The flagellar type III protein export apparatus coordinates protein export with assembly, which allows the flagellum to be efficiently built at the cell surface. Hook completion is an important morphological checkpoint for the sequential flagellar assembly process. The protein export apparatus switches its substrate specificity from the hook protein to the filament protein upon hook completion. FliK, FlhB, and FlhA are involved in the export-switching process, but the mechanism remains a mystery. By analyzing a slow-cleaving flhB(P270A) mutant, we provide evidence that an interaction between FliK and FlhB induces conformational rearrangements in FlhB, followed by a structural remodeling of the FlhA ring structure that terminates hook assembly and initiates filament formation.
KEYWORDS: bacterial flagella, FlhA, FlhB, FliK, hook length control, substrate specificity switching, type III protein secretion
ABSTRACT
FlhA and FlhB are transmembrane proteins of the flagellar type III protein export apparatus, and their C-terminal cytoplasmic domains (FlhAC and FlhBC) coordinate flagellar protein export with assembly. FlhBC undergoes autocleavage between Asn-269 and Pro-270 in a well-conserved NPTH loop located between FlhBCN and FlhBCC polypeptides and interacts with the C-terminal domain of the FliK ruler when the length of the hook has reached about 55 nm in Salmonella. As a result, the flagellar protein export apparatus switches its substrate specificity, thereby terminating hook assembly and initiating filament assembly. The mechanism of export switching remains unclear. Here, we report the role of FlhBC cleavage in the switching mechanism. Photo-cross-linking experiments revealed that the flhB(N269A) and flhB(P270A) mutations did not affect the binding affinity of FlhBC for FliK. Genetic analysis of the flhB(P270A) mutant revealed that the P270A mutation affects a FliK-dependent conformational change of FlhBC, thereby inhibiting the substrate specificity switching. The flhA(A489E) mutation in FlhAC suppressed the flhB(P270A) mutation, suggesting that an interaction between FlhBC and FlhAC is critical for the export switching. We propose that the interaction between FliKC and a cleaved form of FlhBC promotes a conformational change in FlhBC responsible for the termination of hook-type protein export and a structural remodeling of the FlhAC ring responsible for the initiation of filament-type protein export.
IMPORTANCE The flagellar type III protein export apparatus coordinates protein export with assembly, which allows the flagellum to be efficiently built at the cell surface. Hook completion is an important morphological checkpoint for the sequential flagellar assembly process. The protein export apparatus switches its substrate specificity from the hook protein to the filament protein upon hook completion. FliK, FlhB, and FlhA are involved in the export-switching process, but the mechanism remains a mystery. By analyzing a slow-cleaving flhB(P270A) mutant, we provide evidence that an interaction between FliK and FlhB induces conformational rearrangements in FlhB, followed by a structural remodeling of the FlhA ring structure that terminates hook assembly and initiates filament formation.
INTRODUCTION
The bacterial flagellum is responsible for swimming motility in liquid media and for swarming motility on solid surfaces for many bacterial species. The flagellum is composed of a basal body (rotary motor), a hook (universal joint), and a filament (helical propeller). Flagellar assembly begins with the basal body, followed by the hook and finally the filament (1). For assembly of the hook and filament from the cell surface, a type III protein export apparatus transports these component proteins from the cytoplasm to the distal end of the growing structure. The type III protein export apparatus is composed of five transmembrane proteins, FlhA, FlhB, FliP, FliQ, and FliR and three cytoplasmic proteins, FliH, FliI, and FliJ (2).
The length of the hook is controlled, extending about 55 nm from the cell surface in wild-type cells of Salmonella enterica serovar Typhimurium (here referred to Salmonella) (3). The flagellar type III protein export apparatus uses the FliK ruler to measure the hook length and facilitates the switching of export specificity from hook-type substrates (FlgD, FlgE, and FliK) to filament-type ones (FlgK, FlgL, FlgM, FliC, and FliD) upon hook completion. The substrate specificity switch terminates hook assembly and initiates filament assembly. FlhB and FlhA are directly involved in export switching of the type III protein export apparatus (4).
FliK consists of N-terminal (FliKN) and C-terminal (FliKC) domains (5, 6). FliKN acts as a molecular ruler to measure the length of the hook through interactions of FliKN with the hook-capping protein FlgD and the hook protein FlgE (7–11), whereas FliKC interacts with the C-terminal cytoplasmic domain of FlhB (FlhBC) to promote substrate specificity switching of the flagellar type III protein export apparatus (12–15).
FlhBC consists of FlhBCN and FlhBCC polypeptides with molecular weights of 8.5 and 11.5 kDa, respectively (16, 17). A conserved hydrophobic patch formed by Ala-286, Pro-287, Ala-341, and Leu-344 residues in FlhBCC is involved in the interaction of FlhBC with the N-terminal segments of FlgD, FlgE, and FliK, which contain an export signal recognized by the flagellar type III protein export apparatus (18). Autocatalytic cleavage between Asn-269 and Pro-270, which is located within a highly conserved NPTH loop between FlhBCN and FlhBCC, is critical for export switching of the protein export apparatus (19, 20). It has been reported that autocleavage of the C-terminal cytoplasmic domain of SpaS, which is a FlhB homologue of the Salmonella SPI-1 injectisome, allows the proper conformation of SpaSC to render it competent for its switching function (21), but it remains unclear how its cleaved form flips export specificity of the type III protein export apparatus of the Salmonella SPI-1 injectisome.
FlhA forms a homo-nonamer through its C-terminal cytoplasmic domain (FlhAC) in the flagellar type III protein export apparatus (22). FlhAC interacts with flagellar chaperones in complex with their cognate filament-type substrates to promote the export of filament-type proteins to form the flagellar filament at the hook tip (23–26). Interactions of a flexible linker of FlhA (FlhAL) with its neighboring FlhAC subunit in the ring structure induce the remodeling of the FlhAC ring structure upon completion of the hook structure, thereby allowing the flagellar chaperones in complex with their cognate substrates to bind to the FlhAC ring to facilitate the export of filament-type proteins (27, 28). However, it remains unknown which protein triggers the structural remodeling of the FlhAC ring structure.
The flhB(N269A) mutation inhibits not only autocleavage of FlhBC but also substrate specificity switching of the flagellar type III protein export apparatus (19), leading to a plausible hypothesis that an interaction between FliKC and a cleaved form of FlhBC may induce the structural remodeling of the FlhAC ring responsible for the substrate specificity switching. To clarify this hypothesis, we analyzed a slow-cleaving flhB(P270A) mutant in detail and provide evidence suggesting that the binding of FliKC to a cleaved form of FlhBC not only terminates hook polymerization but also induces conformational rearrangements of the FlhAC ring to initiate filament assembly at the hook tip.
RESULTS
Effect of the flhB(N269A) and flhB(P270A) mutations on photo-cross-linking between FliKC and FlhBC.
