Pseudomonas aeruginosa forms multicellular aggregates or biofilms encased in a matrix. We show for the first time here that dispersion by P. aeruginosa requires the endogenous expression of pelA and pslG, leading to the degradation of both Pel and Psl polysaccharides, with PslG-induced dispersion being CdrA dependent. The findings suggested that endogenously induced Psl degradation is a sequential process, initiated by untethering of CdrA-bound Psl or CdrA-dependent cell interactions to enable Psl degradation and ultimately, dispersion. Untethering likely involves CdrA release in a manner independent of c-di-GMP modulation and thus LapG. Our findings not only provide insight into matrix degrading factors contributing to dispersion but also identify key steps in the degradation of structural components of the P. aeruginosa biofilm matrix.
KEYWORDS: CdrA, PelA, PslG, biofilm, dispersion, exopolysaccharide, glycoside hydrolase, matrix, untethering
ABSTRACT
Biofilms are multicellular aggregates of bacteria that are encased in an extracellular matrix. The biofilm matrix of Pseudomonas aeruginosa PAO1 is composed of eDNA, proteins, and the polysaccharides Pel and Psl. This matrix is thought to be degraded during dispersion to liberate cells from the biofilms, with dispersion being apparent not only by single cells escaping from the biofilm but also leaving behind eroded or hollowed-out biofilm. However, little is known of the factors involved in matrix degradation. Here, we focused on the glycoside hydrolases PelA and PslG. We demonstrate that induction of pelA but not pslG expression resulted in dispersion. As Psl is tethered to the matrix adhesin CdrA, we furthermore explored the role of CdrA in dispersion. cdrA mutant biofilms were hyperdispersive, while lapG mutant biofilms were impaired in dispersion in response to glutamate and nitric oxide, indicating the presence of the surface-associated matrix protein CdrA impedes the dispersion response. In turn, insertional inactivation of cdrA enabled pslG-induced dispersion. Lowering of the intracellular c-di-GMP level via induction of PA2133 encoding a phosphodiesterase was not sufficient to induce dispersion by wild-type strains and strains overexpressing pslG, indicating that pslG-induced dispersion is independent of c-di-GMP modulation and, likely, LapG.
IMPORTANCE Pseudomonas aeruginosa forms multicellular aggregates or biofilms encased in a matrix. We show for the first time here that dispersion by P. aeruginosa requires the endogenous expression of pelA and pslG, leading to the degradation of both Pel and Psl polysaccharides, with PslG-induced dispersion being CdrA dependent. The findings suggested that endogenously induced Psl degradation is a sequential process, initiated by untethering of CdrA-bound Psl or CdrA-dependent cell interactions to enable Psl degradation and ultimately, dispersion. Untethering likely involves CdrA release in a manner independent of c-di-GMP modulation and thus LapG. Our findings not only provide insight into matrix degrading factors contributing to dispersion but also identify key steps in the degradation of structural components of the P. aeruginosa biofilm matrix.
INTRODUCTION
Biofilms are structured communities of bacteria that form on surfaces, on interfaces, or in mucus-encased aggregates. Growing as biofilms is the most prevalent and important form of life for bacteria in natural, industrial, and clinical settings. Biofilms are embedded in an extracellular matrix that can make up to 90% of the dry mass (1). The extracellular matrix contributes to biofilm architecture and integrity and protects resident biofilm bacteria against antimicrobial agents and engulfment by phagocytic cells within a mammalian host (2). Additional functions attributed to the matrix components include initial surface adhesion, aggregation of bacterial cells, water retention, sorption and storage of nutrients, binding of enzymes, and protection from environmental stressors, as well as shear forces in fluid environments (1). Bacteria growing as biofilms are well protected from environmental stressors, including the host immune response, and are notoriously difficult to eradicate due to their heightened tolerance to almost all antimicrobials and antibiotic classes (3–6). It is thus not surprising that biofilms pose one of the biggest threats to patients in hospital settings, being responsible for >80% of microbial infections and >60% of all nosocomial infections, affecting approximately 13 million Americans (7).
The biofilm matrix of nonmucoid strains of Pseudomonas aeruginosa is composed of the Psl and Pel exopolysaccharides, extracellular DNA (eDNA), and matrix-stabilizing proteins such as the adhesins CdrA and LecB (1, 8–12). Recent reports indicate that Psl, the primary matrix polysaccharide of P. aeruginosa PAO1, is primarily localized at the periphery and base of biofilms, with Psl interacting with the adhesin proteins CdrA and LecB, forming a “shell around the biofilm aggregates” (13, 14). Pel is limited to the base of the biofilm and is cross-linked to eDNA (13). eDNA, however, is not limited to the biofilm base but has also been detected in the aggregate interior (15, 16). CdrA is a c-di-GMP regulated adhesin that reinforces the biofilm matrix by binding to and cross-linking Psl (11, 12). CdrA can be tethered to the outer membrane at its C terminus or released from the outer membrane via cleavage by the periplasmic protease LapG at low c-di-GMP (11, 17). Similar to CdrA, LecB binds to Psl, likely via the branched side chains present on Psl, and contributes to the localization of Psl within the biofilm (18).
The shell-like structure is formed over the course of biofilm formation, a cyclical process that is initiated by single planktonic cells aggregating and/or attaching to a surface. Once the matrix is formed, the interactions of matrix components render the biofilm matrix incredibly stable and likely contribute to the resistance of biofilm matrix components to extracellular proteases and nucleases (13, 19, 20). Biofilm formation comes full cycle when cells disperse from the mature biofilm to resume a planktonic lifestyle. During dispersion, biofilms have been observed to undergo a “hollowing out” process, leaving a small amount of biofilm biomass that is thought to correspond to the “shell” surrounding the microcolonies after dispersion has occurred. This process has been characterized many times by cells leaving the interior of aggregates resulting in central voids (21–23). Dispersion can also involve an erosion process that results in the active release of biofilm biomass (22, 24, 25). The exact process by which bacteria liberate themselves from the matrix-enmeshed biofilm structure is unclear, but it is thought to involve active matrix degradation (26–31). This is supported by freshly dispersed cells demonstrating increased polysaccharide, protein, and eDNA degrading activity relative to planktonic and biofilm cells that showed little to no degradative activity (26). Consistent with this observation, we recently demonstrated that dispersion coincided with the increased expression of genes encoding secreted nucleases EndA, EddA, and EddB, which are capable of degrading eDNA present in the biofilm matrix (32). Moreover, inactivation of endA encoding a secreted DNase coincided with biofilms that are impaired in dispersion in response to glutamate and nitric oxide, whereas endA expression promoted dispersion (32), with EndA-induced dispersion coinciding with a loss of matrix eDNA content (32). Additional evidence suggests that the release of surface-associated adhesins contributes to the dispersal. Examples of such adhesins are CdrA from Pseudomonas aeruginosa (12), and LapA from P. putida and P. fluorescens (33, 34). While the two adhesins share no sequence homology, both can be tethered to the outer membrane or released from the outer membrane upon proteolytic cleavage by the periplasmic protease LapG at low c-di-GMP (12, 35, 36). LapA is localized at the outer membrane at high c-di-GMP levels, but released from the cell surface upon proteolytic cleavage by the protease LapG at low c-di-GMP (36). Gjermansen et al. demonstrated that starvation‐induced dispersal of P. putida biofilms was dependent on LapG-dependent proteolytic cleavage of LapA (33, 37). The same mechanism was found to contribute to phosphate limitation‐induced dispersal of P. fluorescens biofilms (36). However, the role of the c-di-GMP-regulated adhesin CdrA that reinforces the biofilm matrix in dispersion by P. aeruginosa biofilms has only been inferred, with lapG mutants demonstrating increased attachment, while cdrA mutants demonstrated decreased attachment relative to the parental strain upon extended incubation (11).
In addition, several exogenously added glycoside hydrolases, enzymes that act by hydrolyzing the glycosidic linkages between two or more carbohydrates (38), have been demonstrated to disrupt or disaggregate biofilms. The glycoside hydrolases PelA and PslG are required for the synthesis of their respective exopolysaccharides Pel and Psl (39, 40) but also contribute to their degradation (41). Yu et al. (30) demonstrated that exogenous addition of PslG disassembled established biofilms. Likewise, Baker et al. (41) demonstrated that purified glycoside hydrolases PelA and PslG can be utilized to disrupt and prevent P. aeruginosa biofilms. Fleming et al. (42) used a cocktail of glycoside hydrolases (cellulase and α-amylase) that targeted glycosidic linkages commonly seen within the exopolysaccharides secreted by a wide range of pathogens, specifically the β-1,4 bond present in cellulose and targeted by cellulases and the α-1,4 bond targeted by α-amylases. Exposure of established Staphylococcus aureus and P. aeruginosa biofilms with this cocktail effectively disrupted monoculture and coculture biofilms resulted in significant reductions in biomass and dissolution of the biofilm (42).
