Human gut Bacteroides species exhibit strain-level differences in their physiology, ecology, and impact on human health and disease. However, existing approaches for genetic manipulation generally require construction of genetically modified parental strains for each microbe of interest or defined medium formulations. In this report, we introduce a robust and efficient strategy for targeted genetic manipulation of diverse wild-type Bacteroides species from the human gut. This system enables genetic investigation of members of human and animal microbiomes beyond existing model organisms.
KEYWORDS: Bacteroides, microbiome, antimicrobial peptides, genetics
ABSTRACT
Bacteroides is one of the most prominent genera in the human gut microbiome, and study of this bacterial group provides insights into gut microbial ecology and pathogenesis. In this report, we introduce a negative selection system for rapid and efficient allelic exchange in wild Bacteroides species that does not require any alterations to the genetic background or a nutritionally defined culture medium. In this approach, dual antibacterial effectors normally delivered via type VI secretion are targeted to the bacterial periplasm under the control of tightly regulated anhydrotetracycline (aTC)-inducible promoters. Introduction of aTC selects for recombination events producing the desired genetic modification, and the dual effector design allows for broad applicability across strains that may have immunity to one counterselection effector. We demonstrate the utility of this approach across 21 human gut Bacteroides isolates representing diverse species, including strains isolated directly from human donors. We use this system to establish that antimicrobial peptide resistance in Bacteroides vulgatus is determined by the product of a gene that is not included in the genomes of previously genetically tractable members of the human gut microbiome.
IMPORTANCE Human gut Bacteroides species exhibit strain-level differences in their physiology, ecology, and impact on human health and disease. However, existing approaches for genetic manipulation generally require construction of genetically modified parental strains for each microbe of interest or defined medium formulations. In this report, we introduce a robust and efficient strategy for targeted genetic manipulation of diverse wild-type Bacteroides species from the human gut. This system enables genetic investigation of members of human and animal microbiomes beyond existing model organisms.
INTRODUCTION
The gut microbiome is associated with many aspects of human health and disease. Genomic and metagenomic sequencing efforts are beginning to implicate specific microbial taxa and functions in these interactions, but approaches to describe these communities vastly outpace complementary methods to manipulate their gene contents for research or therapeutic purposes (1, 2). Bacteroides is among the most abundant bacterial genera in the guts of humans and other mammals (3), and members of this genus are associated with health benefits of the microbiome, resistance to pathogens, and other host phenotypes (4, 5). While the functions of most Bacteroides genes are unknown, specific genes have been implicated in dietary responses (6, 7), host inflammation and inflammatory bowel disease (8), resistance to antimicrobial peptides (AMPs) (9), and other functions. As a result, Bacteroides species have emerged as important models for understanding the dynamics of the human gut environment, and tools for genetic manipulation of these species provide valuable means to understand the role of the microbiome in health and disease.
Allelic exchange is a versatile approach that allows repeated introduction of targeted, stable, and unmarked genetic insertions, deletions, and sequence changes (10). This approach often makes use of a nonreplicating (suicide) plasmid that carries an antibiotic resistance cassette, a means for counterselection, and the genomic modification flanked by homologous sequences for recombination at the appropriate location in the recipient genome. After the plasmid is introduced into the recipient strain (typically by conjugation or electroporation), antibiotic selection allows isolation of merodiploids resulting from a single recombination event between one of the homologous regions in the plasmid and the corresponding sequence in the genome. In a second step, the counterselectable marker is used to select for cells that have excised the plasmid (and included antibiotic resistance cassette) via a second recombination event, producing mutant and revertant strains by negative selection.
The most widely used system for allelic exchange in bacteria utilizes the levansucrase (sacB) gene, whose product converts sucrose into the toxic compound levan (11, 12). Because Bacteroides species catabolize levan (13), alternate strategies rely on genetically modified parental strains (14, 15) or selection on chemically defined culture medium, with or without the addition and subsequent removal of an accessory plasmid (16, 17). While these methods have provided many key advances in the field, important limitations exist. For example, approaches that rely on genetically modified parental strains require construction of this mutation (e.g., deletion of the thymidine kinase gene tdk) in each species or strain of interest prior to use of the allelic exchange strategy. In addition, escape mutants in which the integrated allelic exchange vector is not resolved from the genome but instead acquires any single inactivating mutation in the negative selection marker can impede strain construction. Further, experimental studies using these mutants may be complicated by the impact of the parental strain modification on bacterial physiology. Similarly, methods that require selection on chemically defined culture medium can be difficult to apply to fastidious strains that are readily cultured only on undefined medium. Antibacterial toxins have been employed for negative selection in Bacillus subtilis, Clostridium acetobutylicum, Enterobacteriaceae species, Proteobacteria species, and Bacteroidales species (18–22). These approaches, which rely on a tightly regulated inducible expression system, provide an attractive method for negative selection in wild-type bacterial strains.
Here, we describe selection by inducible effectors (SIE), a broadly applicable and modular allelic exchange strategy for Bacteroides that does not require genetic modification of the parental strain or growth on defined culture medium. In this approach, an allelic exchange plasmid encodes multiple antibacterial effectors that are normally delivered by Bacteroides type VI secretion systems but have been modified to include periplasmic targeting signal sequences (23). These toxic effectors are regulated by tightly controlled inducible promoters (23), such that expression occurs only during the counterselection step. This approach complements methods for allelic exchange that rely on expression of a single antibacterial effector (22). We demonstrate functional counterselection in 21 Bacteroides isolates representing 10 species, including multiple strains that were captured directly from human donors and displayed a range of resistance to commonly used antibiotic markers. We constructed targeted genetic deletions in 3 Bacteroides strains, including 1 isolated directly from a human donor. Using this approach, we identified a gene that mediates AMP resistance in Bacteroides vulgatus.
RESULTS
An inducible effector-based selection cassette functions across human gut Bacteroides species.