A well-conserved Ile-304 residue of FliK is critical for substrate specificity switching of the flagellar type III protein export apparatus (14). Recently, it has been shown that Ile-304 of FliKC is in relatively close proximity to FlhBC, allowing FliK to form a photo-cross-linked product with FlhBCC (15). To investigate whether autocleavage of FlhBC is required for the interaction of FlhBC with FliKC, photo-cross-linking experiments were conducted using a photoreactive phenylalanine, p-benzoyl-phenylalanine (pBPA), that was introduced into an amber codon at position 304 of FliK using the amber suppressor tyrosyl tRNA and the engineered tyrosyl-tRNA synthetase (29). In agreement with a previous report (15), UV irradiation of FliK(I304pBPA) reproducibly produced a ca. 53-kDa photo-cross-linked product with FlhBCC when coexpressing with wild-type FlhBC (Fig. 1, lane 2). Photo-cross-linked products were also seen when FliK(I304pBPA) was coexpressed with FlhBC(N269A) and FlhBC(P270A) (Fig. 1, lanes 4 and 6). In contrast to wild-type FlhBC, which is cleaved into FlhBCN and FlhBCC polypeptides in an autocatalytic manner, FlhBC(N269A) and FlhBC(P270A) do not undergo autocleavage. As a result, the molecular mass of the FliK-FlhBC heterodimer is estimated to be ca. 64 kDa, which is the sum of the molecular masses of FliK (42 kDa) and noncleaved FlhBC (22 kDa), respectively. This mass is almost the same as that of the cross-linked products detected on immunoblots. Therefore, we suggest that the flhB(N269A) and flhB(P270A) mutations do not interfere with the interaction of FlhBC with FliK.
FIG 1.
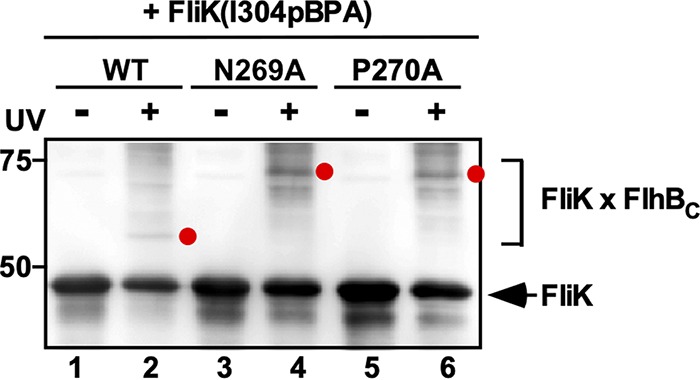
Effect of the flhB(N269A) and flhB(P270A) mutations on photo-cross-linking between FliKC and FlhBC. E. coli BL21 Star(DE3) cells coexpressing FliK(I304pBPA) with FlhBC, FlhBC(N269A), or FlhBC(P270A) were UV irradiated for 5 min (+) or not irradiated (–) and then analyzed by immunoblotting with polyclonal anti-FliK antibody. Photo-cross-linked FliK-FlhBC products are indicated by red dots. The position of free FliK is indicated by an arrow. Molecular mass markers (in kilodaltons) are shown on the left.
Effect of the flhB(N269A) and flhB(P270A) mutations on hook length.
The flhB(N269A) and flhB(P270A) mutants produce polyhooks (10, 19). We found that the flhB(N269A) and flhB(P270A) mutations do not affect the interaction of FlhBC with FliK, raising the question of whether the FliK-FlhBC interaction influences the rate of hook assembly. To clarify this question, we purified polyhook-basal bodies from these two flhB mutants and measured the length of each polyhook. The average hook lengths of the flhB(N269A) and flhB(P270A) mutants were 127.4 ± 85.6 nm (n = 237) and 125.6 ± 77.9 nm (n = 251), respectively, which were much longer than the wild-type length with a wide size distribution (53.5 ± 7.5 nm, n = 103) but shorter than the length of the polyhook produced by the fliK null mutant (418.5 ± 228.5 nm, n = 332) (Fig. 2). When a ΔfliK::tetRA allele was introduced into the flhB(N269A) and flhB(P270A) mutants by P22-mediated transduction, the average lengths of the polyhooks produced by the flhB(N269A) ΔfliK::tetRA and flhB(P270A) ΔfliK::tetRA mutants were 331 ± 198.9 nm (n = 316) and 319.1 ± 169.9 nm (n = 318) (Fig. 2). These results suggest that the FliK-FlhBc interaction reduces the hook polymerization rate significantly.
FIG 2.

Effect of flhB(N269A) and flhB(P270A) mutations on hook length control. Electron micrograms of hook-basal bodies isolated from SJW1103 (wild type, indicated as WT), TH13763 [flhB(N269A), indicated as N269A], TH12499 [flhB(P270A), indicated as P270A], MM1103iK (ΔfliK), TH13763iK (N269A ΔfliK), and TH12499iK (P270A ΔfliK) are shown. The average hook length and standard deviations are shown. N, number of hook-basal bodies and polyhook-basal bodies, which were measured. Scale bars, 100 nm.
Isolation of pseudorevertants form the flhB(P270A) mutant.
To clarify the role of autocleavage of FlhBC on substrate specificity switching of the flagellar type III protein export apparatus, we isolated five pseudorevertants from the flhB(P270A) mutant. The motility of these pseudorevertants was better than that of their flhB(P270A) mutant, although it was not as good as that of the wild-type strain (Fig. 3A). P22-mediated genetic mapping showed that the gain-of-function mutations were located within the flhBAE operon. DNA sequencing identified two missense mutations, T268I (isolated two times) and E314A (isolated two times) in FlhBC, and a missense mutation, A489E, in FlhAC (Fig. 3B).
FIG 3.
Isolation of pseudorevertants from the flhB(P270A) mutant. (A) Motilities of SJW1103 (WT), TH12499 (P270A), MMB12499-SP1 [flhB(P270A/T268I), indicated as P270A/T268I], MMB12499-SP2 [flhB(P270A/E314A), indicated as P270A/E314A], and MMB12499-SP3 [flhB(P270A) flhA(A489E), indicated as P270A/FlhA(A489E)] in soft agar. Plates were incubated at 30°C for 7.5 h. (B) Location of intragenic flhB(T268I) and flhB(E314A) suppressor mutations in the crystal structure of FlhBC (PDB ID 3B0Z) and an extragenic flhA(A489E) suppressor mutation in the crystal structure of FlhAC (PDB ID 3A5I). FlhBC undergoes autocatalytic cleavage between Asn-269 and Pro-270 residues and so is composed of two distinct FlhBCN (magenta) and FlhBCC (rainbow) polypeptides. Well-conserved Asp-456, Phe-459, and Thr-490 residues of FlhA form part of the flagellar chaperone-binding site. The C-α backbone is color-coded from blue to red, going through the rainbow colors from the N terminus to the C terminus. (C) Electron micrograms of hook-basal bodies and polyhook-basal bodies isolated from the MMB12499-SP2 and MMB12499-SP3 strains. The average hook lengths and standard deviations are shown. Scale bars, 100 nm. (D) Secretion assays of FlgD, FlgE, FlgK, and FliC. Immunoblotting using polyclonal anti-FlgD (first row), anti-FlgE (second row), anti-FlgK (third row), or anti-FliC (fourth row) antibody of whole-cell proteins (Cell) and culture supernatants (Sup) from the strains described above. Arrowheads indicate positions of FlgD, FlgE, FlgK, and FliC. The positions of molecular mass markers (in kilodaltons) are given on the left. (E) Effect of flhB(T268I) and flhB(E314A) suppressor mutations on autocleavage of FlhBC(P270A). Coomassie blue-stained SDS-PAGE gels of whole-cell proteins prepared from E. coli BL21 Star(DE3) cells transformed with pET19b (V), pYI124 (N-terminally His-tagged FlhBC, indicated as WT), pYI140 [His-FlhBC(P270A), indicated as P270A], pYI140-SP1 [His-FlhBC(P270A/T268I), indicated as P270A/T268I], or pYI140-SP2 [His-FlhBC(P270A/E314A), indicated as P270A/E314A] are shown. The positions of the intact form and the cleaved His-FlhBCN and FlhBCC polypeptides are indicated by arrowheads.