While the findings underscore exogenous glycoside hydrolases as capable of disrupting established biofilms in a process which has been interpreted as dispersal or dispersion, it is not known whether the dispersion response requires the expression of genes and gene functions that specifically target the matrix polysaccharides. This is supported by the finding of exogenous DNase being unable to disaggregate established P. aeruginosa biofilms (20), whereas activation of intrinsic DNase, by expressing endA encoding an extracellular nuclease, promoted the dispersion by P. aeruginosa biofilms (32). Moreover, no obvious self-produced matrix-degrading enzymes contributing to the degradation of the Pel and Psl polysaccharides, the major polysaccharides of P. aeruginosa PAO1, have been identified to play a role during the dispersion response. These findings not only underscore how little we know about the functions that are required for dispersion and how the process is orchestrated but also raise the question of whether matrix components other than eDNA, particularly matrix polysaccharides, are indeed targeted for degradation upon induction of dispersion by P. aeruginosa PAO1 biofilms.
RESULTS
Overproduction of Pel and Psl polysaccharides affects dispersion.
To begin addressing the question of whether additional matrix components, more specifically matrix polysaccharides, are targeted for degradation upon induction of dispersion, we focused on the main polysaccharides present in the biofilm matrix of P. aeruginosa PAO1, the Pel and Psl exopolysaccharides. We reasoned that if dispersion requires matrix polysaccharide-degrading factors, that overproduction of Pel and Psl polysaccharides would likely overwhelm such factors and coincide with reduced or impaired dispersion response. We therefore made use of two P. aeruginosa strains that overproduce Pel or Psl, referred to here as PAO1::PBAD-pel and PAO1::PBAD-psl.
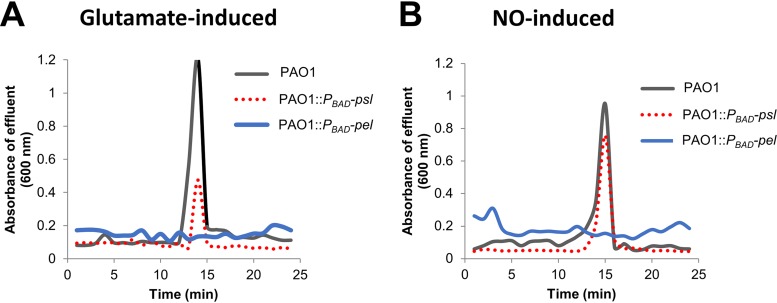
Previous findings indicated that dispersion can be induced by exposure to various exogenous dispersion cues (24, 43), including nitric oxide (NO) and glutamate (23, 44–47). Furthermore, it has been demonstrated that dispersion can be detected by a sharp increase in the absorbance at 600 nm in the effluent within 15 to 20 min upon induction of dispersion, as determined using tube reactors (23, 25, 44, 45, 48, 49). Biofilms by PAO1::PBAD-pel and PAO1::PBAD-psl were grown for 5 days in biofilm tube reactors, and dispersion was subsequently induced by exposing biofilms to the dispersion cues glutamate or nitric oxide. P. aeruginosa wild-type biofilms were used as a control. Wild-type PAO1 biofilms readily dispersed in response to glutamate or nitric oxide. In contrast, PAO1::PBAD-pel biofilms failed to disperse upon challenge with glutamate, while the dispersion response by PAO1::PBAD-psl biofilms was significantly reduced relative to wild-type biofilms (Fig. 1A). Similar results were obtained when the dispersion cue nitric oxide was used, apparent by biofilms by PAO1::PBAD-pel being impaired in dispersion in response to nitric oxide, and biofilms by PAO1::PBAD-psl, demonstrating a reduced dispersion response relative to wild-type biofilms (Fig. 1B).
FIG 1.
Representative dispersion responses of biofilms by P. aeruginosa PAO1 and the polysaccharide-overproducing strains PAO1::PBAD-pel and PAO1::PBAD-psl after exposure to glutamate (A) or nitric oxide (B). Sodium nitroprusside served as a source of nitric oxide. Biofilms were grown for 5 days in 5-fold-diluted VBMM in tube reactors. The absorbance of biofilm tube reactor effluents after the induction of dispersion is shown. Dispersion assays were performed at least three times, with each biological replicate consisting of four technical replicates, but only representative dispersion profiles are shown.
The findings suggested that although manipulation of the quantity of matrix polysaccharide has a negative effect on the dispersion response, increasing the Pel has a more severe negative impact on dispersion than does overproduction of Psl.
Generating P. aeruginosa strains capable of producing PelA and PslG.
Overproduction of matrix polysaccharides impeding or reducing the dispersion response suggested dispersion requires a reduction of one or both polysaccharides, likely by degradation. Two possible candidates contributing to Pel and Psl degradation are PelA and PslG, respectively. PelA has been confirmed to have PEL deacetylase and hydrolase activities (39, 41, 50). Although the PelA deacetylase activity is required for Pel synthesis, the role of PelA’s hydrolase activity in biosynthesis is not known but has been suggested to be involved in chain-length regulation, Pel degradation, and biofilm disassembly (30, 39, 41, 51). PelA was originally predicted to be a periplasmically localized protein (39), and Marmont et al. (50) recently confirmed PelA to be located in the periplasm. Moreover, that study demonstrated that PelA was localized to the outer membrane via its interaction with the outer membrane protein PelB (50). PslG is anchored to the inner membrane, with the majority of the protein predicted to be primarily located in the periplasm (40, 52, 53). Similar to PelA, purified PslG has been shown to be critical for Psl polysaccharide biosynthesis (53) and to contribute to Psl degradation and biofilm disassembly (30, 41, 51). Despite its location in the periplasm, the glycoside hydrolase PslG has recently been demonstrated to hydrolyze surface-associated Psl, with PslG residues E165 and E276 having been implicated in the catalysis (54).
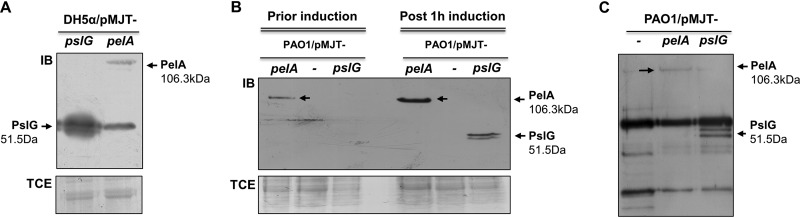
To determine whether endogenously produced PelA and PslG contribute to the polysaccharide matrix degradation, resulting in dispersion, we therefore cloned the pslG and pelA genes into pMJT-1 under the control of an arabinose-inducible promoter. The genes were modified to harbor a C-terminal V5 tag which adds 1.4 kDa, resulting in the calculated molecular mass for PelA being 106.3 kDa and PslG being 51.5 kDa. Immunoblot analysis with anti-V5 antibodies confirmed PelA and PslG to be produced at their expected molecular masses (Fig. 2A). The resulting constructs were subsequently transferred into P. aeruginosa PAO1 to generate strains PAO1/pMJT-pelA and PAO1/pMJT-pslG. Production of PelA and PslG in the respective strains grown as biofilms was confirmed by immunoblot analysis. Although little to no PelA or PslG was detected in biofilms not exposed to arabinose to induce gene expression, both PelA and PslG were detectable in total cell extracts obtained from biofilms by PAO1/pMJT-pelA and PAO1/pMJT-pslG, respectively, after 1 h of induction of gene expression by arabinose (Fig. 2B). In contrast, no protein bands corresponding to PelA or PslG were detectable in in total cell extracts obtained from biofilms by PAO1 harboring the empty vector pMJT-1 (Fig. 2B).
FIG 2.
Confirmation of PelA and PslG protein production. (A) Total cell extracts (TCEs) from E. coli DH5α/pMJT-pslG_V5 and DH5α/pMJT-pelA_V5 cells grown planktonically to exponential phase. Then, pslG and pelA gene expression was induced upon addition of 1% arabinose for 2 h. A representative image of immunoblot (IB) probed for the presence of V5-tagged PelA or PslG using anti-V5 antibodies is shown. (B) TCEs were used as a loading control. TCE, image of Coomassie-stained SDS-gel after immunoblotting. A total of 20 μg of TCE was loaded. The TCEs from PAO1/pMJT-pslG_V5, PAO1/pMJT-pelA_V5, and PAO1/pMJT-1 strains grown as biofilms for 5 days prior to and after the addition of 1% arabinose for 1 h are shown. Representative images of immunoblot (IB) and Coomassie-stained SDS-gel after immunoblotting (TCE) are shown. Arrows point at a band suggestive of PelA. A total of 20 μg of TCE was loaded. (C) Representative images of immunoblots demonstrating the presence of PslG and PelA in culture supernatants of PAO1/pMJT-pslG_V5 and PAO1/pMJT-pelA_V5 strains grown planktonically to the exponential phase, respectively. Culture supernatants were obtained 1 h after the addition of arabinose. Culture supernatants by PAO1/pMJT-1 were used as a control. The arrow points at a band suggestive of PelA. A total of 10 μg of concentrated supernatant protein was loaded.