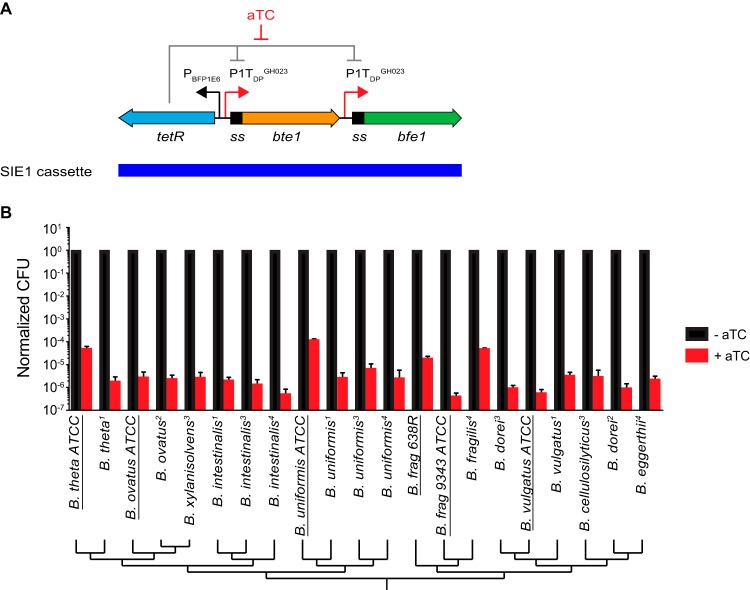
Certain antibacterial effectors secreted by the Bacteroides fragilis type VI secretion system are highly toxic to Bacteroides thetaiotaomicron when targeted to the periplasm (23). To explore the possibility of using these effectors as counterselectable markers, we created a counterselection cassette (SIE1) in which the type VI effector genes bte1 and bfe1 (24, 25) are modified to encode periplasmic localization signals and are independently controlled by the inducible promoter P1TDPGH023 (23) (Fig. 1A). The cassette also includes the repressor gene tetR, which is expressed from the constitutive promoter PBFP1E6 (26). In this design, expression of the effector genes can be activated using anhydrotetracycline (aTC), which binds to TetR and blocks its repressor activity.
FIG 1.
Selection by inducible effectors. (A) Design of the SIE1 dual effector negative selection cassette. Antibacterial effectors are localized to the periplasm via signal sequences (ss), under the control of independent aTC-inducible promoters. (B) Dual effector selection efficiency across human gut Bacteroides species. Type strains are underlined; strains isolated directly from human donors are indicated by superscript donor numbers. The 16S rRNA-based phylogeny is shown at the bottom. Data represent means and standard deviations from triplicate experiments. theta, thetaiotaomicron; frag, fragilis.
To explore the performance of this cassette across a variety of Bacteroides species, we modified the pNBU2_erm plasmid (27) to include this counterselection cassette. Because pNBU2_erm integrates into phage attachment (att) sites that are conserved across Bacteroides species, this allows testing of the counterselection cassette across multiple species with a single construct. We integrated the modified pNBU2_erm_SIE1 plasmid or the empty pNBU2_erm vector control into 21 Bacteroides strains, including 15 isolates cultured directly from humans (23). Integrants were readily obtained in each strain background, confirming that effector expression is tightly controlled in the absence of aTC. Plating these integrants in the presence of aTC reduced CFU by 104- to 106-fold (Fig. 1B); in contrast, the same strains carrying unmodified pNBU2_erm exhibited equivalent viability in the presence and absence of aTC (data not shown). Figure 1B includes strains that are immune to either of the effectors in the SIE1 cassette, indicating that both effectors contribute to counterselection. These results suggest that the SIE1 counterselection cassette could be applied to a wide range of Bacteroides strains, including direct human isolates.
Design of plasmid pSIE1.
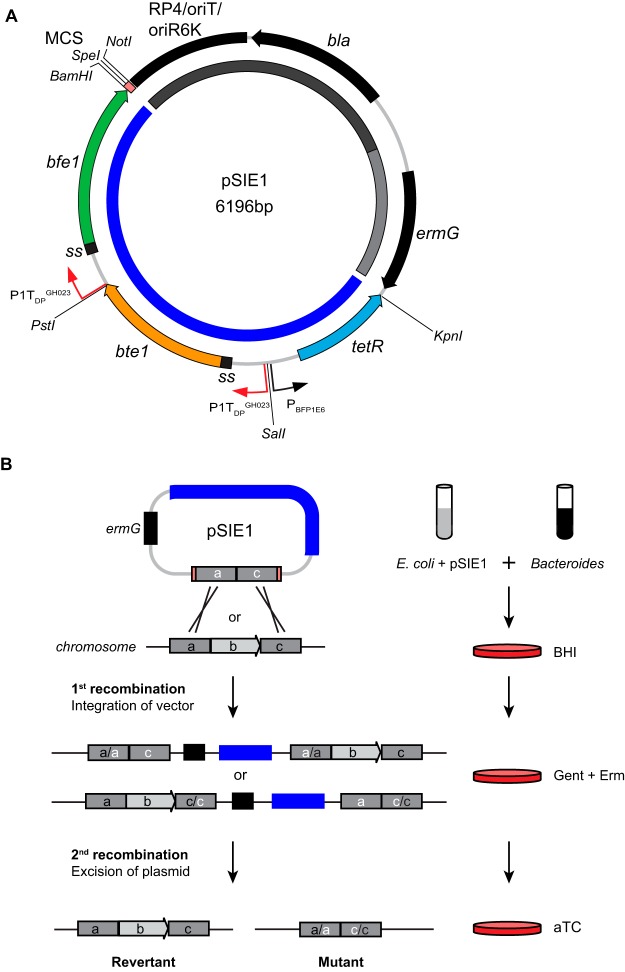
We next designed an allelic exchange vector to test the utility of the counterselection cassette in the genetic modification of wild-type Bacteroides species. The pSIE1 vector (Fig. 2A) consists of a backbone from pExchange-tdk (14) that encodes machinery for Escherichia coli pir-dependent replication, transfer by conjugation, and selection in E. coli and Bacteroides species; the effector-based SIE1 counterselection cassette; and a multiple cloning site (MCS) for insertion of the replacement allele. Restriction sites allow for straightforward exchange of the effector modules. Because the counterselection cassette includes all necessary machinery for conditional toxicity, the plasmid is designed to facilitate genetic manipulation without any requirement for previous modification of the parental strain or a chemically defined culture medium.
