.2 mm of gs3–21 ( Figure 3B ). In recent years, increasing numbers of genes have been cloned and studied, most of which regulate the grain size by controlling cell proliferation and expansion in spikelet hulls, such as FZP, GS5, and GS2 (Li et al., 2011; Hu et al., 2015; Ren et al., 2018). Two genes, CYCT1 and H1, involved in the cell cycle, were selected to measure the effects of the CRISPR/Cas9 system on the GS3 target sites according to previous studies. The qRT-PCR results showed that the expression levels of the marker genes in gs3–9 and gs3–21 were significantly increased compared with the wild-type, which was consistent with expectations ( Figure 3C ). These results demonstrated that GS3 was successfully edited with the CRISPR/Cas9 system, and the gs3 mutants exhibited larger grain sizes in Nipponbare, which is consistent with previous studies (Fan et al., 2006).
Figure 3.
CRISPR/Cas9-induced gs3 mutants and the expression level of cell cycle-related genes. (A) Schematic representation of the GS3 genomic region. The two target sites of GS3 and PCR primers were marked in first and fifth exons. (B) The grain phenotype of gs3–9, gs3–21 and WT, and statistics for the average grain length. Bar = 1 cm. (C) The expression level of marker gene OsH1 and OsCYCT1 in WT, gs3–9 and gs3–21. Data are means ± SD from three biological replicates. Student’s t test, **P < 0.001, *0.001 < P < 0.05.
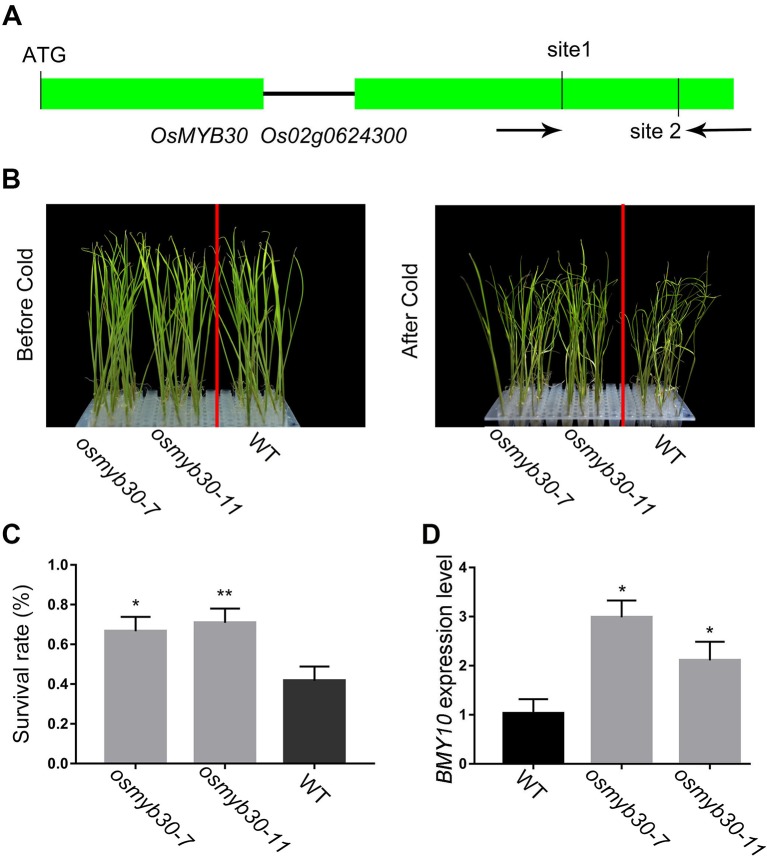
The osmyb30 Mutant Lines Displayed Enhanced Cold Tolerance
OsMYB30 was characterized as a cold-responsive R2R3-type MYB gene. Previous studies showed that the osmyb30 mutant, which was generated by inserting T-DNA in the second exon, exhibited weak cold sensitivity compared with the wild type and the overexpression lines. Therefore, we chose two target sites from the second exon of OsMYB30 for editing with the CRISPR–Cas9 system ( Figure 4A ). Finally, 26 mutant lines were identified among the 38 independent lines, and the editing efficiency reached up to 68% ( Table 1 ). Among the 26 mutant lines, most of them were single-base mutations, which belong to biallelic mutations ( Table S1 ). T2 generation of the osmyb30 mutants, osmyb30-7 and osmyb30-11, in which only OsMYB30 had been edited, were used to do the following experiments. Under the condition of 4°C, the wild type exhibited a wilting phenotype earlier than the two mutants osmyb30-7 and osmyb30-11 ( Figure 4B ), and the plant survival rates of osmyb30-7 and osmyb30-11 (66.7% and 70.8%, respectively) were higher than the wild type (41.7%; Figure 4C ), indicating that the two mutant lines had better cold tolerance compared with