We likewise determined the presence of PelA and PslG in culture supernatants of PAO1/pMJT-pelA and PAO1/pMJT-pslG. Under the conditions tested, PelA and PslG were detectable by 1 h after induction by arabinose. In contrast, no protein bands corresponding to PelA or PslG were detectable in supernatants obtained from planktonic cells by PAO1 harboring the empty vector pMJT-1 (Fig. 2C). However, it is unclear whether the presence of PelA and PslG in supernatants is due to protein secretion or cell lysis.
Given that biofilms by strains PAO1/pMJT-pelA and PAO1/pMJT-pslG were confirmed to produce PelA and PslG, respectively, under the conditions tested, we used the respective strains to determine whether endogenous expression of pelA and pslG is sufficient to induce dispersion.
Induction of pelA but not pslG results in biofilm dispersion.
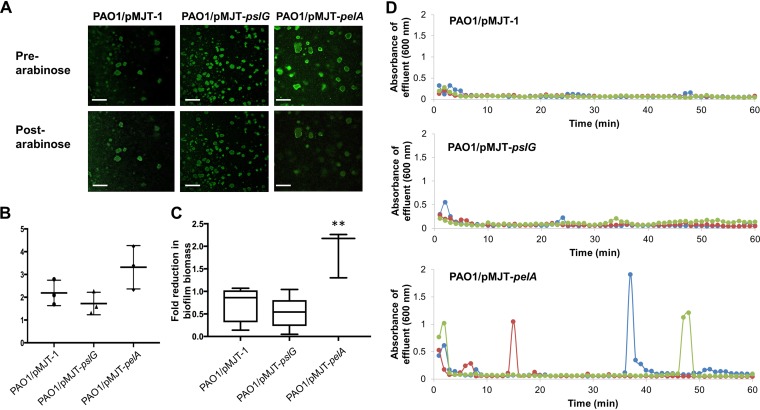
We therefore sought to determine whether endogenous expression of pelA and pslG is sufficient in inducing dispersion. Since biofilm biomass loss is indicative of dispersion, we first wanted to confirm whether induction of pelA or pslG gene expression indeed results in biofilm dispersion. We therefore made use of a strains that harbored pelA and pslG under the control of the arabinose-inducible PBAD promoter (PAO1/pMJT-pelA, PAO1/pMJT-pslG). P. aeruginosa wild type harboring the empty plasmid pMJT-1 was used as a control. Biofilms by the respective mutant strains were grown for 5 days in the absence of arabinose to ensure the establishment of biofilms (Fig. 3A and B). After 5 days of growth, arabinose was added to the growth medium for a period of 60 min to induce the transcription of pelA or pslG. Prior to and following arabinose supplementation, the biofilm architecture was monitored by confocal microscopy (Fig. 3A). Relative to uninduced PAO1/pMJT-1 biofilms, exposure to arabinose appeared to have no effect on the biofilm architecture (Fig. 3A). The findings were supported by the quantitative analysis of fluorescence indicative of the biofilm biomass prior to and after the addition of arabinose (Fig. 3C). In contrast, induction of pelA gene expression, upon exposure to arabinose, resulted in a reduction of the biofilm biomass in biofilms by PAO1/pMJT-pelA (Fig. 3A and C). Induction of pslG gene expression, however, failed to induce a change in the biofilm architecture or to induce a significant reduction in the biofilm biomass (Fig. 3A and C). Our findings suggested that induction of pslG failed to result in the disaggregation of biofilms, while induction of pelA gene expression resulted in dispersion, coinciding with a significant change in the biofilm architecture.
FIG 3.
Induction of pelA but not pslG gene expression coincides with a reduction in biofilm biomass and dispersion events, as evidenced by absorbance increases in the biofilm effluent. (A) Representative confocal images of 5-day-old biofilms by PAO1 prior to and after the induction of pelA and pslG gene expression. Induction of gene expression was accomplished by adding arabinose to the growth medium. Scale bars, 100 μm. (B) Biofilm biomass, as determined using COMSTAT analysis, prior the induction of dispersion. Experiments were performed in triplicate, with each biological replicate consisting of four technical replicates. (C) Fold change in the biofilm biomass after the induction of dispersion by the addition of arabinose. The biofilm biomass prior to and after arabinose addition was quantitated using COMSTAT. Experiments were performed in triplicate, with each biological replicate consisting of 4 technical replicates. **, Significantly different (P < 0.05) relative to PAO1/pMJT-1 biofilms used as controls. (D) Detection of dispersion events after the induction of pelA and pslG gene expression, with gene expression being induced upon addition of 1% arabinose to the growth medium. Different colors represent biological replicates.
However, dispersion not only correlates with a reduction of the biofilm biomass but also correlates with a simultaneous increase of bacteria present in biofilm effluents compared to wild-type biofilms (23, 25, 49). To further confirm that the observed reduction in the biofilm biomass (Fig. 3A and C) was indeed due to dispersion, we next determined whether induction of pelA or pslG gene expression coincides with observable increases in the number of bacteria leaving the biofilm. To do so, we used tube reactor-grown biofilms and collected biofilm effluents after the addition of arabinose to induce gene expression, with sharp increases in the absorbance (600 nm) of the effluent being considered indicative of dispersion events (25, 32).
Wild-type and mutant biofilms were grown similarly to flow cell-grown biofilms for 5 days in the absence of arabinose. The biofilms were then exposed to arabinose to induce pelA or pslG gene expression, and biofilm effluents were collected for 60 min after the addition of arabinose to the growth medium. Effluents by PAO1/pMJT-1 biofilms appeared to have an average absorbance of ∼0.1 over the entire 60-min period (Fig. 3D). In contrast, addition of arabinose to the growth medium resulted in repeated dispersion events of differing intensity by PAO1/pMJT-pelA biofilms over 60 min (Fig. 3D), a response that was absent in control biofilms and in PAO1/pMJT-pslG biofilms after the induction of pslG gene expression (Fig. 3D). Our findings strongly suggested that induction of pelA gene expression, and thus endogenously produced PelA, results in dispersion. It is interesting that the dispersion events noted upon induction of pelA gene expression were similar to those previously observed upon induction of endA (32) and bdlA_G31A gene expression (25) by P. aeruginosa PAO1 biofilms. Moreover, the findings are in agreement with the reduction of the biofilm biomass noted upon induction of pelA and pslG gene expression in flow cell-grown biofilms (Fig. 3A and C). However, our findings also suggested that whereas the induction of pelA gene expression resulted in dispersion, the induction of pslG failed to result in the disaggregation of biofilms. Considering that both PelA and PslG were confirmed to be produced under the conditions tested, the lack of dispersion upon induction of pslG expression was not due to lack of PslG production.
Surface-associated CdrA impedes the dispersion response, while the lack of CdrA enhances dispersion.
The finding of pelA but not pslG expression results in dispersion was unexpected. For example, Psl, not Pel, is the primary matrix polysaccharide of P. aeruginosa PAO1 biofilms (13, 14). Moreover, dispersion frequently results in shell-like structures or voids left behind, suggesting that some of the Psl located in the periphery of biofilms (13, 14) will have to be removed. Since both endogenously produced (54) and purified PslG (41) have been demonstrated to hydrolyze surface-associated Psl, we hypothesized that Psl hydrolase activity may be somewhat hampered due to structural limitation. Because Psl has been described to be tethered by the matrix-stabilizing protein CdrA (12), we next sought to determine whether Psl being tethered impedes Psl degradation and thus dispersion.