FIG 2.
Allelic exchange using pSIE1. (A) pSIE1 plasmid, including the dual-effector negative selection cassette (dark blue), machinery for pir-dependent replication, conjugation, and antibiotic resistance in E. coli (dark gray), and a selectable marker for recipient Bacteroides species (light gray). Relevant restriction sites are indicated. (B) Allelic exchange methodology. Regions of homology included in the pSIE1 plasmid are marked with white letters, while the genomic sequence is marked with black letters. Gent, gentamicin; Erm, erythromycin.
Genetic manipulation of Bacteroides species using pSIE1.
A schematic of pSIE1 integration and gene deletion is presented in Fig. 2B. In the first step, a genetic sequence consisting of 700 to 1,000 bp of the upstream and downstream regions of the target site (joined directly for deletions or separated by the modified allele) is cloned into the pSIE1 MCS. Next, this plasmid is mobilized from an E. coli S17-1 pir donor strain into the Bacteroides recipient by conjugation; plating on gentamicin and erythromycin selects for Bacteroides single-crossover merodiploids that have incorporated the plasmid into the genome by homologous recombination. This merodiploid is then exposed to aTC, selecting for a second recombination event that removes the plasmid backbone (including the antibiotic resistance genes and the toxic effectors). If the first and second recombination events occur in the same region of homology between the genome and the plasmid, then a revertant (to the parental genotype) is generated; if the second recombination event occurs at the other homologous region, then the result is the desired allelic exchange. These outcomes can be distinguished by PCR or other methods.
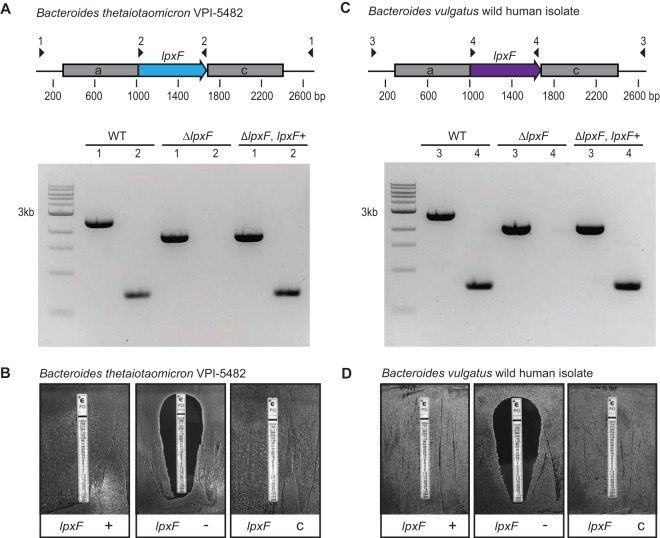
As a proof of concept, we applied this approach to genes involved in AMP resistance in Bacteroides species. AMP resistance is a useful target because this phenotype is conserved across the genus, including species with no established system for targeted genetic manipulation (9), and because the contribution of individual genes can be readily assessed by MIC determinations and related assays. Lipid A modification is one of the primary means by which a wide variety of bacterial species combat AMPs (28–30). Commensal Bacteroides species have been shown to modify lipid A by way of the lpxF gene, which encodes a lipid A phosphatase that is required for AMP resistance (9). Deletion of lpxF (BT_1854) from a B. thetaiotaomicron tdk mutant increases sensitivity to the AMP polymyxin B (PMB) by 3 to 4 orders of magnitude (9). We modified pSIE1 to include genomic sequences of the flanking regions adjacent to the lpxF gene and used this plasmid to delete lpxF from wild-type B. thetaiotaomicron strain VPI-5482 (Fig. 3A). Of the 48 aTC-resistant clones tested, all were sensitive to erythromycin, validating the efficacy of this counterselectable cassette in eliminating unresolved merodiploids and preventing escape mutants (see Table S1 in the supplemental material). Successful deletion was confirmed by PCR screening (Fig. 3A). We next restored lpxF function to the mutant by complementation with this gene, expressed in single copy in a heterologous genomic location. PMB sensitivity of the mutant strain, compared to the parent strain or complemented mutants, establishes that the phenotypes match these genotypes (Fig. 3B; also see Table S2).
FIG 3.
pSIE1-mediated allelic exchange in wild-type Bacteroides species. (A) Deletion of lpxF from wild-type B. thetaiotaomicron VPI-5482. (Top) Regions of homology (gray) and primer locations (arrowheads) are indicated. (Bottom) PCR results from wild-type (WT), mutant, and complemented strains are shown. (B) PMB sensitivity of wild-type (+), mutant (−), and complemented (c) strains. (C) Deletion of lpxF from a B. vulgatus strain isolated directly from a human donor. (D) PMB sensitivity of the wild-type B. vulgatus human isolate (+), its isogenic lpxF mutant (−), and the complemented strain (c). Representative selection efficiencies during mutant construction are provided in Table S1; MICs determined from liquid cultures are shown in Table S2.
Bacteroides vulgatus also contains an lpxF gene implicated in PMB resistance (9), but a genetically modified parent, which would be required for gene deletion with existing methods, has not been available for any strains of this species. We modified the pSIE1 plasmid to delete lpxF from B. vulgatus and introduced this plasmid into a B. vulgatus strain isolated directly from a human donor (23). Selection on aTC again resulted in successful lpxF deletion, with corresponding PMB sensitivity on solid medium and in liquid culture (Fig. 3C and D; also see Table S2). The counterselectable cassette again displayed 100% efficiency in eliminating unresolved merodiploids (Table S1). Together, these proof-of-principle experiments establish the utility of pSIE1 for genetic manipulation of type strains and isolates captured directly from humans.
pSIE1 establishes a novel gene involved in PMB resistance.