To test whether these suppressor mutations shorten the length of the polyhooks produced by the flhB(P270A) mutant, we isolated flagella from the flhB(P270A/E314A) and flhB(P270A) flhA(A489E) suppressor mutants and measured the hook length. The hook lengths of the flhB(P270A/E314A) and flhB(P270A) flhA(A489E) suppressor mutants were 97.7 ± 65.5 nm (n = 129) and 102.6 ± 62.8 nm (n = 138) (Fig. 3C) compared to 53.5 ± 7.5 nm (n = 103) for wild-type cells and 125.6 ± 77.9 nm (n = 251) for the flhB(P270A) cells (Fig. 2). This indicates that these two suppressor mutations slightly shorten the polyhook length. This suggests that these suppressor mutants still cannot properly terminate the export of FlgE. Consistently, there was no significant difference in the secretion levels of hook-type proteins such as FlgE and FlgD between the flhB(P270A) mutant and the suppressor mutants (Fig. 3D, first and second rows).
To investigate the export-switching efficiency of the flhB(P270A/T268I), flhB(P270A/E314A), and flhB(P270A) flhA(A489E) suppressor mutants, we analyzed the secretion levels of FlgK and FliC by immunoblotting with polyclonal anti-FlgK and anti-FliC antibodies, respectively. The levels of FlgK and FliC secreted by these suppressor mutants were higher than those seen in their parent flhB(P270A) mutant and were almost the same as the wild-type levels (Fig. 3D, third and fourth rows). These results suggest that the flhB(T268I), flhB(E314A), and flhA(A489E) suppressor mutations allow the flhB(P270A) mutant to initiate the assembly of the filament efficiently.
We tested whether intragenic flhB(T268I) and flhB(E314A) suppressor mutations promote autocleavage of FlhBC(P270A). Only the intact form of FlhBC was detected in whole-cell lysates prepared from E. coli BL21 Star(DE3) cells overexpressing His-FlhBC(P270A) (Fig. 3E). In contrast, significant amounts of cleaved forms of FlhBC were detected in BL21 Star(DE3) cells overexpressing His-FlhBC(P270A/T268I) or His-FlhBC(P270A/E314A) in a way similar to wild-type FlhBC although their intact forms were also seen (Fig. 3E). These results indicate that the intragenic flhB(T268I) and flhB(E314A) suppressor mutations induce a conformational change of the NPTH loop, allowing FlhBC to undergo autocatalytic cleavage into two distinct FlhBCN and FlhBCC polypeptides. Therefore, we conclude that the autocleavage of FlhBC is required not only for efficient termination of hook-type protein export but also for the efficient export of filament-type proteins.
Effect of second-site flhB and flhA mutations on motility.
To test whether second-site flhB and flhA mutations by themselves affect the export-switching function of the flagellar type III protein export apparatus, we introduced these second-site mutations into wild-type FlhB and FlhA by site-directed mutagenesis and analyzed their motility. The motilities of the flhB(T268A), flhB(E314A), and flhA(E314A) mutants were almost the same as that of wild-type cells (see Fig. S1A and B in the supplemental material), indicating that these second-site mutations display no significant motility phenotype.
Effect of FliK defect on the export-switching function of the flagellar type III protein export apparatus.
To test whether the export-switching function of the flhB(P270A/T268I), flhB(P270A/E314A), and flhB(P270A) flhA(A489E) suppressor mutants is dependent on FliK, we introduced a ΔfliK::tetRA allele into these three suppressor mutants by P22-mediated transduction and analyzed the secretion levels of FlgE and FliC. A deletion of the fliK gene totally inhibited the export of FliC but not that of FlgE (Fig. S2). This indicates that these suppressor mutations do not induce autonomous export switching of the flagellar type III protein export apparatus in the absence of FliK.
Effect of the flhA(A489E) mutation on the interaction of FlhAC with flagellar chaperones in complex with their cognate substrates.
Asp-456, Phe-459, and Thr-490 of FlhA are directly involved in the interaction of the FlgN, FliS, and FliT chaperones in complex with their cognate export substrates, FlgK and FlgL, FliC, and FliD, respectively (24–26). The flhA(A489E) mutation is located in the chaperone-binding site of FlhAC (Fig. 3B), raising the question of whether the flhA(A489E) suppressor mutation affects the interaction of FlhAC with the chaperone/filament-type export substrate complexes. To clarify this question, we carried out pulldown assays by glutathione S-transferase (GST) affinity chromatography. FlhAC coeluted with the GST-FlgN/FlgK and GST-FliS/FliC complexes from a GST column (Fig. 4A, second and fourth rows) but not with GST alone (first row), in agreement with previous reports (24, 25). However, the flhA(A489E) mutation reduced its binding affinities for the FlgN/FlgK and FliS/FliC complexes (third and fifth rows).
FIG 4.

Effect of flhA(A489E) mutation on the export of filament-type proteins. (A) Effect of the flhA(A489E) mutation on the interaction of FlhAC with the FlgN/FlgK and FliS/FliC complexes. Mixtures (L) of purified His-FlhAC or His-FlhAC(A489E) with GST (first row), GST-FlgN/FlgK (second and third rows), or GST-FliS/FliC (fourth and fifth rows) were purified by GST affinity chromatography. The flowthrough fraction (F.T.), wash fractions, and elution fractions were analyzed by Coomassie brilliant blue staining. Molecular mass markers (in kilodaltons) are given on the left. (B) Secretion assays of FlgD and FliC. Immunoblotting with polyclonal anti-FlgD (first row) or anti-FliC (second row) antibody of whole-cell proteins (Cell) and culture supernatants (Sup) prepared from the Salmonella NH001 strain carrying pTrc99AFF4 (indicated as ΔflhA), pMM130 (indicated as WT), or pYI130-SP3 [indicated as flhA(A489E)] and the Salmonella NH004 strain (ΔfliHI flhB* ΔflhA) transformed with pTrc99AFF4 (indicated as ΔfliHI flhB* ΔflhA), pMM130 (indicated as ΔfliHI flhB*), or pYI130-SP3 [indicated as ΔfliHI flhB* flhA(A489E)] was performed. Arrowheads indicate the positions of FlgD and FliC.