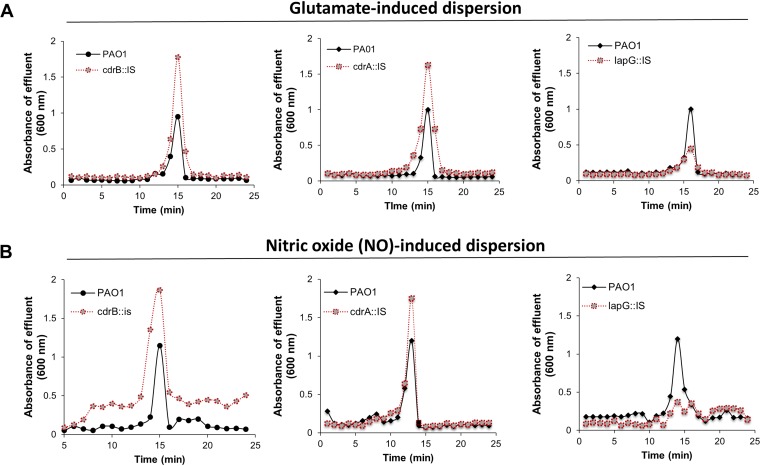
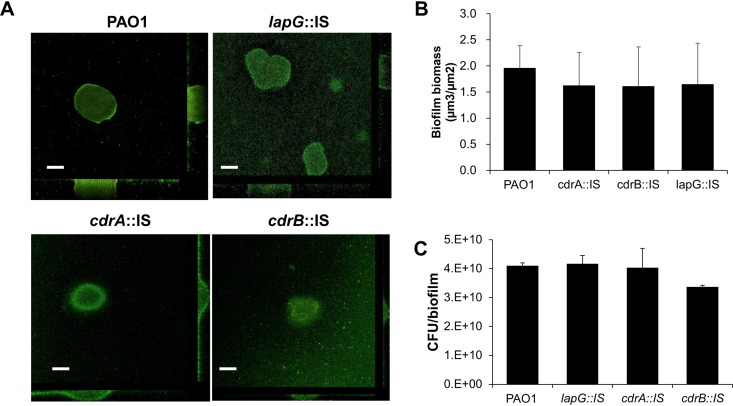
To address this question, we first evaluated the dispersion phenotype of P. aeruginosa mutants in which either CdrA remains affixed to the outer membrane (CdrA is not cleaved in a lapG mutant) or CdrA is lacking (by using a cdrA mutant strain). We also analyzed the role of cdrB inactivation, since cdrB encodes an outer membrane transporter responsible for transporting CdrA across the outer membrane (12). We used mutant strains harboring transposon insertions in lapG, cdrA, and cdrB. The respective mutant strains are referred to as lapG::IS, cdrA::IS, and cdrB::IS. Biofilms of the respective mutant strains were grown for 5 days under flowing conditions in tube reactors after which time biofilms were exposed to glutamate or nitric oxide to induce dispersion. Effluents from the biofilm reactor were collected for 20 min after induction of dispersion, and the absorbance was determined at 600 nm. Under the conditions tested, insertional inactivation of cdrA and cdrB significantly enhanced the dispersion response to glutamate relative to wild-type biofilms (Fig. 4A). This was apparent by the absorbance of the effluents within ∼15 min upon induction of dispersion being consistently higher than the absorbance noted for effluents from wild-type biofilms. Similar results were obtained when dispersion was induced by the addition of nitric oxide (Fig. 4B). This is in contrast to insertional inactivation of lapG, which has previously been demonstrated to result in CdrA remaining tethered to the outer membrane (12, 19). Biofilms by lapG::IS failed to disperse in response to nitric oxide (Fig. 4B) and demonstrated significantly reduced dispersion responses (Fig. 4A). It is interesting that insertional inactivation had no effect on the overall biofilm architecture, as determined by confocal microscopy (Fig. 5A). Subsequent COMSTAT analysis furthermore confirmed that insertional inactivation of the genes of interest had no significant effect on the overall biofilm biomass (Fig. 5B). This was confirmed by the CFU counts of biofilms grown for 5 days under the conditions tested (Fig. 5C).
FIG 4.
Representative dispersion responses of biofilms by P. aeruginosa PAO1, lapG::IS, cdrA::IS, and cdrB::IS strains. Biofilms were grown for 5 days in 5-fold-diluted VBMM in tube reactors. The absorbance of biofilm tube reactor effluents after the induction of dispersion is shown. (A) Absorbance of biofilm effluents after the induction of dispersion by glutamate. (B) Absorbance of biofilm effluents after the induction of dispersion by nitric oxide (NO). Sodium nitroprusside served as a source of nitric oxide. Dispersion assays were performed at least three times, with each biological replicate consisting of four technical replicates, but only representative dispersion profiles are shown.
FIG 5.
Insertional inactivation of lapG, cdrA, and cdrB does not affect the biofilm architecture. (A) Representative confocal images of 5-day-old biofilms by PAO1, lapG::IS, cdrA::IS, and cdrB::IS strains. Scale bars, 50 μm. (B) Biofilm biomass, as determined using COMSTAT analysis, by PAO1 and the respective lapG::IS, cdrA::IS, and cdrB::IS mutant strains. Experiments were performed in triplicate, with each biological replicate consisting of four technical replicates. (C) Biofilm biomass, as determined by using the CFU count. Biofilms were grown for 5 days in 5-fold-diluted VBMM in tube reactors. Biofilms were harvested, homogenized, serially diluted, and spread plated onto LB agar. Experiments were performed in triplicate, with each biological replicate consisting of two technical replicates. Error bars represent the standard deviations.
Our data indicate that the inactivation of lapG coincided with impaired or reduced dispersion, suggesting that cell-associated CdrA impedes or reduces the dispersion response, whereas the release of CdrA from the surface enhances dispersion.
Dispersion requires untethering of the Psl polysaccharide.
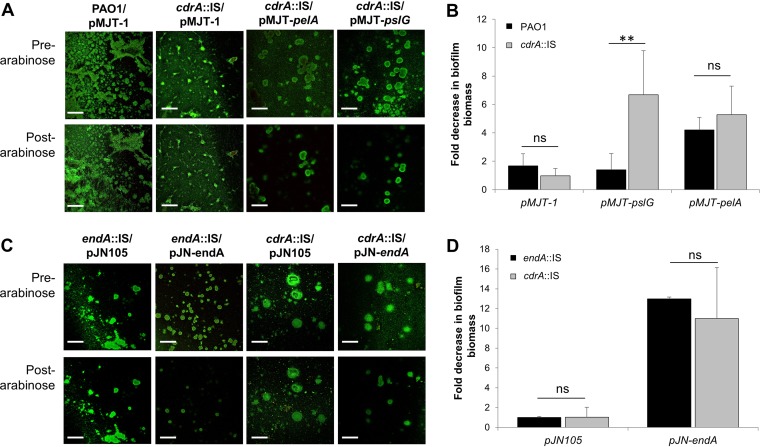
The finding of CdrA, when bound to the outer membrane, impeding dispersion suggested the possibility that dispersion is enhanced by the release of CdrA from the surface, which is likely to untether the matrix as a first step to enable Psl polysaccharide degradation. If so, we reasoned that we can induce dispersion upon induction of pslG only when CdrA is absent and thus incapable of tethering Psl to the outer membrane. To test our hypothesis, we generated pslG-inducible strains in a wild-type and cdrA mutant background. Strains harboring the empty pMJT-1 vector or pelA under the control of the arabinose-inducible PBAD promoter were used as controls. The respective strains were grown in flow cells for 5 days in the absence of arabinose to ensure the establishment of biofilms (Fig. 6A). Arabinose was then added to the growth medium for 60 min to induce the transcription of pslG or pelA. Prior to and after arabinose supplementation, the biofilm architecture was monitored by confocal microscopy (Fig. 6A). No difference in the biofilm architecture or biofilm biomass was noted for PAO1/pMJT-1 prior to and after the addition of arabinose, whereas the induction of pelA gene expression resulted in a reduction in the biofilm biomass (Fig. 6A). Quantitative analysis of the biofilm architecture prior to and after the addition of arabinose suggested that the induction of pelA gene expression correlates with a 4-fold reduction in the biofilm biomass in a PAO1 background (Fig. 6B). A similar reduction was noted when pelA was expressed in a cdrA mutant background (Fig. 6B), indicating that pelA expression induces dispersion regardless of the absence or presence of CdrA. While biofilms by PAO1/pMJT-pslG failed to disperse (Fig. 3A and C and Fig. 6B), induction of pslG gene expression in a cdrA mutant background coincided with the dispersal of the mutant biofilm architecture (Fig. 6A). Quantitative analysis of the cdrA::IS/pMJT-pslG biofilm architecture prior to and after the addition of arabinose indicated that induction of pslG expression correlates with a 6-fold reduction in the biofilm biomass (Fig. 6B). The quantitative analysis further indicated that dispersion induced by pslG exceeded the dispersion response due to pelA expression (Fig. 6B).
FIG 6.