Transposon mutant screens for PMB sensitivity in nonmodel Bacteroides species identified multiple genes that appeared to be required for PMB resistance in those species, despite being absent from the PMB-resistant species B. thetaiotaomicron (9). However, this observation was complicated by known polar effects of the transposon, assessment of mutant phenotypes only in the context of a mixed pool of thousands of mutant strains, and the lack of methods for targeted deletion of candidate genes in these nonmodel species. One example is BVU_1078 and its homologs, which are significantly (q values of <10−8) required for fitness in a PMB-dependent manner, in B. vulgatus ATCC 8482 and Bacteroides eggerthii ATCC 27754, despite being absent from B. thetaiotaomicron (9). These proteins are putative glycosyltransferases that show greatest similarity to the ArnT protein of Cupriavidus metallidurans, according to a PHYRE2 search (31, 32). ArnT functions to reduce cell surface negative charge by attaching the cationic sugar 4-amino-4-deoxy-l-arabinose (l-Ara4N) to lipopolysaccharide (LPS)-lipid A (33). ArnT is typically encoded within an operon encoding seven enzymes (arnBCADTEF) involved in synthesizing and transporting l-Ara4N (34). Notably, C. metallidurans arnT and BVU_1078 do not share this typical genetic organization. However, C. metallidurans ArnT functions to attach l-Ara4N to lipid A at the 1-phosphate position (32), suggesting a possible role for BVU_1078 in lipid A modification.
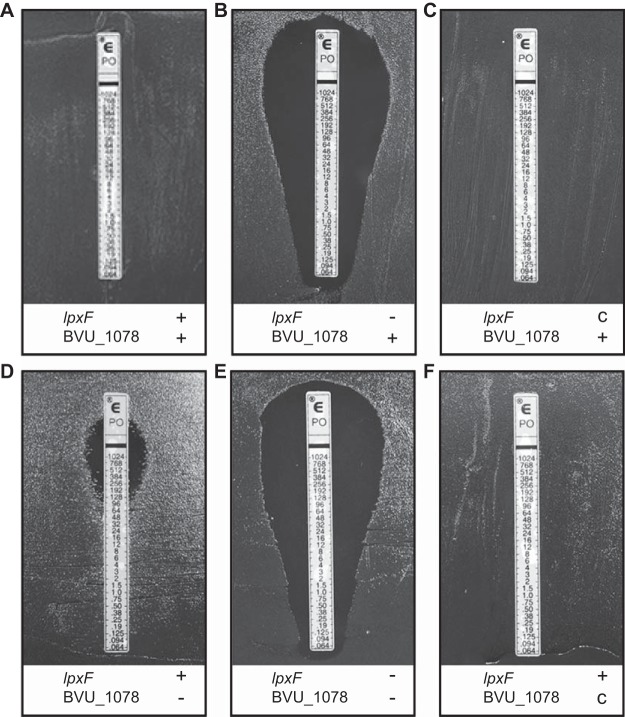
We used pSIE1 to delete BVU_1078 and lpxF (BVU_3293), separately and in combination, from Bacteroides vulgatus ATCC 8482. As expected, deletion of lpxF significantly increased PMB sensitivity (Fig. 4A to C). Notably, deletion of BVU_1078 also significantly increased PMB sensitivity, and complementation restored the wild-type phenotype, directly implicating this gene in AMP resistance (Fig. 4; also see Table S2). Mass spectrometric analysis of lipid A from the wild-type strain and ΔlpxF, ΔBVU_1078, and double deletion mutants revealed the expected lpxF-dependent dephosphorylation at the 4′ position but no BVU_1078-dependent differences in lipid A structure (Fig. S1), suggesting an alternative role for this protein in PMB resistance. This proof-of-concept study demonstrates how pSIE1 can be used successively to engineer multiple modifications in wild-type Bacteroides strains.
FIG 4.
BVU_1078 determination of PMB resistance in B. vulgatus. Strains containing the native copy of each gene (+), deletions (−), and complementations (C) are indicated. MICs determined from liquid cultures are shown in Table S2.
DISCUSSION
Current tools for the genus Bacteroides, including high-throughput genetic screens, engineered promoters, and gnotobiotic animal models, have provided valuable insights about the human gut microbiome (3, 26). In this study, we introduce an approach for genetic modification of Bacteroides species utilizing an engineered cassette that provides selection through the induction of multiple toxic effectors. This system complements related techniques utilizing a single effector (Bfe1) (22). Given the prevalence of type VI effector-immunity pairs and orphan immunity genes across human gut Bacteroides species (25, 35, 36) and the observation that immunity proteins confer specific resistance to individual effectors (25), a counterselection system utilizing multiple effectors reduces the likelihood that a recipient strain would be resistant to effector-mediated selection. No orphan Bfe1 immunity genes were found by Garcia-Bayona et al. (22), although another study did identify candidate orphan Bfe1 immunity genes in both human metagenomic and microbial genome sequences (36). The modular design of the plasmid (Fig. 2A) allows for the exchange of effectors in the event that a target strain has immunity to both effectors encoded by pSIE1. In addition, counterselection systems that rely on the expression and activity of a single gene to confer toxicity can be subject to inactivating missense or nonsense mutations that produce colonies that have evaded the desired recombination event, a phenomenon known as “counterselection escape” (37–39). Robust tetR expression is critical to repress effector expression prior to counterselection; because ribosomal promoters and their derivatives can show reduced activity in stationary phase (26), the ribosomal P2A21 promoter driving tetR in the original construct (22, 23) was also replaced.