FlhA requires the support of FliH and FliI to fully exert its export function (30–32). When FliH and FliI are missing, much larger amounts of FliC molecules are leaked out into the culture media because the hook-filament junction structure is not formed at the hook tip properly by FlgK and FlgL (32). Although the flhA(A489E) mutation reduced the binding affinities of FlhAC for the FlgN/FlgK and FliS/FliC complexes, the flhA(A489E) mutation increased the secretion levels of FlgK and FliC significantly even in the presence of the fllhB(P270A) mutation, raising the possibility that FlhA(A489E) requires FliH and FliI to exert its export function. Therefore, we analyzed the effect of the flhA(A489E) mutation on the filament-type protein export in a ΔfliH-fliI flhB(P28T) (ΔfliH-fliI flhB*) mutant background, of which second-site flhB(P28T) mutation considerably increases the probability of flagellar formation in the absence of FliH and FliI (33). The motility of the ΔfliH-fliI flhB(P28T) flhA(A489E) mutant was worse than that of the ΔfliH-fliI flhB(P28T) mutant (Fig. S1C). Consistently, the secretion level of FliC was lower in the ΔfliH-fliI flhB(P28T) flhA(A489E) mutant than in the ΔfliH-fliI flhB(P28T) mutant (Fig. 4B, right panel, second row). However, there was no difference in the secretion level of FlgD between these two mutant strains (Fig. 4B, right panel, first row). When FliH and FliI were present, the flhA(A489E) mutation does not affect the secretion level of FliC at all (Fig. 4C, left panel, second row). These results indicate that FlhA(A489E) requires FliH and FliI to efficiently promote the export of filament-type proteins upon completion of hook assembly. Because the flagellar chaperones in complex with their cognate filament-type substrates bind to the open form of FlhAC in the FlhAC ring structure but not to the closed form (26, 28), we propose that the FlhAC(A489E) ring structure may adopt the open form to allow the flagellar type III protein export apparatus to promote the export of filament-type proteins in the presence of the flhB(P270A) mutation.
Effect of flhB(G293R), flhB(G293V), and flhB(A298V) mutations on export switching.
The flhB(G293R), flhB(G293V), and flhB(A298V) mutations in FlhBCC were originally isolated as extragenic fliK suppressor mutations to support filament assembly even in the absence of FliK (13). Motility of the flhB(G293R), flhB(G293V), and flhB(A298V) mutants is worse than the wild-type level and much better than that of the flhB(N269A) and flhB(P270A) mutants (Fig. 5A). The flhB(G293R), flhB(G293V), and flhB(A298V) mutations significantly affect autocleavage of FlhBC (16), raising a question about the impact of these flhB mutations on the export switching of the flagellar type III protein export apparatus. To clarify this question, we analyzed the levels of filament-type proteins secreted by the flhB(G293R), flhB(G293V), and flhB(A298V) mutants. These three flhB mutations reduced the secretion levels of FlgK and FliC by approximately 60% of the wild-type levels (Fig. 5B), confirming that FlhBC cleavage is required for the efficient export of filament-type proteins.
FIG 5.

Effect of flhB(G293R), flhB(G293V), and flhB(A298V) mutations on substrate specificity switching of the flagellar type III protein export apparatus. (A) Motility of SJW1103 (WT), MKM50 (ΔflhB), MMB2714 [flhB(G293R), indicated as G293R], MMB3201 [flhB(G293V), indicated as G293V], MMB2701 [flhB(A298V), indicated as A298V], TH13763 [flhB(N269A), indicated as N269A], and TH12499 [flhB(P270A), indicated as P270A] in soft agar. Plates were incubated at 30°C for 6 h. (B) Secretion assays of FlgE, FlgK, and FliC. Immunoblotting with polyclonal anti-FlgE (first row), anti-FlgK (second row), or anti-FliC (third row) antibody of whole-cell proteins (Cell) and culture supernatants (Sup) prepared from the above strains was performed. Arrowheads indicate the positions of FlgE, FlgK, and FliC. Molecular mass markers (in kilodaltons) are given on the left. (C) Electron micrograms of hook-basal bodies and polyhook-basal bodies isolated from the MMB2714, MMB3201, and MMB2701 strains. The average hook length and standard deviations are shown. Scale bars, 100 nm.
To investigate whether the flhB(G293R), flhB(G293V), and flhB(A298V) mutations affect hook length control, we prepared hook-basal bodies from these flhB mutants and measured their hook length. The average hook lengths of the flhB(G293R) and flhB(A298V) mutants were 63.4 ± 20.2 nm (n = 107) and 66.4 ± 28.1 nm (n = 128) (Fig. 5C) compared to (53.5 ± 7.5 nm, n = 103) for the wild-type (Fig. 2), suggesting that the flhB(G293R) and flhB(A298V) mutants cannot terminate the export of FlgE at an appropriate timing of hook assembly, thereby causing a loose hook length control. Consistently, the levels of FlgE secreted by the flhB(G293R) and flhB(A298V) mutants were about 1.4-fold higher than the wild-type level (Fig. 5B). In contrast, hook length control was not affected by the flhB(G293V) mutation (Fig. 5C), suggesting that the flhB(G293V) mutation affects the initiation of filament formation but not the termination of hook assembly.
DISCUSSION
An interaction between FliKC and FlhBC triggers substrate specificity switching of the flagellar type III protein export apparatus from the hook protein to the filament protein when the length of the hook has reached about 55 nm in Salmonella (4). Autocatalytic cleavage between Asn-269 and Pro-270 within the conserved NPTH loop occurs through a chemical reaction involving cyclization of Asp-269 (20, 34). A conformational change of FlhBCC through a remodeling of hydrophobic interaction networks in FlhBCC is postulated to be responsible for the substrate specificity switching (35). However, little is known about the role of the autocleavage of FlhBC in the export-switching mechanism. Here, we showed that the flhB(N269A) and flhB(P270A) mutations did not affect the interaction of FlhBC with FliKC (Fig. 1). We also found that the lengths of polyhooks produced by the flhB(N269A) and flhB(P270A) mutants were shorter than that of the fliK null mutant (Fig. 2). Introduction of the fliK null mutation into these two flhB mutants increased the length of the polyhook considerably (Fig. 2). Therefore, we propose that the binding of FliKC to FlhBC induces conformational rearrangements of FlhBCC to terminate the export of hook-type proteins and that the autocleavage of FlhBC is required for proper conformational changes of FlhBC in a FliK-dependent manner.