Lack of CdrA enhances dispersion upon induction of pslG gene expression. (A) Representative confocal images of 5-day-old biofilms by PAO1 and cdrA::IS strains harboring the empty vector pMJT-1 or expressing pelA or pslG. Images were acquired prior to and after treatment with arabinose to induce pelA or pslG gene expression. Scale bars, 100 μm. (B) Fold change in the biofilm biomass after the induction of dispersion by the addition of arabinose. Biofilm biomass prior to and after arabinose addition was quantitated by using COMSTAT. Experiments were performed in triplicate, with each biological replicate consisting of four technical replicates. **, significantly different (P < 0.05) relative to biofilms by PAO1 tested under identical conditions. ns, not significant. (C) Representative confocal images of 5-day-old biofilms by endA::IS and cdrA::IS strains harboring the empty vector pJN105 or expressing endA. Images were acquired prior to and after treatment with arabinose to induce endA gene expression. Scale bars, 100 μm. (D) Fold change in the biofilm biomass after the induction of dispersion by the addition of arabinose. The biofilm biomass prior to and after arabinose addition was quantitated using COMSTAT. Experiments were performed in triplicate, with each biological replicate consisting of four technical replicates. ns, not significant.
To further ensure that pslG-induced dispersion was CdrA specific, we determined the effect of endA encoding an endonuclease capable of degrading eDNA present in the biofilm matrix (32) on dispersion. Induction of endA gene expression resulted in dispersion when expressed in an endA or cdrA mutant background (Fig. 6C). However, no significant difference in the extents to which endA gene expression reduced the biofilm biomass was noted (Fig. 6D).
Our findings support the notion of CdrA impeding or reducing the dispersion response, likely by preventing efficient degradation of Psl after the induction of pslG expression. The findings furthermore suggest that dispersion requires untethering of CdrA-bound Psl prior to Psl degradation.
Reduced c-di-GMP levels are not sufficient in enabling pslG-dependent dispersion.
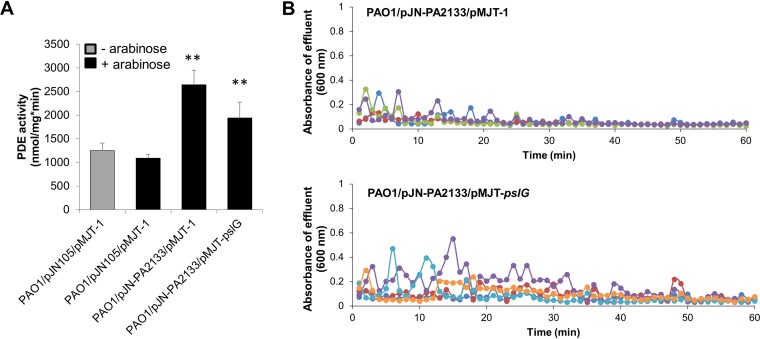
Previous findings indicated that a dispersion response by P. putida and P. fluorescens biofilms is linked to the release of the large adhesin LapA from the cell surface via cleavage by protease LapG, with the release being linked to the reduction in cellular c-di-GMP levels upon induction of dispersion (33). Similar to LapA, CdrA is a c-di-GMP regulated adhesin (11, 12) that is cleaved from the cell surface by LapG at low c-di-GMP levels. To determine whether untethering of CdrA-bound Psl to enable pslG-dependent dispersion is c-di-GMP dependent, we next sought to determine whether pslG-dependent dispersion can be enabled at a low c-di-GMP level. We previously demonstrated that P. aeruginosa PAO1 expressing PA2133 harbors significantly reduced cellular c-di-GMP levels compared to strains harboring an empty vector (55), whereas Hickman et al. demonstrated that overexpression of PA2133 significantly inhibited biofilm formation by PAO1 (56). We therefore used PA2133 and generated strains PAO1/pJN-PA2133/pMJT-1 and PAO1/pJN-PA2133/pMJT-pslG to enable the reduction of c-di-GMP levels via the expression of PA2133 encoding a phosphodiesterase in the absence or presence of inducible pslG expression. To ensure that under the conditions tested, PA2133 indeed affects cellular c-di-GMP levels, we determined the overall phosphodiesterase activity using total cell extracts obtained from PAO1/pJN-PA2133/pMJT-pslG, PAO1/pJN-PA2133/pMJT-1, and PAO1/pJN105/pMJT-1. No significant difference in phosphodiesterase activity was noted in cell extracts by PAO1/pJN105/pMJT-1 prior to and after the induction of gene expression by arabinose (Fig. 7A). Relative to the total extracts obtained from PAO1/pJN105/pMJT-1, we detected significantly increased phosphodiesterase activity in cell extracts by PAO1/pJN-PA2133/pMJT-pslG and PAO1/pJN-PA2133/pMJT-1 after induction by arabinose for 1 h (Fig. 7A).
FIG 7.
Dispersion profile upon induction of PA2133 and/or pslG gene expression. (A) Specific phosphodiesterase (PDE) activity. The indicated strains were grown planktonically to exponential phase in VBMM and subsequently left untreated (– arabinose) or exposed to 1% arabinose for 1 h to induce gene expression (+ arabinose). Phosphodiesterase activity assays were carried out using the chromogenic substrate bis-pNPP and 100 μg of total cell extracts. Total cell extracts obtained from PAO1 harboring the empty vectors pMJT1 and pJN105 (with or without arabinose) acted as a control. Error bars represent the standard deviations. **, significantly different (P < 0.05) relative to the vector control in the presence of arabinose. (B) Absorbance of biofilm tube reactor effluents after the induction of PA2133 and/or pslG gene expression. Induction of gene expression was accomplished by adding 1% arabinose to the growth medium. Graphs shown are representative of four independent biofilm replicates. Different colors represent biological replicates.
To determine whether pslG-dependent dispersion can be enabled at low c-di-GMP, the respective strains were grown for 5 days in tube reactors in the absence of arabinose to ensure the establishment of biofilms, and then arabinose was added to the growth medium for a period of 60 min to induce the transcription of PA2133 and/or pslG. In agreement with previous findings (55), no dispersion events were noted upon lowering c-di-GMP levels by induction of PA2133 expression (Fig. 7B). Likewise, no dispersion events were noted when the expression of PA2133 and that of pslG were induced simultaneously (Fig. 7B). However, although we failed to note dispersion events as for PAO1/pMJT-pelA biofilms (Fig. 3C and D), we noticed an increase in the overall increase in the absorbance of biofilm effluents (Fig. 7B).
DISCUSSION
Biofilm dispersion has been characterized by an overall erosion of the biofilm's biomass, sometimes resulting in central void formation (21–23). The overall change in the biofilm architecture is due to cells actively evacuating the matrix-enmeshed biofilm structure. Thus, it is not surprising that dispersion is thought to involve active matrix degradation. However, with the exception of eDNA degradation (32), evidence supporting degradation of the matrix polysaccharides Pel and Psl leading to the dispersal of P. aeruginosa biofilms stems from exogenously added matrix-degrading factors (30, 41, 42, 51, 57). We therefore sought to determine in this study whether endogenously induced degradation of the matrix polysaccharides Pel and Psl contributes to the dispersion response. Our findings indicate that induction of pelA and pslG gene expression coincides with dispersion, suggesting matrix polysaccharide degradation is an essential step in the dispersion by P. aeruginosa biofilms. Our findings are in agreement with observations by Ma et al. (14) demonstrating that in dispersing P. aeruginosa biofilms, Psl-matrix-enmeshed bacteria are present in the periphery of microcolonies surrounding an interior area with swimming cells and little Psl.
However, these findings also raise the question of how the activity of the two glycoside hydrolases, PelA and PslG, is modulated in vivo to enable dispersion. For one, in vivo, pelA and pslG are not differentially expressed relative to the remaining genes that comprise the pel and psl operons. Second, PelA and PslG are bifunctional enzymes that contribute not only to the synthesis of their respective exopolysaccharides but also to their degradation (39–41). What contributes to PelA and PslG switching from synthesis to degradation of exopolysaccharides? Previous findings indicated that PelA directly interacts with PelB, with the interaction positively affecting the deacetylase activity of PelA (39, 50). In contrast, however, the interaction between PelA and PelB was found to decrease the hydrolase activity of PelA (39, 50). It is interesting that these protein interactions were observed in developing, 24- and 48-h-old P. aeruginosa biofilms, suggesting that the presence of the PelAB complex corresponds with matrix assembly. It is thus possible that the PelAB complex starts to dissociate as biofilms mature and matrix productions decreases, with complex dissociation enhancing PelA’s glycoside hydrolase activity and ultimately, Pel degradation. Considering that both PelA and PslG were detected in culture supernatants (Fig. 2C), it is likewise possible that increased glycoside hydrolase activity is due to translocation (actively or passively, through cell lysis) to the extracellular space. Support for PelA and PslG likely being present in culture supernatant stems from observations by Qin et al. (58) indicating the presence of extracellular products in P. aeruginosa PAO1 supernatant cultures capable of disrupting established Staphylococcus epidermidis biofilms, whereas cellulase- or heat-treated P. aeruginosa supernatant and supernatant from ΔpelA and ΔpelA ΔpslBCD mutants diminished the disruption of S. epidermidis biofilms. Although that study did not identify the extracellular products contributing to biofilm disruption, the authors suggested a role of exopolysaccharides or related factors (58). Likewise, Li et al. (26) noted increased Psl degradative activities in culture supernatants of P. aeruginosa dispersed cells. Future investigations are needed to determine the mechanism contributing to the switch in activity.