the wild type. Previous study has proven that OsMYB30 is a transcription factor that interacts with OsJAZ9 to regulate the downstream BMY genes expression under a cold environment (Lv et al., 2017). Hence, we detected the expression level of BMY10 in leaves grown at 4°C and found it was increased in the two mutant lines compared with the wild type ( Figure 4D ). Thus, the phenotypes of the osmyb30 mutants indicated that the OsMYB30 target sites were efficiently edited in rice, and the osmyb30 mutant lines have similar effects in Nipponbare, which is consistent with the previous studies (Lv et al., 2017).
Figure 4.
The phenotype analysis of osmyb30 mutants and the expression level of downstream BMY10 gene. (A) Schematic representation of the OsMYB30 genomic region. The two target sites of OsMYB30 and PCR primers were marked in second exon. (B) The seedlings of osmyb30-7, osmyb30-11 and WT after being treated in 4°C chamber. (C) The survival rate of osmyb30 mutants after 4°C treatment. (D) The expression level of marker gene OsBMY10 in WT, osmyb30-7 and osmyb30-11. Data are means ± SD from three biological replicates. Student’s t test, **P < 0.001, *0.001 < P < 0.05.
The Phenotype Analysis of Double Mutants
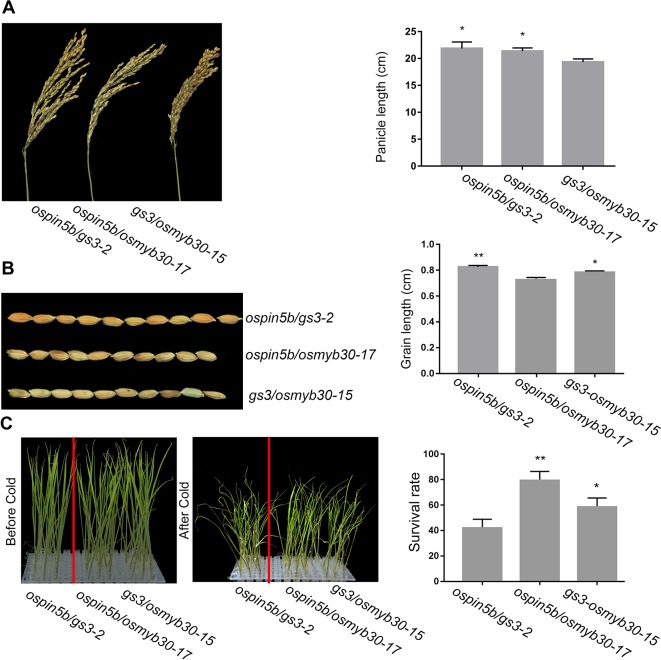
When we detected the efficiency of the system, several double mutants of the three genes were identified: nine transgenic lines of the ospin5b/gs3, six lines of ospin5b/osmyb30 and six lines of gs3/osmyb30 ( Table 3 ). To figure out whether the double mutants will show the corresponding phenotypes, ospin5b/gs3-2, ospin5b/osmyb30-17 and gs3/osmyb30-15 were chosen to measure their yield related traits and cold tolerance. As shown in Table 4 and Figure S3 , the panicle number/plant and grain number/panicle of the ospin5b/gs3-2 and ospin5b/osmyb30-17 double mutants were improved compared with the wild type and the gs3/osmyb30-15 mutants while the thousand grain weight of the ospin5b/gs3-2 and gs3/osmyb30-15 double mutants were increased compared with the wild type and the ospin5b/osmyb30-17 mutants. The three double mutants were transferred into a chamber at 4°C at the three-leaf stage to test the cold tolerance and the ospin5b/osmyb30-17 and gs3/osmyb30-15 showed a wilting phenotype later than ospin5b/gs3-2 ( Figure 5 ). The plant survival rates of ospin5b/osmyb30-17 and gs3/osmyb30-15 (79.1% and 58.3%) were higher than ospin5b/gs3-2 (47.6%) as showed in Figure 5 . The data of the three double mutants indicated that the traits could be added together in the same mutants and we could study the new and elite varieties with multiple genes were edited simultaneously.