The C-terminal cytoplasmic domain EscU (EscUC), which is a FlhB homologue of the injectisome of pathogenic E. coli, undergoes autocleavage into EscUCN and EscUCC in a way similar to FlhBC (34). Structural comparison between EscUC and EscUC(P263A) has revealed that the escU(P263A) mutation does not induce a large conformational change of the entire EscUC structure (Fig. S3A) (34). However, Ala-263 makes hydrophobic contacts with Lys-261, Asn-262, and Glu-307, whereas Pro-263 does not (see Fig. S3A in the supplemental material), suggesting that the conformational flexibility of the conserved NPTH loop connecting between EscUCN and EscUCC could be restricted by a replacement of Pro-270 by Ala, thereby suppressing the autocleavage of EscUC. Here, we found that the intragenic flhB(T268I) and flhB(E314A) suppressor mutations facilitated the autocleavage of FlhBC(P270A) to a significant degree (Fig. 3E). Thr-268 and Glu-314 of FlhBC correspond to Lys-261 and Glu-307 of EscUC, suggesting that the flhB(T268I) and flhB(E314A) suppressor mutations affect hydrophobic interactions of Ala-270 with Thr-268, Asn-269, and Glu-307 to allow Asn-269 to be exposed to solvent to induce the autocleavage of FlhBC (Fig. 3B). Recently, it has been shown that the flhB(R320A) mutation affecting hydrophobic interaction networks in FlhBCC inhibits the autocleavage of FlhBC significantly, causing a loose hook length control (35). The flhB(G293R), flhB(G293V), and flhB(A298V) mutations, which reduce the rate of the autocleavage process of FlhBC considerably (16), seem to affect hydrophobic interaction networks in FlhBCC in a way similar to the flhB(R320A) mutation (Fig. S3B). The flhB(293R), flhB(G293V), and flhB(A298V) mutations reduced the secretion levels of filament-type proteins (Fig. 5B). Interestingly, the flhB(G293R) and flhB(A298V) mutants produced longer hooks compared to wild-type cells (Fig. 5C). Because we found that the flhB(T268I) and flhB(E314A) suppressor mutations significantly increased the probability of filament assembly in the flhB(P270A) mutant (Fig. 3), we propose that the autocleavage of FlhBC induces a conformational flexibility of FlhBCN-FlhBCC boundary and that such a flexibility is important not only for the initiation of the export of filament-type proteins but also for the termination of the export of hook-type proteins at an appropriate timing of hook assembly.
How does the FliKC-FlhBC interaction terminate the export of hook-type proteins? The C-terminal domain of YscP (YscPC), which is a FliK homologue of the Yersinia injectisome, directly binds to the C-terminal domain of YscU (YscUC), which is a FlhB homologue. Ala-335 of YscUC, which corresponds to Ala-341 of FlhBC, is critical for the binding to YscPC, and Leu-280 of YscUC, which corresponds to Ala-286 of FlhBC, contributes to this binding (36). Ala-286, Pro-287, Ala-341, and Leu-344 form a well-conserved hydrophobic patch on the molecular surface of FlhBCC, and this hydrophobic patch is directly involved in the recognition of the export signals of hook-type proteins (18). Therefore, we propose that the binding of FliKC to FlhBCC may induce a conformational change of the hydrophobic patch, thereby terminating the export of the hook-type proteins.
Flk, which is a transmembrane protein with a large cytoplasmic domain, interferes with premature switching of the substrate specificity of the flagellar type III protein export apparatus during hook-basal body assembly, and this inhibitory effect of Flk on the export of filament-type proteins is released after completion of hook assembly (9, 37–39). The flhB(N269A) and flhB(P270A) mutations interfere with the export of filament-type proteins (19), raising the possibility that these two mutations affect the negative impact of Flk on filament-type protein export. A disruption of the flk gene significantly increased the secretion level of FliC in the flgE mutant (Fig. S4), in agreement with a previous report (9). However, deletion of the flk gene did not increase the secretion level of FliC in the flhB(N269A) and flhB(P270A) mutants (Fig. S4), suggesting that the inability of FlhB(N269A) and FlhB(P270A) to switch export specificity does not result from failure to release the inhibitory effect of Flk on the switching. Therefore, we conclude that the autocleavage of FlhBC is critical for the flagellar type III protein export apparatus to promote the initiation of filament-type protein export.
Asp-456, Phe-459, and Thr-490 of FlhAC are involved in the interactions with the FlgN, FliS, and FliT chaperones in complex with their cognate substrates (Fig. 3B) (24–26). FlhAC adopts two distinct, open and closed conformations (40, 41), and the flagellar chaperones in complex with their cognate substrates bind to the open form of FlhAC but not to the closed form (26, 28). Interactions of FlhAL with its neighboring FlhAC subunit convert the entire FlhAC ring structure from a closed to an open form to facilitate the export of filament-type proteins (27, 28). Although it has been shown FlhBC binds to FlhAC with a micromolar affinity in solution (42), it remained unknown how FlhBC could induce a conformational change of FlhAC and promote filament-type protein export upon completion of hook assembly. Here, we found that the extragenic flhA(A489E) suppressor mutation facilitated the export of filament-type proteins even in the presence of the flhB(P270A) mutation (Fig. 3D). Interestingly, the flhA(A489E) mutation is located in the chaperone-binding site of FlhAC (Fig. 3B). Pulldown assays by GST affinity chromatography revealed that the flhA(A489E) mutation reduced the binding affinities of FlhAC for the FlgN/FlgK and FliS/FliC complexes (Fig. 4A). However, FliH and FliI overcame the defect of FlhAC(A489E) in the interaction with these chaperone/substrate complexes (Fig. 4B), thereby allowing the flhB(P270A) flhA(A489E) mutant to transport FlgK and FliC at wild-type levels (Fig. 3D). Therefore, we suggest that the flhA(A489E) mutation allows FlhAC to adopt a certain conformation mimicking a chaperone-bound state of FlhAC even in the presence of the flhB(P270A) mutation. Introduction of the fliK null mutation into the flhB(P270A) flhA(A489E) suppressor mutant abolished the export of FliC but not that of FlgE (Fig. S2), suggesting that FliK is required for conformational rearrangements of the FlhAC(A489E) ring structure to initiate the export of filament-type proteins. Therefore, we propose that the binding of FliKC to a cleaved form of FlhBC may promote the FlhBC-FlhAC interaction to allow the FlhAC ring structure to undergo a structural transition from a closed to an open form, thereby promoting efficient docking of flagellar chaperones in complex with their cognate substrates to the FlhAC ring structure to facilitate subsequent export of filament-type proteins (Fig. 6).
FIG 6.
Model for export switching of the flagellar type III protein export apparatus. During hook assembly, the flagellar type III protein export apparatus transports the hook protein (FlgE) from the cytoplasm to the distal end of the nascent flagellar structure. The export apparatus also secretes the FliK ruler to measure the hook length during hook assembly. FlhBCC provides a binding site for FlgE and FliK for their efficient export. When the hook length has reached about 55 nm, the C-terminal domain of FliK (FliKC) binds to FlhBCC to induce a conformational change of FlhBC, thereby terminating the export of FlgE. Then, cleaved FlhBC binds to FlhAC to induce a structural remodeling of the FlhAC ring structure, allowing the FlgN, FliT, and FliS chaperones in complex with their cognate substrates to bind to the FlhAC ring to initiate the export of filament-type proteins to form the filament at the hook tip.