Regardless of the mechanisms, our findings suggested Psl-induced dispersion to be dependent on CdrA. This is supported by the finding that the inactivation of lapG coincided with reduced dispersion, suggesting that cell-associated CdrA impedes or reduces the dispersion response, whereas the release of CdrA from the surface enhances dispersion. This was further confirmed by the inactivation of cdrA or cdrB enhancing the dispersion response. The findings suggested that the presence of cell-associated CdrA impedes or reduces the dispersion response, while release of CdrA from the surface enhances dispersion. A similar reciprocal relationship was noted by Rybtke et al. (11) for attachment by P. aeruginosa, with lapG mutants demonstrating increased attachment, while cdrA mutants demonstrated reduced attachment relative to the parental strain. Likewise, in P. putida, inactivation of lapA encoding a cell-associated adhesin coincided with decreased surface adhesion (33).
The finding of CdrA impeding dispersion, however, has additional implications. This is because our data suggested that the Psl polysaccharide likely needs to be untethered to enable PslG to effectively degrade the Psl polysaccharide. This was supported by biofilms by cdrA::IS/pMJT-pslG dispersing upon induction of pslG expression, while biofilms by PAO1/pMJT-pslG failed to do so (Fig. 6). The interaction between CdrA and Psl imparting protection from degradation has recently been shown for CdrA, with CdrA bound to Psl being protected from proteolysis to both exogenous and self-produced proteases (19). Moreover, lack of CdrA enhancing dispersion further suggests that degradation of the polysaccharide matrix to induce dispersion is probably an orchestrated two-stage process, requiring first untethering, likely by the release of CdrA from the surface, followed by Psl degradation to enable the egress of cells from the biofilm.
Untethering may be linked to the modulation of c-di-GMP, since dispersion, as well as the release of surface-associated CdrA, has been linked to cleavage by LapG at low c-di-GMP levels. In P. putida, lowering the intracellular c-di-GMP level via induction of a phosphodiesterase led to dispersal of wild-type biofilms but failed to disperse lapG mutant biofilms, indicating that LapG exerts its activity on LapA in response to a decrease in the intracellular c-di-GMP level (33). However, overexpression of the phosphodiesterase PA2133 to reduce cellular c-di-GMP levels was not sufficient to induce dispersion, even when combined with the overexpression of PslG (Fig. 7). It is thus likely that factors targeting CdrA other than LapG may contribute to the release of CdrA from the surface to enable dispersion. This is in agreement with findings by Reichhardt et al. (19) demonstrating that CdrA is sensitive to self-produced proteases other than LapG. Future work will focus on elucidating the protease contributing to CdrA release during the dispersion response. Moreover, our findings imply that while dispersion has been linked to the modulation of c-di-GMP and dispersed cells are characterized by low levels of c-di-GMP, dispersion by P. aeruginosa biofilms and, more specifically, matrix degradation require both c-di-GMP-dependent and -independent processes. This is supported by dispersion being induced upon overproduction of the endonuclease 1 EndA (32) and PelA (Fig. 3), with both endA and pelA being directly regulated by AmrZ, the central regulator of biofilm formation by P. aeruginosa (59–61).
Collectively, our data indicate that dispersion requires gene functions that target specific matrix components for degradation. In addition to eDNA degradation, dispersion is linked to the degradation of the matrix polysaccharides Pel and Psl. Moreover, given the requirement of CdrA release, matrix degradation is likely a coordinated process involving several degradative enzymes that work in concert.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
All bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa strain PAO1 was used as the parental strain. All planktonic cultures were grown in flasks at 220 rpm at 37°C using Luria-Bertani (LB) medium. For plasmid maintenance, antibiotics were used at the following concentrations: 250 μg/ml carbenicillin and 50 μg/ml gentamicin for P. aeruginosa and 50 μg/ml ampicillin for E. coli. Unless indicated otherwise, arabinose was used at 1% to induce gene expression.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | F– ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK– mK+) phoA supE44 thi-1 gyrA96 relA1 tonA | Life Technologies |
| P. aeruginosa | ||
| PAO1 | Wild-type strain | B. H. Holloway |
| PAO1::PBAD-pel | Chromosomal replacement of native promoter with araC-PBAD promoter, arabinose-inducible expression of pel genes | 66 |
| PAO1::PBAD-psl | Chromosomal replacement of native promoter with araC-PBAD promoter, arabinose-inducible expression of psl genes | 67 |
| endA::IS | PAO1 PA2749::ISlacZ Tetr | 68 |
| cdrA::IS | PAO1 PA4625::ISlacZ Tetr | 68 |
| cdrB::IS | PAO1 PA4624::ISphoA Tetr | 68 |
| lapG::IS | PAO1 PA1434::ISlacZ Tetr | 68 |
| Plasmids | ||
| pJN105 | Arabinose-inducible gene expression vector; pBRR-1 MCS; araC-PBAD Gmr | 69 |
| pJN-endA-V5 | Arabinose-inducible expression of C-terminal V5/6×His-tagged endA (PA2749) cloned in pJN105, Gmr | 32 |
| pJN-PA2133 | Arabinose-inducible expression of PA2133 cloned in pJN105, Gmr | 56 |
| pMJT-1 | Arabinose-inducible gene expression vector; pBRR-1 MCS; araC-PBAD Gmr | 70 |
| pMJT-pslG-V5 | C-terminal V5-tagged PA3910 cloned into pMJT-1 XbaI and SacI; araC-PBAD Carbr | This study |
| pMJT-pelA-V5 | C-terminal V5-tagged PA3909 cloned into pMJT-1 XbaI and SacI; araC-PBAD Carbr | This study |
Gmr, gentamicin resistance; Tetr, tetracycline resistance; Carbr, carbenicillin resistance.
Strain construction.
C-terminal V5 tagging of PelA and PslG was accomplished by amplifying pelA and pslG with primers harboring the sequence for V5 (Table 2). The tagged constructs were cloned into pMJT-1. The identity of all vector inserts was confirmed by PCR and sequencing. Plasmids were introduced into P. aeruginosa via electroporation or conjugation for pMJT-1 and pJN105, respectively. In addition, transposon insertional inactivation of cdrA, cdrB, and lapG was confirmed by PCR and sequencing. The primers used for strain construction and confirmation are listed in Table 2.
TABLE 2.
Primers used in this study
| Purpose and oligonucleotide | Sequence (5′–3′)a |
|---|---|
| Transposon insertional inactivation check | |
| lapG_FOR | TGGGACCTGGAGTCGATACT |
| lapG_REV | GCATCTTGGTCAGCAGGTC |
| cdrA_FOR | TCAACTGGAAGGGCTTCGAC |
| cdrA_REV | TCGTTGGTAGGGAAACTGGC |
| cdrB_FOR | CTGTACAGCCATCCCAGCTC |
| cdrB_REV | GAGGTTGTAGGTCTTGCCCC |
| Cloning | |
| pMJT1 MCS _f | GACCGCGAATGGTGAG |
| pMJT1 MCS _r | GAGCTGATACCGCTCG |
| Cloning into pMJT-1 for C-terminal V5-tagging | |
| pslG_For_NheI | gctagcATGGCACGTAAGGGACTCTAT |
| pslG_REV_SacI V5 | GCGCGCGCgagctcTCAcgtagaatcgagaccgaggagagggttagggataggcttaccCTCCCAGACCAGCATCTG |
| pelA_For_NheI | gctagcATGCGGTTCAGCAAGAAAGGA |
| pelA_REV_SacI _V5 | GCGCGCGCgagctcTCAcgtagaatcgagaccgaggagagggttagggataggcttaccGCAGACGAGTTGGCC |
Restriction sites are indicated in lowercase letters; the sequence of the V5 tag is underlined.
Biofilm growth.
Biofilms were grown for 5 days under continuous flow conditions in biofilm tube reactors or flow cells. The flow rate was 0.2 ml/min using 5-fold diluted Vogel-Bonner minimal medium (VBMM). For plasmid maintenance, 10 μg/ml carbenicillin and 2 μg/ml gentamicin were added. Where indicated, the growth medium was supplemented with 0.1% arabinose to induce expression of genes of interest. Flow cell-grown biofilms were stained using a LIVE/DEAD BacLight viability stain kit (Invitrogen, Carlsbad, CA) to visualize the biofilms. The biofilm architecture was visualized via confocal laser scanning microscopy (CLSM) using a Leica TCS SP5 confocal microscope. The CLSM images were processed using LAS AF software. Quantitative analysis of biofilm architecture was accomplished using MATLAB with the COMSTAT software package (62).