Table 3.
The number of single mutants, double mutants and triple mutants.
| OsPIN5b | ospin5b | ospin5b/gs3 | ospin5b/osmyb30 | ospin5b/gs3/osmyb30 | |
| No. | 24 | 1 | 9 | 6 | 8 |
| GS3 | gs3 | ospin5b/gs3 | gs3/osmyb30 | ospin5b/gs3/osmyb30 | |
| No. | 25 | 2 | 9 | 6 | 8 |
| OsMYB30 | osmyb30 | ospin5b/osmyb30 | gs3/osmyb30 | ospin5b/gs3/osmyb30 | |
| No. | 26 | 6 | 6 | 6 | 8 |
Table 4.
The phenotype of wild type and edited lines.
| Characters | Plant Height (cm) | Panicle number/plant | Grain number/panicle | Thousand grain weight (g) | Yield/plant (g) | Pollen fertility (%) | Seed-setting rate (%) | Heading date (d) |
|---|---|---|---|---|---|---|---|---|
| Wild type | 97.8 ± 1.0 | 9.7 ± 1.5 | 92.6 ± 6.0 | 23.8 ± 0.4 | 19.3 ± 2.0 | 100 ± 0 | 89.1 ± 1.7 | 77 ± 2.6 |
| ospin5b-1 | 98.1 ± 1.3 | 13.6 ± 0.5* | 115.0 ± 7.5* | 23.5 ± 0.7 | 27.0 ± 1.7** | 100 ± 0 | 89.6 ± 2.8 | 77 ± 0.5 |
| gs3-9 | 97.7 ± 1.4 | 9.3 ± 1.1 | 90.6 ± 3.0 | 26.8 ± 0.3** | 22.8 ± 1.3* | 100± 0 | 87.0 ± 3.3 | 76 ± 1.5 |
| ospin5b/gs3-2 | 97.6 ± 0.6 | 13.6 ± 0.5* | 117.0 ± 7* | 26.4 ± 0.2* | 28.5 ± 0.7** | 100 ± 0 | 89.4 ± 1.8 | 77 ± 0.3 |
| ospin5b/osmyb30-17 | 98.3 ± 1.1 | 13.3 ± 1.1* | 115.0 ± 6.9* | 23.4 ± 1.3 | 27.0 ± 0.6** | 100 ± 0 | 87.5 ± 0.5 | 77 ± 1.1 |
| gs3/osmyb30-15 | 96.7 ± 0.4 | 10.3 ± 0.5 | 93.6 ± 5.5 | 25.2 ± 0.7* | 22.8 ± 0.7 | 100 ± 0 | 89.6 ± 2.6 | 78 ± 1.5 |
| ospin5b/gs3/osmyb30-4 | 97.3 ± 0.9 | 12.3 ± 0.5* | 116 ± 4.5** | 25.3 ± 0.4* | 30.4 ± 0.9** | 100 ± 0 | 90.4 ± 2.6 | 78 ± 0.5 |
| ospin5b/gs3/osmyb30-25 | 99.1 ± 0.5 | 14.3 ± 1.1* | 113.6 ± 7.2* | 24.9 ± 0.3* | 28.7 ± 2.3** | 100 ± 0 | 89.7 ± 1.3 | 80 ± 1.1 |
Data are means ± SD from three biological replicates. Student’s t test, **P < 0.001, *0.001 < P < 0.05.