MATERIALS AND METHODS
Bacterial strains, plasmids, transduction, and media.
Bacterial strains and plasmids used in this study are listed in Table 1 . P22-mediated transduction was carried out as described previously (43). L broth and 0.35% soft agar plates were prepared as described previously (44, 45). Ampicillin was added at a final concentration of 100 μg/ml if needed.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli BL21 Star(DE3) | Overexpression of proteins | Novagen |
| Salmonella | ||
| SJW1103 | Wild type for motility and chemotaxis | 43 |
| SJW1103RflH | Δflk::tetRA | 9 |
| MKM50 | ΔflhB | 19 |
| MMB2701 | flhB(A298V) | 20 |
| MMB2714 | flhB(G293R) | 20 |
| MMB3201 | flhB(G293V) | 20 |
| MM1103iK | ΔfliK::tetRA | This study |
| NH001 | ΔflhA | 30 |
| NH004 | ΔfliH-fliI flhB(P28T) ΔflhA | 30 |
| NME001 | ΔflgE | 9 |
| NME001RflH | ΔflgE Δflk::tetRA | 9 |
| TH8426 | ΔfliK | 14 |
| TH8426RflH | ΔfliK Δflk::tetRA | 9 |
| TH13763 | flhB(N269A) | Kelly T. Hughes |
| TH13763iK | flhB(N269A) ΔfliK::tetRA | This study |
| TH13763RflH | flhB(N269A) Δflk::tetRA | This study |
| TH12499 | flhB(P270A) | Kelly T. Hughes |
| TH12499iK | flhB(P270A) ΔfliK::tetRA | This study |
| TH12499RflH | flhB(P270A) Δflk::tetRA | This study |
| MMB12499-SP1 | flhB(P270A/T268I) | This study |
| MMB12499-SP1iK | flhB(P270A/T268I) ΔfliK::tetRA | This study |
| MMB12499-SP2 | flhB(P270A/E314A) | This study |
| MMB12499-SP2iK | flhB(P270A/E314A) ΔfliK::tetRA | This study |
| MMA12499-SP3 | flhB(P270A) flhA(A489E) | This study |
| MMA12499-SP3iK | flhB(P270A) flhA(A489E) ΔfliK::tetRA | This study |
| Plasmids | ||
| pTrc99AFF4 | Modified pTrc expression vector | 48 |
| pEVOL | For incorporation of pBPA into the amber codon | 29 |
| pGEX-6p-1 | Expression vector | GE Healthcare |
| pMKGK2 | pTrc99A/FlgK | 49 |
| pMKM1002 | pGEX-6p-1/GST-FliS | 25 |
| pMM104 | pET19b/His-FlhAC (residues 211 to 692) | 45 |
| pMM130 | pTrc99AFF4/FlhA | 50 |
| pMMGN101 | pGEX-6p-1/GST-FlgN | 24 |
| pMMK521 | pETDuet-1/FliK(I304amber) + FlhBC | 15 |
| pMMK536 | pETDuet-1/FliK(I304amber) + FlhBC(N269A) | This study |
| pMMK537 | pETDuet-1/FliK(I304amber) + FlhBC(P270A) | This study |
| pYI101 | pTrc99AFF4/FlhB | 35 |
| pYI101-SP1 | pTrc99AFF4/FlhB(T268I) | This study |
| pYI101-SP2 | pTrc99AFF4/FlhB(E314A) | This study |
| pYI124 | pET19b/His-FlhBC (residues 211 to 383) | 35 |
| pYI140 | pET19b/His-FlhBC(P270A) | 35 |
| pYI140-SP1 | pET19b/His-FlhBC(P270A/T268I) | This study |
| pYI140-SP2 | pET19b/His-FlhBC(P270A/E314A) | This study |
| pYI130-SP3 | pTrc99A/FlhA(A489E) | This study |
| pYI104-SP3 | pET19b/His-FlhAC(A489E) | This study |
Site-directed mutagenesis.
Site-directed mutagenesis was carried out using the QuikChange site-directed mutagenesis method (Stratagene). DNA sequencing reactions were carried out using BigDye v3.1 (Applied Biosystems); the reaction mixtures were analyzed by a 3130 Genetic Analyzer (Applied Biosystems).
Photo-cross-linking.
E. coli BL21 Star(DE3) cells harboring pEVOL (29) and a pETDuet-based plasmid encoding both FliK(I304amber) and wild-type FlhBC, FlhBC(N269A), or FlhBC(P270A) were exponentially grown at 30°C in L broth containing 1 mM p-benzoyl-phenylalanine (pBPA) and 100 μg/ml ampicillin. Then, 100 μM IPTG (isopropyl-β-d-thiogalactopyranoside) and 0.02% arabinose were added, and the incubation was continued until the culture density had reached an optical density at 600 nm (OD600) of ca. 1.4 to 1.5. Photo-cross-linking experiments were performed as described previously (46). After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), immunoblotting with polyclonal anti-FliK antibody was carried out as described previously (44). Detection was done with an ECL Prime Western blotting detection reagent (GE Healthcare). Chemiluminescence signals were captured by a Luminoimage analyzer LAS-3000 (GE Healthcare). At least three independent experiments were performed.
Electron microscopy.
Salmonella cells were grown in L broth at 30°C with shaking until the cell density had reached an OD600 of ca. 1.0 to 1.3. The flagella were isolated by 20 to 50% (wt/wt) sucrose density gradient ultracentrifugation as described previously (32). After ultracentrifugation at 60,000 × g for 60 min, the pellets were suspended in 50 mM glycine (pH 2.5) and 0.1% Triton X-100, followed by incubation at room temperature for 30 min to depolymerize flagellar filaments. After ultracentrifugation, the pellets were resuspended in 50 μl of 10 mM Tris-HCl (pH 8.0), 5 mM EDTA, and 0.1% Triton X-100. Samples were negatively stained with 2% (wt/vol) uranyl acetate. Electron micrograms were recorded with a JEM-1011 transmission electron microscope (JEOL, Tokyo, Japan) operated at 100 kV and equipped with a F415 charge-coupled device camera (TVIPS, Gauting, Germany). Hook length was measured by ImageJ, version 1.48 (National Institutes of Health).
Motility assay.
Fresh colonies were inoculated onto 0.35% soft agar plates and incubated at 30°C. At least six independent measurements were carried out.
Flagellar protein export assay.
Details of sample preparations have been described previously (47). Both whole-cell proteins and culture supernatants were normalized to a cell density of each culture to give a constant number of Salmonella cells. After heating at 95°C for 3 min, these protein samples were assessed by SDS-PAGE, followed by immunoblotting with polyclonal anti-FlgD, anti-FlgE, anti-FlgK, or anti-FliC antibody. More than five independent experiments were carried out.
Preparation of whole-cell proteins.