Biofilm biomass determination.
To determine biofilm biomass accumulation over the course of biofilm formation, biofilms were harvested, homogenized, serially diluted, and spread plated onto LB agar. The biofilm biomass was determined via CFU counts. The biofilm biomass was determined daily for up to 5 days.
Biofilm dispersion.
Dispersion assays were performed using biofilms grown for 5 days in flow cells or tube reactors. Image acquisition using flow cell-grown biofilms was done so that the same biofilm microcolonies were observed prior to and after the addition of 1% arabinose. To quantify the amount of biofilm biomass pre- and postdispersion, confocal images were acquired before and after the addition of arabinose. Each image was subsequently analyzed for the relative fluorescence intensity indicative of biofilm biomass using Intensity Luminance V1 software (63). For tube reactor-grown biofilms, dispersion was induced by the sudden addition of l-glutamate (18 mM) or sodium nitroprusside (500 μM) to the growth medium, as previously described (44, 47). Sodium nitroprusside was used as a source of nitric oxide (NO). In addition, biofilms were exposed to 1% arabinose to induce endA, pslG, pelA, or PA2133 gene expression to determine whether induction of gene expression resulted in dispersion events. Regardless of the dispersion cue used, dispersed cells were collected from the tube reactor effluents at 1-min intervals for a total of 24 or 60 min using 96-well microtiter plates. The absorbance of the biofilm effluents was assessed by spectrophotometry at 600 nm. Dispersion events were characterized by an increase in the effluent optical density with the optical density being at least two times greater than the baseline.
Immunoblot analysis.
Confirmation of V5-tagged PslG and PelA production was assessed by SDS-PAGE and immunoblot analysis. For confirmation of PslG and PelA production in Escherichia coli, strains DH5α/pMJT-pelA-V5 and DH5α/pMJT-pslG-V5 were grown planktonically in LB medium containing ampicillin for plasmid maintenance to exponential phase. pelA and pslG gene expression was then induced by arabinose for 2 h. Empty vector pMJT-1 was used as a control. For confirmation of PslG and PelA production in P. aeruginosa PAO1, strains PAO1/pMJT-pelA and PAO1/pMJT-pslG were grown as biofilms for 5 days under continuous-flow conditions using 5-fold-diluted VBMM, as described above. pelA and pslG gene expression was then induced by arabinose for 1 h. Strains harboring the empty vector pMJT-1 were used as a control. Cells were subsequently harvested by centrifugation for 5 min at 16,000 × g and resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) containing 0.3 mg of phenylmethanesulfonyl fluoride; the cells were then sonicated on ice with six 10-s bursts at 4 W, followed by centrifugation for 5 min at 21,200 × g to remove cell debris and unbroken cells. To determine whether PelA and PslG are detectable in culture supernatants, strains PAO1/pMJT-pelA and PAO1/pMJT-pslG were grown planktonically in LB medium to exponential phase and subsequently exposed to arabinose for 1 h. The culture supernatant was collected and concentrated using Vivaspin 20 columns (Sartorius Stedim Biotech, Gottingen, Germany) according to the manufacturer’s protocol. The protein concentrations were determined by using a modified Lowry assay (Thermo Scientific, Waltham, MA) and bovine serum albumin as a standard. The samples (10 or 20 μg) were resolved on an 11% polyacrylamide gel and subsequently transferred onto a polyvinylidene difluoride membrane using a TurboTransblot apparatus (Bio-Rad, Hercules, CA). Western blots were first probed with anti-V5, followed by a secondary anti-mouse IgG antibody (Cell Signaling Technologies, Danvers, MA). The blots were subsequently developed using Immun-Star WesternC chemiluminescence reagents (Bio-Rad, Hercules, CA). After transfer, SDS-PAGE gels were Coomassie stained to ensure equal loading.
Phosphodiesterase activity assay.
Phosphodiesterase activity was determined using the synthetic chromogenic substrate bis(p-nitrophenyl) phosphate (bis-pNPP; Sigma-Aldrich) essentially as previously described (64, 65). Total cell extracts were obtained from PAO1/pJN-PA2133/pMJT-1 and PAO1/pJN-PA2133/pMJT-pslG which were grown to exponential phase in 1× VBMM, prior to and after the addition of arabinose for 1 h. Total cell extracts obtained from PAO1/pJN105/pMJT-1 cells prior to and after the addition of arabinose for 1 h were used as controls. Briefly, total cell extracts (100 μg) were added to reaction buffer consisting of 50 mM Tris (pH 9.1), 50 mM NaCl, and 50 mM MgCl2. Reaction mixtures were incubated at 25°C for 240 min. The specific phosphodiesterase activity was determined by measuring the release of p-nitrophenol (pNP) at 405 nm. An extinction coefficient for p-nitrophenol of 1.78 × 104/(M × cm) was used. Controls without extracts were included to account for any nonenzymatic bis-pNPP hydrolysis.
Statistical analysis.
All experiments were carried out at least in triplicate. A Student t test was performed for pairwise comparisons of groups, and multivariant analyses were performed a one-way analysis of variance, followed by Tukey’s test a posteriori to compare the means of all treatment groups. All statistical analyses were performed using the Prism5 software (GraphPad, La Jolla, CA).
REFERENCES
- 1.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 2.Mishra M, Byrd MS, Sergeant S, Azad AK, Parsek MR, McPhail L, Schlesinger LS, Wozniak DJ. 2012. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol 14:95–106. doi: 10.1111/j.1462-5822.2011.01704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 4.Lewis K. 2008. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol 322:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- 5.Spoering AL, Lewis K. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol 183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancio LH, Howard PA, McManus AT, Kim SH, Goodwin CW, Pruitt BA Jr. 2001. Burn wound infections, p 671–683. In Holzheimer RG, Mannick JA (ed). Surgical treatment: evidence-based and problem-oriented. Zuckschwerdt, Munich, Germany. [PubMed] [Google Scholar]
- 7.Wolcott R, Dowd S. 2011. The role of biofilms: are we hitting the right target? Plastic Reconstruct Surg 127:28S–35S. doi: 10.1097/PRS.0b013e3181fca244. [DOI] [PubMed] [Google Scholar]
- 8.Flemming H-C. 2016. EPS: then and now. Microorganisms 4:41. doi: 10.3390/microorganisms4040041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flemming H-C, Neu TR, Wozniak DJ. 2007. The EPS matrix: the “house of biofilm cells. J Bacteriol 189:7945–7947. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryder C, Byrd M, Wozniak DJ. 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol 10:644–648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rybtke M, Berthelsen J, Yang L, Høiby N, Givskov M, Tolker-Nielsen T. 2015. The LapG protein plays a role in Pseudomonas aeruginosa biofilm formation by controlling the presence of the CdrA adhesin on the cell surface. Microbiologyopen 4:917. doi: 10.1002/mbo3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I, Secor PR, Tseng BS, Scian M, Filloux A, Wozniak DJ, Howell PL, Parsek MR. 2015. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci U S A 112:11353–11358. doi: 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog 5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrova OE, Schurr JR, Schurr MJ, Sauer K. 2011. The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol Microbiol 81:767–783. doi: 10.1111/j.1365-2958.2011.07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand JJ, Klausen M, Givskov M, Whitchurch CB, Engel JN, Tolker-Nielsen T. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol 10:2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 17.Cooley RB, Smith TJ, Leung W, Tierney V, Borlee BR, O’Toole GA, Sondermann H. 2016. Cyclic di-GMP-regulated periplasmic proteolysis of a Pseudomonas aeruginosa type Vb secretion system substrate. J Bacteriology 198:66–76. doi: 10.1128/JB.00369-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Passos da Silva D, Matwichuk ML, Townsend DO, Reichhardt C, Lamba D, Wozniak DJ, Parsek MR. 2019. The Pseudomonas aeruginosa lectin LecB binds to the exopolysaccharide Psl and stabilizes the biofilm matrix. Nat Commun 10:2183. doi: 10.1038/s41467-019-10201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reichhardt C, Wong C, Passos da Silva D, Wozniak DJ, Parsek MR. 2018. CdrA interactions within the Pseudomonas aeruginosa biofilm matrix safeguard it from proteolysis and promote cellular packing. mBio 9:e01376-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 21.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies DG, Marques C. 2009. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol 191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauer K, Cullen MC, Rickard AH, Zeef LAH, Davies DG, Gilbert P. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol 186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrova OE, Sauer K. 2016. Escaping the biofilm in more than one way: desorption, detachment or dispersion. Curr Opin Microbiol 30:67–78. doi: 10.1016/j.mib.