Figure 5.
The phenotypes of ospin5b/gs3-2, ospin5b/osmyb30-17 and gs3/osmyb30-15 mutants. (A) The panicles and data statistics of ospin5b/gs3-2, ospin5b/osmyb30-17 and gs3/osmyb30-15 mutants, Bar = 5 cm. (B) The grain phenotype of ospin5b/gs3-2, ospin5b/osmyb30-17 and gs3/osmyb30-15, and statistics for the average grain length. Bar = 1 cm. (C) The seedlings and survival rate of ospin5b/gs3-2, ospin5b/osmyb30-17 and gs3/osmyb30-15 after being treated in 4°C chamber. Data are means ± SD from three biological replicates. Student’s t test, **P < 0.001, *0.001 < P < 0.05.
The ospin5b/gs3/osmyb30 Triple Mutants Exhibited High Yield and Good Cold Tolerance
We have generated the ospin5b mutant, the gs3 mutant, and the osmyb30 mutant with the CRISPR–Cas9 system, and their phenotypes were consistent with the previous studies. To investigate the phenotypes of plants with all three genes edited simultaneously, we obtained two triple mutants, named ospin5b/gs3/osmyb30-4 and ospin5b/gs3/osmyb30-25, which did not experience off-target events ( Figure S2 ). T2 generations of ospin5b/gs3/osmyb30-4 and ospin5b/gs3/osmyb30-25 had normal characteristics, including plant height, pollen fertility, and seed-setting rate, compared with the wild type ( Table 4 ). However, they exhibited increased panicle length, less greenness, more tillers, larger grain size, and decreased cold sensitivity in the experimental field ( Figure 6A ). The yield-related traits are presented in Table 4 , which showed the panicle number/plant, grain number/panicle and thousand grain weight. Under the condition of 4 °C, the plant survival rates of ospin5b/gs3/osmyb30-4 and ospin5b/gs3/osmyb30-25 were 70.8% and 79.1%, respectively, while that of the wild type was only 45.8% ( Figure 6A ).
Figure 6.
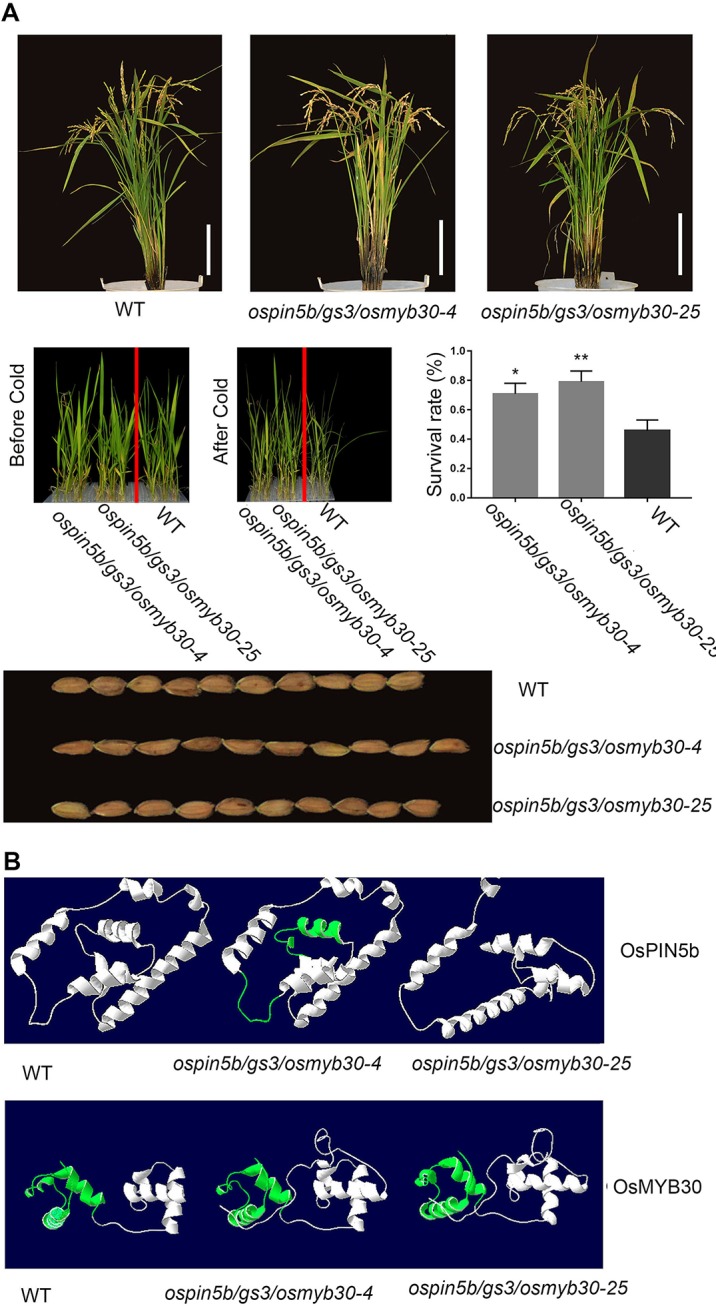
The phenotypes of ospin5b/gs3/osmyb30-4, ospin5b/gs3/osmyb30-25 and the predicted protein structure of OsPIN5b and OsMYB30 in ospin5b/gs3/osmyb30-4, ospin5b/gs3/osmyb30-25. (A) Whole plant morphology, survival rate after cold treatment and grain size in WT, ospin5b/gs3/osmyb30-4, ospin5b/gs3/osmyb30-25. Bar = 20 cm. (B) The OsPIN5b and OsMYB30 protein structures of WT, ospin5b/gs3/osmyb30-4 and ospin5b/gs3/osmyb30-25 were predicted by SWISS-MODEL. The observed differential regions were highlighted in green.