E. coli BL21 Star(DE3) cells harboring a pET19b-based plasmid encoding N-terminally His-tagged FlhBC or its mutant variants were grown overnight in 5 ml of L broth containing ampicillin at 30°C with shaking. Then, 500 μl of each culture was transferred into a 1.5-ml Eppendorf tube. After centrifugation at 20,000 × g for 5 min, each cell pellet was suspended in 1× SDS loading buffer (the amount of loading buffer was equal to OD600 × 250 μl) containing 1 μl of 2-mercaptoethanol and then heated at 95°C for 3 min. Proteins in whole-cell lysates were separated by SDS-PAGE, followed by Coomassie blue staining and immunoblotting with polyclonal anti-FlhBC antibody.
Pulldown assays by GST chromatography.
Detailed protocols for pulldown assays by GST affinity chromatography have been described previously (25). At least three independent experiments were carried out.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kelly T. Hughes for the kind gift of the flhB(N269A) and flhB(P270A) mutants and for critical reading of the manuscript.
This study was supported in part by the Japan Society for the Promotion of Science (JSPS KAKENHI grants JP26293097 and JP19H03182 to T.M., JP18K14638 to M.K., and JP25000013 to K.N.). This study was also partially supported by JEOL Yokogushi Research Alliance Laboratories of Osaka University to K.N.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Nakamura S, Minamino T. 2019. Flagellum-driven motility of bacteria. Biomolecules 9:279. doi: 10.3390/biom9070279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Minamino T. 2014. Protein export through the bacterial flagellar type III export pathway. Biochim Biophys Acta 1843:1642–1648. doi: 10.1016/j.bbamcr.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Hirano T, Yamaguchi S, Oosawa K, Aizawa SI. 1994. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella Typhimurium. J Bacteriol 176:5439–5449. doi: 10.1128/jb.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minamino T. 2018. Hierarchical protein export mechanism of the bacterial flagellar type III protein export apparatus. FEMS Microbiol Lett 365:fny117. [DOI] [PubMed] [Google Scholar]
- 5.Minamino T, Saijo-Hamano Y, Furukawa Y, González-Pedrajo B, Macnab RM, Namba K. 2004. Domain organization and function of Salmonella FliK, a flagellar hook-length control protein. J Mol Biol 341:491–502. doi: 10.1016/j.jmb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Kodera N, Uchida K, Ando T, Aizawa SI. 2015. Two-ball structure of the flagellar hook-length control protein FliK as revealed by high-speed atomic force microscopy. J Mol Biol 427:406–414. doi: 10.1016/j.jmb.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Moriya N, Minamino T, Hughes KT, Macnab RM, Namba K. 2006. The type III flagellar export specificity switch is dependent on FliK ruler and a molecular clock. J Mol Biol 359:466–477. doi: 10.1016/j.jmb.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 8.Shibata S, Takahashi N, Chevance FFV, Karlinsey JE, Hughes KT, Aizawa SI. 2007. FliK regulates flagellar hook length as an internal ruler. Mol Microbiol 64:1404–1415. doi: 10.1111/j.1365-2958.2007.05750.x. [DOI] [PubMed] [Google Scholar]
- 9.Minamino T, Moriya N, Hirano T, Hughes KT, Namba K. 2009. Interaction of FliK with the bacterial flagellar hook is required for efficient export specificity switching. Mol Microbiol 74:239–251. doi: 10.1111/j.1365-2958.2009.06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erhardt M, Hirano T, Su Y, Paul K, Wee DH, Mizuno S, Aizawa SI, Hughes KT. 2010. The role of the FliK molecular ruler in hook-length control in Salmonella enterica. Mol Microbiol 75:1272–1284. doi: 10.1111/j.1365-2958.2010.07050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT. 2011. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J 30:2948–2961. doi: 10.1038/emboj.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kutsukake K, Minamino T, Yokoseki T. 1994. Isolation and characterization of FliK-independent flagellation mutants from Salmonella Typhimurium. J Bacteriol 176:7625–7629. doi: 10.1128/jb.176.24.7625-7629.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams AW, Yamaguchi S, Togashi F, Aizawa SI, Kawagishi I, Macnab RM. 1996. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella Typhimurium. J Bacteriol 178:2960–2970. doi: 10.1128/jb.178.10.2960-2970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minamino T, Ferris HU, Moriya N, Kihara M, Namba K. 2006. Two parts of the T3S4 domain of the hook-length control protein FliK are essential for the substrate specificity switching of the flagellar type III export apparatus. J Mol Biol 362:1148–1158. doi: 10.1016/j.jmb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Kinoshita M, Aizawa SI, Inoue Y, Namba K, Minamino T. 2017. The role of intrinsically disordered C-terminal region of FliK in substrate specificity switching of the bacterial flagellar type III export apparatus. Mol Microbiol 105:572–588. doi: 10.1111/mmi.13718. [DOI] [PubMed] [Google Scholar]
- 16.Minamino T, Macnab RM. 2000. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J Bacteriol 182:4906–4919. doi: 10.1128/jb.182.17.4906-4914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meshcheryakov VA, Kitao A, Matsunami H, Samatey FA. 2013. Inhibition of a type III secretion system by the deletion of a short loop in one of its membrane proteins. Acta Crystallogr D Biol Crystallogr 69:812–820. doi: 10.1107/S0907444913002102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans LD, Poulter S, Terentjev EM, Hughes C, Fraser GM. 2013. A chain mechanism for flagellum growth. Nature 504:287–290. doi: 10.1038/nature12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser GM, Hirano T, Ferris HU, Devgan LL, Kihara M, Macnab RM. 2003. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol Microbiol 48:1043–1057. doi: 10.1046/j.1365-2958.2003.03487.x. [DOI] [PubMed] [Google Scholar]
- 20.Ferris HU, Furukawa Y, Minamino T, Kroetz MB, Kihara M, Namba K, Macnab RM. 2005. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem 280:41236–41242. doi: 10.1074/jbc.M509438200. [DOI] [PubMed] [Google Scholar]
- 21.Monjarás Feria JV, Lefebre MD, Stierhof YD, Galán JE, Wagner S. 2015. Role of autocleavage in the function of a type III secretion specificity switch protein in Salmonella enterica serovar Typhimurium. mBio 6:e01459. doi: 10.1128/mBio.01459-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrusci P, Vergara-Irigaray M, Johnson S, Beeby MD, Hendrixson DR, Roversi P, Friede ME, Deane JE, Jensen GJ, Tang CM, Lea SM. 2013. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol 20:99–104. doi: 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bange G, Kümmerer N, Engel C, Bozkurt G, Wild K, Sinning I. 2010. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc Natl Acad Sci U S A 107:11295–11300. doi: 10.1073/pnas.1001383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minamino T, Kinoshita M, Hara N, Takeuchi S, Hida A, Koya S, Glenwright H, Imada K, Aldridge PD, Namba K. 2012. Interaction of a bacterial flagellar chaperone FlgN with FlhA is required for efficient export of its cognate substrates. Mol Microbiol 83:775–788. doi: 10.1111/j.1365-2958.2011.07964.x. [DOI] [PubMed] [Google Scholar]