As a proof of concept, we deleted multiple genes in multiple Bacteroides species, which recapitulated known phenotypes and established a new role for a gene that is contained only in species with no previously established genetic system. Targeted deletion of this gene, represented by BVU_1078 in B. vulgatus, significantly reduces PMB resistance in this species; restoring expression in single copy fully complements this defect. The mechanism of BVU_1078 activity is unknown. PMB resistance in wild-type B. thetaiotaomicron is significantly greater than resistance in a B. vulgatus ΔBVU_1078 mutant, although both of these strains contain an lpxF homolog and lack a BVU_1078 homolog. This finding suggests that BVU_1078 could target a substrate that is present in B. vulgatus but absent in B. thetaiotaomicron. Notably, this gene is located adjacent to a predicted LPS biosynthesis gene cluster, and the LPS architectures of B. thetaiotaomicron and B. vulgatus are distinct (40). Furthermore, LPS can vary between strains of B. vulgatus (41), which could also affect the contribution of BVU_1078. While further studies are required to define how BVU_1078 determines antimicrobial resistance, these results highlight the diversity of AMP resistance mechanisms that can be accessed through genetic manipulation of nonmodel gut Bacteroides species.
This genetic system does have limitations. First, this strategy uses conjugation to transfer pSIE1 to the recipient strain. While conjugation is generally feasible across Bacteroides species, restriction modification and other factors can limit the efficiency of incorporation of foreign DNA. Donor strains with appropriate DNA methylation systems may partially address this challenge. Second, Bacteroides genetics are limited by a paucity of selectable markers; if the strain of interest is naturally resistant to erythromycin, tetracycline, chloramphenicol and other commonly used antibiotics, then a different approach can be used to select for recombinants after conjugation (22). The pNBU2_erm_SIE1 plasmid (Fig. 1) provides a straightforward means of assessing effector function and these other concerns prior to construction of an allelic exchange vector. The 21 strains tested in this study display wide ranges of susceptibility to erythromycin (0.34 to 12 μg/ml) and tetracycline (0.09 to 16 μg/ml) (see Table S3 in the supplemental material), but the SIE1 cassette allows robust selection in each case, demonstrating that erythromycin selection and aTC-dependent effector expression are still functional across this range of antibiotic sensitivities (Fig. 1B). As with any genetic manipulation, complementation of mutants is an important step; for example, if a recipient strain contains a conjugative transposon that mobilizes spontaneously or in response to aTC exposure, then comparison of parental, mutant, and complemented strains will directly connect the intended mutation with the phenotype.
Despite these limitations, selection by inducible effector expression offers some unique advantages. First, this approach provides the ability to perform targeted genetic modification in a wide variety of wild-type Bacteroides species, avoiding the requirement for construction of a genetically modified parent strain for each bacterial strain of interest and allowing study of pathways that may be affected by the parental genetic modification. As an example, we deleted multiple genes in type strains of B. thetaiotaomicron and B. vulgatus, as well as in a B. vulgatus strain isolated directly from a human donor. The demonstrated functionality of the counterselection cassette across 21 Bacteroides strains, including 15 direct human isolates and representing 10 species, suggests that this approach could be general (Fig. 1B). This could allow the genetic study of patient isolates associated with clinical disease states. For example, human clinical isolates of Bacteroides species exhibit distinct antibiotic resistance profiles, capsular polysaccharides, and LPS, compared to type strains (42–44), and this system could potentially be used to elucidate the mechanisms underlying the associations of these phenotypes with colitis. Other recent studies used high-throughput culturing to describe the evolution of Bacteroides species within an individual (45); pSIE1 and derivatives could enable direct genetic manipulation of ancestral and evolved isolates. Second, this strategy does not require specific nutritional requirements or growth conditions. The ability to use rich undefined culture media may help overcome some of the challenges in applying a genetic system to a wide variety of species with various growth requirements. Last, the allelic exchange vector is modular in design; the effectors can be exchanged for others selected from the broad repertoire encoded in various Bacteroides strains (25, 46) and the inducible promoters can be readily exchanged for others with different dynamic ranges (23). The inclusion of two effectors independently expressed through a tightly regulated promoter enhances the functional capability of this approach by facilitating genetic manipulation of strains that harbor an immunity gene or other resistance mechanisms for one of the selected effectors and also prevents any single inactivating mutation that silences one effector from disabling the system. Together, these features should facilitate mechanistic study of one of the most abundant genera in the human gut microbiome and may provide a strategy that could be adapted for other commensal microbes from humans and other animals, pathogens, and microbes from the environment.
MATERIALS AND METHODS
Primers, plasmids, and strains.
Full lists of primers, plasmids, and strains are provided in Tables S3 and S4 in the supplemental material. Plasmids are available from Addgene (identification numbers 136355 and 136356).
Bacterial culture conditions.
Escherichia coli was grown aerobically at 37°C in Luria-Bertani (LB) medium. All Bacteroides species, including human-gut-derived isolates, were grown anaerobically in tryptone-yeast-glucose (TYG) medium or on brain heart infusion (BHI) agar supplemented with 10% horse blood (Quad Five Co.). For selection, ampicillin (100 μg/ml), gentamicin (200 μg/ml), erythromycin (25 or 12.5 μg/ml), and aTC (100 ng/ml) (Cayman Chemicals) were added as indicated. A flexible anaerobic chamber (Coy Laboratory Products) containing 20% CO2, 10% H2, and 70% N2 was used for all anaerobic microbiology steps.
Construction of pSIE1.
The pSIE1 plasmid was constructed using the backbone of the pExchange plasmid (14). The constitutive PBFP1E6 promoter (26) was fused to the tetR gene using splicing by overlap extension (SOE) PCR (47); this PBFP1E6-driven tetR construct was PCR amplified (primers 1 and 2) using Q5 high-fidelity DNA polymerase (NEB) and cloned into pExchange at the KpnI and SalI sites using standard restriction cloning. The antibacterial effectors ss-bte1 and ss-bfe1 were PCR amplified from pNBU2_erm_P1T_DP-GH023-ss-bte1 and pNBU2_erm_P1T_DP-GH023-ss-bfe1 (23), respectively, and cloned into the vector with aTC-inducible promoters and periplasmic signal sequences using primers 3 to 6. The periplasmic signal sequence is amino acids 1 to 21 (base pairs 1 to 63) from BT_4676.