The function of proteins is known to be based on their three-dimensional structures, so it was necessary for us to determine the structures of the mutants, which can be achieved by SWISS-MODEL. Therefore, we modeled the structures of OsPIN5b and OsMYB30 both in mutated form and original form, respectively. From Figure 6B we could clearly observe that there was a slight change in the distortion angle of the OsPIN5b protein in ospin5b/gs3/osmyb30-4 and a completely different form in ospin5b/gs3/osmyb30-25 compared with the wild type. For OsMYB30, the conserved Myb-like DNA-binding domain in the two mutants had a looser structure compared with that in the wild type ( Figure 6B ). Furthermore, the two mutated forms of OsMYB30 had other obvious changes compared with wild type OsMYB30 ( Figure 6B ). The mutations of ospin5b/gs3/osmyb30-4 and ospin5b/gs3/osmyb30-25 caused a truncation in the OSR domain of GS3, leading to the functional loss of this protein. Therefore, there was no need to predict its structures.
Taken together, we can conclude that the CRISPR–Cas9 system can efficiently edit the three genes alone or simultaneously. Breeders may be able to simultaneously manipulate multiple agriculture traits with this system to cultivate new varieties.
Discussion
To date, the CRISPR/Cas9 system has been used to solve challenges that have not been overcome with traditional approaches in rice, such as the coordination between disease resistance and high yield. With the development of this technology, many genes in a good number of crops have been edited with this system, such as sorghum, maize, rice, wheat, soybean, potato, cotton, grapefruit, and watermelon (Shan et al., 2013; Zhang et al., 2016). In this study, we gained new rice mutants with both high yield and good cold tolerance by simultaneously editing the yield-related genes (OsPIN5b and GS3) and the cold stress related gene (OsMYB30) with the CRISPR–Cas9 system, which decreased many manpower and material resources compared with the traditional breeding method. We obtained single mutants, double mutants and triple mutants together to study the relationship of high yield and abiotic resistance, which has been an obstacle to phenotype improvement.