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrova OE, Sauer K. 2012. PAS domain residues and prosthetic group involved in BdlA-dependent dispersion response by Pseudomonas aeruginosa biofilms. J Bacteriol 194:5817–5828. doi: 10.1128/JB.00780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Petrova OE, Su S, Lau GW, Panmanee W, Na R, Hassett DJ, Davies DG, Sauer K. 2014. BdlA, DipA and induced dispersion contribute to acute virulence and chronic persistence of Pseudomonas aeruginosa. PLoS Pathog 10:e1004168. doi: 10.1371/journal.ppat.1004168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan JB. 2010. Biofilm dispersal: mechanisms, clinical Implications, and potential therapeutic uses. J Dent Res 89:205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chua SL, Liu Y, Yam JKH, Chen Y, Vejborg RM, Tan BGC, Kjelleberg S, Tolker-Nielsen T, Givskov M, Yang L. 2014. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyle. Nat Commun 5:4462. doi: 10.1038/ncomms5462. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan JB, Ragunath C, Ramasubbu N, Fine DH. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. J Bacteriol 185:4693–4698. doi: 10.1128/jb.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu S, Su T, Wu H, Liu S, Wang D, Zhao T, Jin Z, Du W, Zhu M-J, Chua SL, Yang L, Zhu D, Gu L, Ma LZ. 2015. PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res 25:1352–1367. doi: 10.1038/cr.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleming D, Rumbaugh K. 2017. Approaches to dispersing medical biofilms. Microorganisms 5:15. doi: 10.3390/microorganisms5020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cherny KE, Sauer K. 2019. Pseudomonas aeruginosa requires the DNA-specific endonuclease EndA to degrade eDNA to disperse from the biofilm. J Bacteriol 201:e00059-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. 2010. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol 75:815–826. doi: 10.1111/j.1365-2958.2009.06793.x. [DOI] [PubMed] [Google Scholar]
- 34.Hinsa SM, Espinosa-Urgel M, Ramos JL, O’Toole GA. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol 49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 35.Newell PD, Boyd CD, Sondermann H, O’Toole GA. 2011. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol 9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monds RD, Newell PD, Gross RH, O’Toole GA. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol Microbiol 63:656–679. doi: 10.1111/j.1365-2958.2006.05539.x. [DOI] [PubMed] [Google Scholar]
- 37.Gjermansen M, Ragas P, Sternberg C, Molin S, Tolker-Nielsen T. 2005. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ Microbiol 7:894–904. doi: 10.1111/j.1462-2920.2005.00775.x. [DOI] [PubMed] [Google Scholar]
- 38.Naumoff D. 2011. Hierarchical classification of glycoside hydrolases. Biochemistry (Mosc) 76:622–635. doi: 10.1134/S0006297911060022. [DOI] [PubMed] [Google Scholar]
- 39.Colvin KM, Alnabelseya N, Baker P, Whitney JC, Howell PL, Parsek MR. 2013. PelA deacetylase activity is required for Pel polysaccharide synthesis in Pseudomonas aeruginosa. J Bacteriol 195:2329–2339. doi: 10.1128/JB.02150-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Byrd MS, Sadovskaya I, Vinogradov E, Lu H, Sprinkle AB, Richardson SH, Ma L, Ralston B, Parsek MR, Anderson EM, Lam JS, Wozniak DJ. 2009. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol 73:622–638. doi: 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker P, Hill PJ, Snarr BD, Alnabelseya N, Pestrak MJ, Lee MJ, Jennings LK, Tam J, Melnyk RA, Parsek MR, Sheppard DC, Wozniak DJ, Howell PL. 2016. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci Adv 2:e1501632. doi: 10.1126/sciadv.1501632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fleming D, Chahin L, Rumbaugh K. 2017. Glycoside hydrolases degrade polymicrobial bacterial biofilms in wounds. Antimicrob Agents Chemother 61:e01998-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marques CN, Davies DG, Sauer K. 2015. Control of biofilms with the fatty acid signaling molecule cis-2-decenoic acid. Pharmaceuticals (Basel) 8:816–835. doi: 10.3390/ph8040816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgan R, Kohn S, Hwang S-H, Hassett DJ, Sauer K. 2006. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol 188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basu Roy A, Sauer K. 2014. Diguanylate cyclase NicD-based signaling mechanism of nutrient-induced dispersion by Pseudomonas aeruginosa. Mol Microbiol 94:771–793. doi: 10.1111/mmi.12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barraud N, Kelso MJ, Rice SA, Kjelleberg S. 2015. Nitric oxide: a key mediator of biofilm dispersal with applications in infectious diseases. Curr Pharm Des 21:31–42. doi: 10.2174/1381612820666140905112822. [DOI] [PubMed] [Google Scholar]
- 47.Barraud N, Hassett DJ, Hwang S-H, Rice SA, Kjelleberg S, Webb JS. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol 188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Heine S, Entian M, Sauer K, Frankenberg-Dinkel N. 2013. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by a MHYT-domain coupled phosphodiesterase. J Bacteriol 195:3531–3542. doi: 10.1128/JB.01156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petrova OE, Sauer K. 2012. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc Natl Acad Sci U S A 109:16690–16695. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marmont LS, Whitfield GB, Rich JD, Yip P, Giesbrecht LB, Stremick CA, Whitney JC, Parsek MR, Harrison JJ, Howell PL. 2017. PelA and PelB proteins form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa. J Biol Chem 292:19411–19422. doi: 10.1074/jbc.M117.812842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pestrak MJ, Baker P, Dellos-Nolan S, Hill PJ, Passos da Silva D, Silver H, Lacdao I, Raju D, Parsek MR, Wozniak DJ, Howell PL. 2019. Treatment with the Pseudomonas aeruginosa glycoside hydrolase PslG combats wound infection by improving antibiotic efficacy and host innate immune activity. Antimicrob Agents Chemother 63:e00234-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franklin MJ, Nivens DE, Weadge JT, Howell PL. 2011. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol 2:167. doi: 10.3389/fmicb.2011.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baker P, Whitfield GB, Hill PJ, Little DJ, Pestrak MJ, Robinson H, Wozniak DJ, Howell PL. 2015. Characterization of the Pseudomonas aeruginosa glycoside hydrolase PslG reveals that its levels are critical for Psl polysaccharide biosynthesis and biofilm formation. J Biol Chem 290:28374–28387. doi: 10.1074/jbc.M115.674929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu H, Wang D, Tang M, Ma LZ. 2019. The advance of assembly of exopolysaccharide Psl biosynthesis machinery in Pseudomonas aeruginosa. Microbiologyopen 8:e857. doi: 10.1002/mbo3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chambers JR, Cherny KE, Sauer K. 2017. Susceptibility of Pseudomonas aeruginosa dispersed cells to antimicrobial agents is dependent on the dispersion cue and class of the antimicrobial agent used. Antimicrob Agents Chemother 61:e00846-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fleming D, Rumbaugh K. 2018. The consequences of biofilm dispersal on the host. Sci Rep 8:10738. doi: 10.1038/s41598-018-29121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qin Z, Yang L, Qu D, Molin S, Tolker-Nielsen T. 2009. Pseudomonas aeruginosa extracellular products inhibit staphylococcal growth, and disrupt established biofilms produced by Staphylococcus epidermidis. Microbiology 155:2148–2156. doi: 10.1099/mic.0.028001-0. [DOI] [PubMed] [Google Scholar]
- 59.Jones CJ, Newsom D, Kelly B, Irie Y, Jennings LK, Xu B, Limoli DH, Harrison JJ, Parsek MR, White P, Wozniak DJ. 2014. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog 10:e1003984. doi: 10.1371/journal.ppat.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu B, Ju Y, Soukup RJ, Ramsey DM, Fishel R, Wysocki VH, Wozniak DJ. 2016. The Pseudomonas aeruginosa AmrZ C-terminal domain mediates tetramerization and is required for its activator and repressor functions. Environ Microbiol Rep 8:85–90. doi: 10.1111/1758-2229.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jones CJ. 2013. AmrZ is a central regulator of biofilm formation in Pseudomonas aeruginosa. PhD dissertation. The Ohio State University, ProQuest Dissertation Publishing, Columbus, OH. [Google Scholar]
- 62.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 63.Marques CNH, Craver SA. 2015. Quantification of respiratory activity in biofilms. bio-protocol 5:e1591. doi: 10.21769/BioProtoc.1591. [DOI] [Google Scholar]
- 64.Bobrov AG, Kirillina O, Perry PD. 2005. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol Lett 247:123–130. doi: 10.1016/j.femsle.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 65.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O’Toole GA. 2007. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GCL, Parsek MR. 2011. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog 7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. 2006. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol 188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203. doi: 10.1016/s0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- 70.Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. 2007. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J Clin Invest 117:877–888. doi: 10.1172/JCI30783. [DOI] [PMC free article] [PubMed] [Google Scholar]