Allelic exchange.
The protocol developed in this study for counterselectable allelic exchange using the pSIE1 plasmid was modified from previously published work (14). Briefly, ∼700-bp regions flanking each gene of interest were amplified from pEX:LpxFKO (9) for BT_1854 and from B. vulgatus strains for BVU_3293 (primers 11 to 14) and BVU_1078 (primers 17 to 20) using high-fidelity Q5 polymerase (NEB). PCR products were purified (QIAquick PCR purification kit; Qiagen), cloned into the pSIE1 plasmid MCS using Gibson assembly (NEB), and transformed into E. coli S17 pir using standard protocols (48). Candidate clones were verified by sequencing.
Verified E. coli S17 pir clones were conjugated to appropriate Bacteroides species using previously published methods (14), with some modifications. Briefly, Bacteroides and E. coli strains were grown for 16 to 24 h in TYG medium (Bacteroides species) or LB medium with ampicillin (E. coli), subcultured in triplicate to an optical density at 600 nm (OD600) of 0.3 to 0.6 in the same medium (10 ml for Bacteroides species and 5 ml for E. coli), pelleted, resuspended, and combined in a total volume of 1 ml of phosphate-buffered saline (PBS). This volume was plated on BHI plates, which were incubated aerobically for 16 to 24 h. Biomass was collected and resuspended in 1 ml of PBS, and dilutions were plated on BHI plates with gentamicin and erythromycin to select for transconjugants. Colonies were screened by PCR to determine whether the pSIE1 plasmid integrated upstream or downstream of the region of interest. Validated merodiploids (approximately one to three) were then grown overnight in TYG medium, plated on BHI plates containing aTC, and screened by PCR (primers 7 to 10, 15, 16, 21, 22, and 25 to 28) and antibiotic selection to identify the desired mutants.
Counterselection sensitivity testing.
The pNBU2_erm plasmid (27) was modified (primers 23 and 24) to include the SIE counterselection cassette (encoding PBFP1E6-tetR, P1TDPGHO23-ssbte1, and P1TDPGH023-ssbfe1) and integrated into the genome of Bacteroides strains (Table S3) by conjugation. Strains were grown in triplicate and plated on BHI plates with gentamicin and erythromycin, in the presence or absence of aTC; recovered CFU were normalized to the average from the non-aTC condition. Sensitivity to aTC was also tested for each strain carrying an unmodified pNBU2_erm plasmid. Sensitivity to erythromycin and tetracycline was determined with Etest strips (bioMérieux) for each Bacteroides strain. For these measurements, liquid cultures (OD600 of 0.1) of each strain were spread evenly on a BHI agar plate and allowed to dry for 10 min, followed by placement of Etest strips in triplicate for each antibiotic. Plates were incubated anaerobically at 37°C for 16 to 24 h.
Construction of complementation vectors.
Complementation of B. thetaiotaomicron ΔlpxF was performed using a previously described vector (9). Vectors for complementation of B. vulgatus were created in the pNBU2_erm integration plasmid. Primers 29 and 30 were used to amplify the PBFP5E4 promoter from pWW3821 (26), which was cloned by Gibson assembly into NotI- and SpeI-digested pNBU2_erm. The promoter region was sequence verified and designated pNBU2_erm_P5E4. Genes were cloned downstream of the PBFP5E4 promoter (primers 25 to 28), using Gibson assembly cloning, in NcoI- and SalI-digested pNBU2_erm_P5E4.
Determination of efficiency of recombination steps.
Conjugations were performed as described above. First recombination CFU were determined after the 16- to 24-h aerobic conjugation step by plating serial dilutions from the collected biomass on BHI plates containing gentamicin or gentamicin plus erythromycin. Each biological replicate was plated on antibiotic selection plates in triplicate. Second recombination CFU were determined by growing confirmed merodiploids in TYG medium overnight and plating them in triplicate on BHI plates containing gentamicin or aTC. All replicate values were averaged.
PMB sensitivity assays.
Liquid MIC values were determined by the Hancock laboratory microtiter broth dilution method (49), with minor modifications. Briefly, strains were cultured for 24 h in TYG medium and inoculated into 96-well microtiter plates at a starting OD600 of 0.01, in a total volume of 150 μl of medium supplemented with selected concentrations of the AMP PMB (Sigma); 96-well plates were incubated at 37°C and monitored continuously using a microplate reader (BioTek Eon). Each experiment was performed in triplicate, with a negative control (no bacteria) and a positive control (no PMB) on each plate. The MIC was calculated as the lowest concentration of PMB that reduced growth (OD600) by more than 50%, compared to the positive control, at 24 h.
Visualization of PMB sensitivity was performed with Etest strips. For these measurements, 200 μl of liquid culture (OD600 of 0.1) of each strain was spread evenly onto a BHI agar plate and allowed to dry for 10 min, followed by placement of a PMB Etest strip (bioMérieux) at the center of the plate. Plates were incubated anaerobically at 37°C for 16 to 24 h before imaging.
Lipid A extraction and mass spectrometry analysis.
Triplicate cultures of each strain were grown to an OD600 of 1.0 in 25 ml of TYG medium, washed with 1× PBS, pelleted, and frozen. Cell pellets were washed in PBS. Lipid A was extracted using mild acid hydrolysis, as described previously (50, 51). Lipid A samples were subjected to matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). All solvents were high-performance liquid chromatography (HPLC)-grade quality. Lipid A samples were dissolved in a mixture of chloroform and methanol (4:1 [vol/vol]); 5-chloro-2-mercaptobenzothiazole (CMBT) (Sigma) was dissolved in methanol/chloroform/water (4:4:1 [vol/vol/vol]) to a concentration of 25 mg/ml. The matrix was prepared by mixing CMBT with saturated tribasic ammonium citrate (20:1 [vol/vol]). Samples and matrix were mixed (1:1 [vol/vol]), and 1 μl was loaded onto the target plate. MALDI-TOF MS data were calibrated using a peptide calibration standard (Bruker Daltonics), in negative-ion mode. MALDI-TOF MS mass spectra of lipid A extracts were acquired in reflector mode with an Autoflex Speed mass spectrometer (Bruker Daltonics). A total of 500 single laser shots were averaged from each mass spectrum. Data were acquired and processed using FlexControl 3.4 and FlexAnalysis 3.4 software (Bruker Daltonics).