However, the CRISPR–Cas9 system has the weakness of unavoidable off-target events, and improving the accuracy of this system is a challenge that scientists have been working to solve. Here, we obtained eight transgenic plants where all three genes were edited simultaneously with the CRISPR–Cas9 system. However, six of them had off-target events at the first off-target site of OsMYB30-site1. By analyzing the sequence of these off-target sites, we suggest that the selected target sites should have a GC content of 50–70% with fewer than four consecutive Ts. Furthermore, those sites should avoid forming hairpin or stem structures with the sg-RNA sequences. In addition, they should contain more mismatches in the seed sequence and target sequence (Ma and Liu, 2016).
Nipponbare is a model plant that has been widely used and studied in rice breeding. Therefore, we adopted it as the material to study how to improve its shorter panicles and cold tolerance compared with other rice cultivars. Since yield and resistance related genes have pleiotropic effects on the phenotypes of rice in different rice varieties backgrounds (Fan et al., 2006), it was necessary to determine the effects of each single gene in Nipponbare before studying the comprehensive phenotypes of these three genes together.
First, the PIN protein family has 12 members in rice and most of them participate in the regulatory network of plant hormones, exhibiting functional redundancy (Xu et al., 2005; Wang et al., 2009; Zhang et al., 2012). In previous studies, the panicle length was significantly increased in ospin5b knockdown lines. In this study, the ospin5b mutants showed less greenness, which may affect the absorption of light energy and thus the yield, but the panicle number/plant and grain number/panicle in the mutants still increased ( Table 4 ), indicating that the defect of less greenness was not enough to affect the yield. In addition, the PIN protein family has 12 members in rice and most of them participate in the regulatory network of plant hormone with exhibiting functional redundancy, so there were no significant changes of other agricultural traits in the knock out mutants.
Second, it was reported that GS3 negatively regulates grain length, and its nonsense mutant allele exists in many large grain rice varieties. In Nipponbare, the thousand grain weight and grain size were significantly increased in the gs3 edited lines, while no other phenotypes changed significantly. This result indicated that the GS3 gene has a positive impact on yield in the Nipponbare background.
Third, OsMYB30 is a nuclear protein and acts as a negative regulator of rice cold tolerance. In the previous study, survival rate of the mutant was significantly higher (80%) than the wild type of ZH11 (less than 20%) (Lv et al., 2017). In this study, the plant survival rates of osmyb30-7 and osmyb30-11 (66.7% and 70.8%, respectively) were higher than the wild type (41.7%) under the condition of 4 ℃. Compared with previous study, survival rates of the mutants in this study were not as high as previous study, possibly because the smaller seedlings (three-leaf stage seedlings) in this study was more cold sensitive than the seedlings (four-leaf stage seedlings) used in the previous study. Because the cold treatment was performed at the three-leaf stage, the surviving plants were not influenced in the following growth periods. Together, these results indicated that the CRISPR–Cas9 system can be used to improve rice yield by editing OsPIN5b and GS3 and enhance cold tolerance by editing OsMYB30 in Nipponbare.
Since the single mutants of the three genes had phenotypes consistent with those reported in previous studies and the other phenotypes were not influenced, we then identified the comprehensive phenotypes of the double mutants and triple mutants. After analyzing the phenotypes of double mutants, we concluded that these mutants showed the desired phenotypes which corresponded to the edited genes. The success of editing two genes together made us sure that we could edit three genes simultaneously to obtain the elite varieties. As shown in Figure 6A , Table 4 and Table S3 , several agricultural traits of the ospin5b/gs3/osmyb30 triple mutants were improved, while the length of the grain did not achieve the desired effect like the single gs3 mutants. We can attribute this phenomenon to the overlapping or influence of the three regulation networks, the incompatibility between high yield and high resistance, the limitations of production growth and so on. Nevertheless, we confirmed that the ospin5b/gs3/osmyb30 triple mutants exhibited both higher yield and better cold tolerance compared with the wild type.
Together, these results indicated that the CRISPR–Cas9 system had high efficiency in editing these three genes both individually and simultaneously. At the same time, the successful transformation of phenotypes in ospin5b/gs3/osmyb30-4 and ospin5b/gs3/osmyb30-25 has provided a new method to solve problems encountered in traditional rice breeding. According to our findings, we highly recommend that preferable rice materials should be obtained by rationally manipulating those genes which control different agricultural traits in rice.