- 25.Kinoshita M, Hara N, Imada K, Namba K, Minamino T. 2013. Interactions of bacterial chaperone-substrate complexes with FlhA contribute to coordinating assembly of the flagellar filament. Mol Microbiol 90:1249–1261. doi: 10.1111/mmi.12430. [DOI] [PubMed] [Google Scholar]
- 26.Xing Q, Shi K, Portaliou A, Rossi P, Economou A, Kalodimos CG. 2018. Structure of chaperone-substrate complexes docked onto the export gate in a type III secretion system. Nat Commun 9:1773. doi: 10.1038/s41467-018-04137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terahara N, Inoue Y, Kodera N, Morimoto YV, Uchihashi T, Imada K, Ando T, Namba K, Minamino T. 2018. Insight into structural remodeling of the FlhA ring responsible for bacterial flagellar type III protein export. Sci Adv 4:eaao7054. doi: 10.1126/sciadv.aao7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inoue Y, Ogawa Y, Kinoshita M, Terahara N, Shimada M, Kodera N, Ando T, Namba K, Kitao A, Imada K, Minamino T. 2019. Structural insight into the substrate specificity switch mechanism of the type III protein export apparatus. Structure 27:965–976. doi: 10.1016/j.str.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 29.Young TS, Ahmad I, Yin JA, Schultz PG. 2010. An enhanced system for unnatural amino acid mutagenesis in Escherichia coli. J Mol Biol 395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 30.Hara N, Namba K, Minamino T. 2011. Genetic characterization of conserved charged residues in the bacterial flagellar type III export protein FlhA. PLoS One 6:e22417. doi: 10.1371/journal.pone.0022417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minamino T, Kinoshita M, Inoue Y, Morimoto YV, Ihara K, Koya S, Hara N, Nishioka N, Kojima S, Homma M, Namba K. 2016. FliH and FliI ensure efficient energy coupling of flagellar type III protein export in Salmonella. MicrobiologyOpen 5:424–435. doi: 10.1002/mbo3.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue Y, Morimoto YV, Namba K, Minamino T. 2018. Novel insights into the mechanism of well-ordered assembly of bacterial flagellar proteins in Salmonella. Sci Rep 8:1787. doi: 10.1038/s41598-018-20209-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minamino T, Namba K. 2008. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451:485–488. doi: 10.1038/nature06449. [DOI] [PubMed] [Google Scholar]
- 34.Zarivach R, Deng W, Vuckovic M, Felise HB, Nguyen HV, Miller SI, Finlay BB, Strynadka NC. 2008. Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature 453:124–127. doi: 10.1038/nature06832. [DOI] [PubMed] [Google Scholar]
- 35.Inoue Y, Kinoshita M, Namba K, Minamino T. 2019. Mutational analysis of the C-terminal cytoplasmic domain of FlhB, a transmembrane component of the flagellar type III protein export apparatus in Salmonella. Genes Cells 24:408–421. doi: 10.1111/gtc.12684. [DOI] [PubMed] [Google Scholar]
- 36.Ho O, Rogne P, Edgren T, Wolf-Watz H, Login FH, Wolf-Watz M. 2017. Characterization of the ruler protein interaction interface on the substrate specificity switch protein in the Yersinia type III secretion system. J Biol Chem 292:3299–3311. doi: 10.1074/jbc.M116.770255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aldridge P, Karlinsey JE, Becker E, Chevance FF, Hughes KT. 2006. Flk prevents premature secretion of the anti-sigma factor FlgM into the periplasm. Mol Microbiol 60:630–642. doi: 10.1111/j.1365-2958.2006.05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirano T, Mizuno S, Aizawa SI, Hughes KT. 2009. Mutations in Flk, FlgG, FlhA, and FlhE that affect the flagellar type III secretion specificity switch in Salmonella enterica. J Bacteriol 181:3938–3949. doi: 10.1128/JB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kutsukake K. 1997. Hook-length control of the export-switching machinery involves a double-locked gate in Salmonella Typhimurium flagellar morphogenesis. J Bacteriol 179:1268–1273. doi: 10.1128/jb.179.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore SA, Jia Y. 2010. Structure of the cytoplasmic domain of the flagellar secretion apparatus component FlhA from Helicobacter pylori. J Biol Chem 285:21060–21069. doi: 10.1074/jbc.M110.119412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saijo-Hamano Y, Imada K, Minamino T, Kihara M, Shimada M, Kitao A, Namba K. 2010. Structure of the cytoplasmic domain of FlhA and implication for flagellar type III protein export. Mol Microbiol 76:260–268. doi: 10.1111/j.1365-2958.2010.07097.x. [DOI] [PubMed] [Google Scholar]
- 42.McMurry JL, Minamino T, Furukawa Y, Francis JW, Hill SA, Helms KA, Namba K. 2015. Weak interactions between Salmonella enterica FlhB and other flagellar export apparatus proteins govern type III secretion dynamics. PLoS One 10:e0134884. doi: 10.1371/journal.pone.0134884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi S, Fujita H, Sugata K, Taira T, Iino T. 1984. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J Gen Microbiol 130:255–265. doi: 10.1099/00221287-130-2-255. [DOI] [PubMed] [Google Scholar]
- 44.Minamino T, Macnab RM. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol 181:1388–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minamino T, Macnab RM. 2000. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol Microbiol 35:1052–1064. doi: 10.1046/j.1365-2958.2000.01771.x. [DOI] [PubMed] [Google Scholar]
- 46.Hara N, Morimoto YV, Kawamoto A, Namba K, Minamino T. 2012. Interaction of the extreme N-terminal region of FliH with FlhA is required for efficient bacterial flagellar protein export. J Bacteriol 194:5353–5360. doi: 10.1128/JB.01028-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minamino T, Kinoshita M, Namba K. 2017. Fuel of the bacterial flagellar type III protein export apparatus. Methods Mol Biol 1593:3–16. doi: 10.1007/978-1-4939-6927-2_1. [DOI] [PubMed] [Google Scholar]
- 48.Ohnishi K, Fan F, Schoenhals GJ, Kihara M, Macnab RM. 1997. The FliO, FliP, FliQ, and FliR proteins of Salmonella Typhimurium: putative components for flagellar assembly. J Bacteriol 179:6092–6099. doi: 10.1128/jb.179.19.6092-6099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furukawa Y, Imada K, Vonderviszt F, Matsunami H, Sano K, Kutsukake K, Namba K. 2002. Interactions between bacterial flagellar axial proteins in their monomeric state in solution. J Mol Biol 318:889–900. doi: 10.1016/S0022-2836(02)00139-0. [DOI] [PubMed] [Google Scholar]
- 50.Kihara M, Minamino T, Yamaguchi S, Macnab RM. 2001. Intergenic suppression between the flagellar MS ring protein FliF of Salmonella and FlhA, a membrane component of its export apparatus. J Bacteriol 183:1655–1662. doi: 10.1128/JB.183.5.1655-1662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