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Goodman laboratory for helpful discussions.
This work was supported by NIH grants GM118159, AI124275, and DK114793 and a Howard Hughes Medical Institute Faculty Scholars Program award to A.L.G. and NIH grants AI138576, AI129940, and AI076322 to M.S.T.
N.A.B.-B., B.L., and A.L.G. conceived and initiated the project, N.A.B.-B. and C.M.H. performed experiments, N.A.B.-B., M.S.T., and A.L.G. analyzed the data, and N.A.B.-B. and A.L.G. wrote the paper.
N.A.B.-B., B.L., and A.L.G. have filed a patent application, based on these studies, with the U.S. Patent and Trade Office (application no. 62/486,526; filed 18 April 2017).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Salyers AA, Bonheyo G, Shoemaker NB. 2000. Starting a new genetic system: lessons from bacteroides. Methods 20:35–46. doi: 10.1006/meth.1999.0903. [DOI] [PubMed] [Google Scholar]
- 2.Bober JR, Beisel CL, Nair NU. 2018. Synthetic biology approaches to engineer probiotics and members of the human microbiota for biomedical applications. Annu Rev Biomed Eng 20:277–300. doi: 10.1146/annurev-bioeng-062117-121019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wexler AG, Goodman AL. 2017. An insider’s perspective: Bacteroides as a window into the microbiome. Nat Microbiol 2:17026. doi: 10.1038/nmicrobiol.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wexler HM. 2007. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev 20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 6.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem 284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Townsend GE II, Han W, Schwalm ND III, Raghavan V, Barry NA, Goodman AL, Groisman EA. 2019. Dietary sugar silences a colonization factor in a mammalian gut symbiont. Proc Natl Acad Sci U S A 116:233–238. doi: 10.1073/pnas.1813780115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown EM, Ke X, Hitchcock D, Jeanfavre S, Avila-Pacheco J, Nakata T, Arthur TD, Fornelos N, Heim C, Franzosa EA, Watson N, Huttenhower C, Haiser HJ, Dillow G, Graham DB, Finlay BB, Kostic AD, Porter JA, Vlamakis H, Clish CB, Xavier RJ. 2019. Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and symbiosis. Cell Host Microbe 25:668–680.E7. doi: 10.1016/j.chom.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, Degnan PH, Booth CJ, Yu H, Goodman AL. 2015. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyrat JM, Pelicic V, Gicquel B, Rappuoli R. 1998. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect Immun 66:4011–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Recorbet G, Robert C, Givaudan A, Kudla B, Normand P, Faurie G. 1993. Conditional suicide system of Escherichia coli released into soil that uses the Bacillus subtilis sacB gene. Appl Environ Microbiol 59:1361–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado CI. 1985. Positive selection procedure for entrapment of insertion sequence elements in Gram-negative bacteria. J Bacteriol 164:918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. 2010. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. 2008. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baughn AD, Malamy MH. 2002. A mitochondrial-like aconitase in the bacterium Bacteroides fragilis: implications for the evolution of the mitochondrial Krebs cycle. Proc Natl Acad Sci U S A 99:4662–4667. doi: 10.1073/pnas.052710199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patrick S, Houston S, Thacker Z, Blakely GW. 2009. Mutational analysis of genes implicated in LPS and capsular polysaccharide biosynthesis in the opportunistic pathogen Bacteroides fragilis. Microbiology 155:1039–1049. doi: 10.1099/mic.0.025361-0. [DOI] [PubMed] [Google Scholar]
- 17.Kino Y, Nakayama-Imaohji H, Fujita M, Tada A, Yoneda S, Murakami K, Hashimoto M, Hayashi T, Okazaki K, Kuwahara T. 2016. Counterselection employing mutated pheS for markerless genetic deletion in Bacteroides species. Anaerobe 42:81–88. doi: 10.1016/j.anaerobe.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Al-Hinai MA, Fast AG, Papoutsakis ET. 2012. Novel system for efficient isolation of Clostridium double-crossover allelic exchange mutants enabling markerless chromosomal gene deletions and DNA integration. Appl Environ Microbiol 78:8112–8121. doi: 10.1128/AEM.02214-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khetrapal V, Mehershahi K, Rafee S, Chen S, Lim CL, Chen SL. 2015. A set of powerful negative selection systems for unmodified Enterobacteriaceae. Nucleic Acids Res 43:e83. doi: 10.1093/nar/gkv248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazarus JE, Warr AR, Kuehl CJ, Giorgio RT, Davis BM, Waldor MK. 2019. A new suite of allelic-exchange vectors for the scarless modification of proteobacterial genomes. Appl Environ Microbiol 85:e00990-19. doi: 10.1128/AEM.00990-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang XZ, Yan X, Cui ZL, Hong Q, Li SP. 2006. mazF, a novel counter-selectable marker for unmarked chromosomal manipulation in Bacillus subtilis. Nucleic Acids Res 34:e71. doi: 10.1093/nar/gkl358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Bayona L, Comstock LE. 2019. Streamlined genetic manipulation of diverse Bacteroides and Parabacteroides isolates from the human gut microbiota. mBio 10:e01762-19. doi: 10.1128/mBio.01762-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim B, Zimmermann M, Barry NA, Goodman AL. 2017. Engineered regulatory systems modulate gene expression of human commensals in the gut. Cell 169:547–558.E15. doi: 10.1016/j.cell.2017.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. 2016. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci U S A 113:3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wexler AG, Bao Y, Whitney JC, Bobay LM, Xavier JB, Schofield WB, Barry NA, Russell AB, Tran BQ, Goo YA, Goodlett DR, Ochman H, Mougous JD, Goodman AL. 2016. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci U S A 113:3639–3644. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitaker WR, Shepherd ES, Sonnenburg JL. 2017. Tunable expression tools enable single-cell strain distinction in the gut microbiome. Cell 169:538–546.E12. doi: 10.1016/j.cell.2017.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Shoemaker NB, Wang GR, Salyers AA. 2000. Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J Bacteriol 182:3559–3571. doi: 10.1128/jb.182.12.3559-3571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raetz CR, Reynolds CM, Trent MS, Bishop RE. 2007. Lipid A modification systems in Gram-negative bacteria. Annu Rev Biochem 76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baron S, Hadjadj L, Rolain JM, Olaitan AO. 2016. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents 48:583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 30.Simpson BW, Trent MS. 2019. Pushing the envelope: LPS modifications and their consequences. Nat Rev Microbiol 17:403–416. doi: 10.1038/s41579-019-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrou VI, Herrera CM, Schultz KM, Clarke OB, Vendome J, Tomasek D, Banerjee S, Rajashankar KR, Belcher Dufrisne M, Kloss B, Kloppmann E, Rost B, Klug CS, Trent MS, Shapiro L, Mancia F. 2016. Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation. Science 351:608–612. doi: 10.1126/science.aad1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CR. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J Biol Chem 276:43122–43131. doi: 10.1074/jbc.M106961200. [DOI] [PubMed] [Google Scholar]
- 34.Yan A, Guan Z, Raetz CR. 2007. An undecaprenyl phosphate-aminoarabinose flippase required for polymyxin resistance in Escherichia coli. J Biol Chem 282:36077–36089. doi: 10.1074/jbc.M706172200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verster AJ, Ross BD, Radey MC, Bao Y, Goodman AL, Mougous JD, Borenstein E. 2017. The landscape of type VI secretion across human gut microbiomes reveals its role in community composition. Cell Host Microbe 22:411–419.E4. doi: 10.1016/j.chom.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross BD, Verster AJ, Radey MC, Schmidtke DT, Pope CE, Hoffman LR, Hajjar AM, Peterson SB, Borenstein E, Mougous J. 2019. Human gut bacteria contain acquired interbacterial defence systems. Nature 575:224–228. doi: 10.1038/s41586-019-1708-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. 2015. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc 10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gregg CJ, Lajoie MJ, Napolitano MG, Mosberg JA, Goodman DB, Aach J, Isaacs FJ, Church GM. 2014. Rational optimization of tolC as a powerful dual selectable marker for genome engineering. Nucleic Acids Res 42:4779–4790. doi: 10.1093/nar/gkt1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tominaga M, Kawai-Noma S, Kawagishi I, Sowa Y, Saito K, Umeno D. 2015. Liquid-based iterative recombineering method tolerant to counter-selection escapes. PLoS One 10:e0119818. doi: 10.1371/journal.pone.0119818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobson AN, Choudhury BP, Fischbach MA. 2018. The biosynthesis of lipooligosaccharide from Bacteroides thetaiotaomicron. mBio 9:e02289-17. doi: 10.1128/mBio.02289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breeling JL, Onderdonk AB, Cisneros RL, Kasper DL. 1988. Bacteroides vulgatus outer membrane antigens associated with carrageenan-induced colitis in guinea pigs. Infect Immun 56:1754–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veloo ACM, Baas WH, Haan FJ, Coco J, Rossen JW. 2019. Prevalence of antimicrobial resistance genes in Bacteroides spp. and Prevotella spp. Dutch clinical isolates. Clin Microbiol Infect 25:1156.e9–1156.e13. doi: 10.1016/j.cmi.2019.02.017. [DOI] [PubMed] [Google Scholar]
- 43.Hashimoto M, Kirikae F, Dohi T, Adachi S, Kusumoto S, Suda Y, Fujita T, Naoki H, Kirikae T. 2002. Structural study on lipid A and the O-specific polysaccharide of the lipopolysaccharide from a clinical isolate of Bacteroides vulgatus from a patient with Crohn’s disease. Eur J Biochem 269:3715–3721. doi: 10.1046/j.1432-1033.2002.03062.x. [DOI] [PubMed] [Google Scholar]
- 44.Rouyan GS, Meisel-Mikołajczyk F, Rumin W. 1994. The toxicity of antigens extracted from strains of Bacteroides vulgatus from different origins to chicken embryos. Acta Microbiol Pol 43:97–101. [PubMed] [Google Scholar]
- 45.Zhao S, Lieberman TD, Poyet M, Kauffman KM, Gibbons SM, Groussin M, Xavier RJ, Alm EJ. 2019. Adaptive evolution within gut microbiomes of healthy people. Cell Host Microbe 25:656–667.E8. doi: 10.1016/j.chom.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coyne MJ, Roelofs KG, Comstock LE. 2016. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics 17:58. doi: 10.1186/s12864-016-2377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lefebvre B, Formstecher P, Lefebvre P. 1995. Improvement of the gene splicing overlap (SOE) method. Biotechniques 19:186–188. [PubMed] [Google Scholar]
- 48.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (ed). 1987. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience John Wiley and Sons, New York, NY. [Google Scholar]
- 49.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 50.Henderson JC, Herrera CM, Trent MS. 2017. AlmG, responsible for polymyxin resistance in pandemic Vibrio cholerae, is a glycyltransferase distantly related to lipid A late acyltransferases. J Biol Chem 292:21205–21215. doi: 10.1074/jbc.RA117.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Z, Lin S, Cotter RJ, Raetz CR. 1999. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12: detection of 4-amino-4-deoxy-l-arabinose, phosphoethanolamine and palmitate. J Biol Chem 274:18503–18514. doi: 10.1074/jbc.274.26.18503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.