Conclusion
In this study, we successfully edited OsPIN5b, GS3 and OsMYB30 individually and simultaneously with high efficiency in rice through use of the CRISPR–Cas9 system. We obtained several novel rice mutants with high yield, good cold tolerance or both that may be applied to rice breeding. Additionally, our studies demonstrated that the simultaneous improvement of multiple agronomic traits can be achieved by the CRISPR–Cas9 system.
Data Availability Statement
All datasets generated for this study are included in the article/ Supplementary Material .
Author Contributions
WH designed the study. YZ performed the entire experiment. JW performed vector construction and rice transformation. WZ and QW performed the RNA extraction.
Funding
This research was supported by the National Key R&D Program of China (2017YFD0100400), National Natural Science Foundation of China (31771746) and the National Rice Industry Technology System (CARS-01-07).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Ruifeng He and Dr. Chunlan He for critical comments on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01663/full#supplementary-material
References
- Boch J., Bonas U. (2010). Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436. 10.1146/annurev-phyto-080508-081936 [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Xing Y., Mao H., Lu T., Han B., Xu C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. 10.1007/s00122-006-0218-1 [DOI] [PubMed] [Google Scholar]
- Fang H., Meng Q., Xu J., Tang H., Tang S., Zhang H., et al. (2015). Knock-down of stress inducible OsSRFP1 encoding an E3 ubiquitin ligase with transcriptional activation activity confers abiotic stress tolerance through enhancing antioxidant protection in rice. Plant Mol. Biol. 87, 441–458. 10.1007/s11103-015-0294-1 [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. 10.1046/j.1365-313x.1994.6020271.x [DOI] [PubMed] [Google Scholar]
- Hu J., Wang Y., Fang Y., Zeng L., Xu J., Yu H., et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 8, 1455–1465. 10.1016/j.molp.2015.07.002 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing, P. (2005). The map-based sequence of the rice genome. Natur 436, 793–800. 10.1038/nature03895 [DOI] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanday I., Skinner D., Yang B., Mercier R., Sundaresan V. (2019). A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Natur 565, 91–95. 10.1038/s41586-018-0785-8 [DOI] [PubMed] [Google Scholar]
- Khush G. S. (1999). Green revolution: preparing for the 21st century. Genome 42, 646–655. 10.1139/g99-044 [DOI] [PubMed] [Google Scholar]
- Kim Y. G., Cha J., Chandrasegaran S. (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U.S.A. 93, 1156–1160. 10.1073/pnas.93.31156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrenson T., Shorinola O., Stacey N., Li C., Ostergaard L., Patron N., et al. (2015). Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 16, 258. 10.1186/s13059-015-0826-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. F., Norville J. E., Aach J., McCormack M., Zhang D., Bush J., et al. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. 10.1038/nbt2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Li X., Zhou Z., Wu P., Fang M., Pan X., et al. (2016). Reassessment of the Four Yield-related Genes Gn1a, DEP1, GS3, and IPA1 in Rice Using a CRISPR/Cas9 System. Front. Plant Sci. 7, 377. 10.3389/fpls.2016.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Fan C., Xing Y., Jiang Y., Luo L., Sun L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. 10.1038/ng.977 [DOI] [PubMed] [Google Scholar]
- Li X., Zhou W., Ren Y., Tian X., Lv T., Wang Z., et al. (2017). High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing. J. Genet. Genomics 44, 175–178. 10.1016/j.jgg.2017.02.001 [DOI] [PubMed] [Google Scholar]
- Liu W., Xie X., Ma X., Li J., Chen J., Liu Y. G. (2015). DSDecode: A web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol. Plant 8 (9), 1431–1433. 10.1016/j.molp.2015.05.009 [DOI] [PubMed] [Google Scholar]
- Lu G., Coneva V., Casaretto J. A., Ying S., Mahmood K., Liu F., et al. (2015). OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 83, 913–925. 10.1111/tpj.12939 [DOI] [PubMed] [Google Scholar]
- Lv Y., Yang M., Hu D., Yang Z., Ma S., Li X., et al. (2017). The OsMYB30 Transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing beta-amylase expression. Plant Physiol. 173, 1475–1491. 10.1104/pp.16.01725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Liu Y. G. (2016). CRISPR/Cas9-Based multiplex genome editing in monocot and dicot plants. Curr. Protoc. Mol. Biol. 115, 31 36 31–31 36 21. 10.1002/cpmb.10 [DOI] [PubMed] [Google Scholar]
- Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., et al. (2015. a). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. 10.1016/j.molp.2015.04.007 [DOI] [PubMed] [Google Scholar]
- Ma Y., Dai X., Xu Y., Luo W., Zheng X., Zeng D., et al. (2015. b). COLD1 confers chilling tolerance in rice. Cell 160, 1209–1221. 10.1016/j.cell.2015.01.046 [DOI] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H., Sun S., Yao J., Wang C., Yu S., Xu C., et al. (2010). Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. U.S.A. 107, 19579–19584. 10.1073/pnas.1014419107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Hu J., Xu Q., Cui Y., Zhang Y., Zhou T., et al. (2018). FZP determines grain size and sterile lemma fate in rice. J. Exp. Bot. 69, 4853–4866. 10.1093/jxb/ery264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q., Wang Y., Li J., Zhang Y., Chen K., Liang Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688. 10.1038/nbt2650 [DOI] [PubMed] [Google Scholar]
- Shen L., Wang C., Fu Y., Wang J., Liu Q., Zhang X., et al. (2018). QTL editing confers opposing yield performance in different rice varieties. J. Integr. Plant Biol. 60, 89–93. 10.1111/jipb.12501 [DOI] [PubMed] [Google Scholar]
- Tsai M. Y., Lu Y. F., Liu Y. H., Lien H. W., Huang C. J., Wu J. L., et al. (2015). Modulation of p53 and met expression by Kruppel-like factor 8 regulates zebrafish cerebellar development. Dev. Neurobiol. 75, 908–926. 10.1002/dneu.22258 [DOI] [PubMed] [Google Scholar]
- Wang Y., Li J. (2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59, 253–279. 10.1146/annurev.arplant.59.032607.092902 [DOI] [PubMed] [Google Scholar]
- Wang J. R., Hu H., Wang G. H., Li J., Chen J. Y., Wu P. (2009). Expression of PIN genes in rice (Oryza sativa L.): tissue specificity and regulation by hormones. Mol. Plant 2, 823–831. 10.1093/mp/ssp023 [DOI] [PubMed] [Google Scholar]
- Wang D., Wang Y., Long W., Niu M., Zhao Z., Teng X., et al. (2017). SGD1, a key enzyme in tocopherol biosynthesis, is essential for plant development and cold tolerance in rice. Plant Sci. 260, 90–100. 10.1016/j.plantsci.2017.04.008 [DOI] [PubMed] [Google Scholar]
- Wang C., Liu Q., Shen Y., Hua Y., Wang J., Lin J., et al. (2019). Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol. 37, 283–286. 10.1038/s41587-018-0003-0 [DOI] [PubMed] [Google Scholar]
- Xing Y., Zhang Q. (2010). Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442. 10.1146/annurev-arplant-042809-112209 [DOI] [PubMed] [Google Scholar]
- Xu M., Zhu L., Shou H., Wu P. (2005). A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 46, 1674–1681. 10.1093/pcp/pci183 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Li J., Zhang W., Yan S., Wang R., Zhao J., et al. (2012). The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance. Plant J. 72, 805–816. 10.1111/j.1365-313X.2012.05121.x [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhang J., Wei P., Zhang B., Gou F., Feng Z., et al. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. 10.1111/pbi.12200 [DOI] [PubMed] [Google Scholar]
- Zhang D., Li Z., Li J. F. (2016). Targeted gene manipulation in plants using the CRISPR/Cas technology. J. Genet. Genomics 43, 251–262. 10.1016/j.jgg.2016.03.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/ Supplementary Material .