Understanding the evolutionary dynamics of antimicrobial resistance is vital to curb its emergence and spread. Being fundamentally similar to natural selection, the fitness of resistant mutants is a key parameter to consider in the evolutionary dynamics of antimicrobial resistance (AMR). Various intrinsic and extrinsic factors modulate the fitness of resistant bacteria. This study demonstrated that Lon, a bacterial master regulator protease, influences drug tolerance and resistance. Lon is a key regulator of several fundamental processes in bacteria, including cytokinesis. I demonstrated that Lon deficiency produces highly contingent phenotypes in E. coli challenged with trimethoprim and can expand the mutational repertoire available to E. coli to evolve resistance. This multipronged influence of Lon on drug resistance provides an illustrative instance of how master regulators shape the response of bacteria to antibiotics.
KEYWORDS: trimethoprim, antimicrobial resistance, drug tolerance, Lon protease, DHFR, mutational potential, protein stability, dihydrofolate reductase
ABSTRACT
Evolutionary trajectories and mutational landscapes of drug-resistant bacteria are influenced by cell-intrinsic and extrinsic factors. In this study, I demonstrated that loss of the Lon protease altered susceptibility of Escherichia coli to trimethoprim and that these effects were strongly contingent on the drug concentration and genetic background. Lon, an AAA+ ATPase, is a bacterial master regulator protease involved in cytokinesis, suppression of transposition events, and clearance of misfolded proteins. I show that Lon deficiency enhances intrinsic drug tolerance at sub-MIC levels of trimethoprim. As a result, loss of Lon, though disadvantageous under drug-free conditions, has a selective advantage at low concentrations of trimethoprim. At high drug concentrations, however, Lon deficiency is detrimental for E. coli. I show that the former is explained by suppression of drug efflux by Lon, while the latter can be attributed to SulA-dependent hyperfilamentation. On the other hand, deletion of lon in a trimethoprim-resistant mutant E. coli strain (harboring the Trp30Gly dihydrofolate reductase [DHFR] allele) directly potentiates resistance by enhancing the in vivo stability of mutant DHFR. Using extensive mutational analysis at 3 hot spots of resistance, I show that many resistance-conferring mutations render DHFR prone to proteolysis. This trade-off between gaining resistance and losing in vivo stability limits the number of mutations in DHFR that can confer trimethoprim resistance. Loss of Lon expands the mutational capacity for acquisition of trimethoprim resistance. This paper identifies the multipronged action of Lon in trimethoprim resistance in E. coli and provides mechanistic insight into how genetic backgrounds and drug concentrations may alter the potential for antimicrobial resistance evolution.
IMPORTANCE Understanding the evolutionary dynamics of antimicrobial resistance is vital to curb its emergence and spread. Being fundamentally similar to natural selection, the fitness of resistant mutants is a key parameter to consider in the evolutionary dynamics of antimicrobial resistance (AMR). Various intrinsic and extrinsic factors modulate the fitness of resistant bacteria. This study demonstrated that Lon, a bacterial master regulator protease, influences drug tolerance and resistance. Lon is a key regulator of several fundamental processes in bacteria, including cytokinesis. I demonstrated that Lon deficiency produces highly contingent phenotypes in E. coli challenged with trimethoprim and can expand the mutational repertoire available to E. coli to evolve resistance. This multipronged influence of Lon on drug resistance provides an illustrative instance of how master regulators shape the response of bacteria to antibiotics.
INTRODUCTION
The evolution of antimicrobial resistance (AMR) in bacteria follows Darwinian logic. Mutants that are able to survive in the presence of antibiotics outcompete their drug-sensitive ancestor in the face of drug challenge. Competition for survival, hence, forms the basis of our understanding of the evolutionary dynamics of drug resistance. Though conceptually straightforward, this idea is rendered complex in practice since the fitness of drug-resistant strains is influenced by several cell-intrinsic and extrinsic factors. For instance, recent studies have demonstrated that the outcomes of selection for resistance can be influenced by environmental factors such as drug concentration (1–4), drug exposure duration (2, 5), and drug interactions (6, 7). Among cell-intrinsic factors, genetic background is an important determinant of how drug resistance evolution proceeds. For instance, genetic backgrounds prone to hypermutation alter the mutational spectrum observed for rifampin-resistant Escherichia coli and Pseudomonas aeruginosa (8, 9). Similarly, efflux pump-overproducing Gram-negative bacteria act as primers for the development of high-level multidrug resistance (10, 11).
A growing body of literature indicates that perturbation of the Lon protease in bacteria such as Escherichia coli alters survival upon antibiotic challenge. Lon is an ATP-dependent serine protease that belongs to the type AAA+ ATPase family. Though initially identified as a mediator of filamentation under conditions of UV exposure (12), Lon is now recognized as a master regulator of bacterial physiology. Through its ATP-dependent protease activity, Lon modulates the levels of short-lived proteins in bacteria and hence influences cellular functions as diverse as cytokinesis (13, 14), suppression of DNA transposition (15), and activation of toxins (16). Proteomics studies have identified several additional substrates for Lon in E. coli, hinting at a far greater influence of this protease on bacterial physiology than was previously understood (17). Lon deletion in E. coli suppresses persistence to antibiotics such as ciprofloxacin, indicating that Lon activity enhances persister populations in bacteria (18, 19). Deletion of the lon gene also results in enhanced levels of MarA, an activator of drug efflux through the AcrAB-TolC pump (20). As a result, Lon-deficient E. coli can tolerate higher concentrations of several antibiotics such as tetracycline, kanamycin, and erythromycin than Lon-producing bacteria (21, 22). These findings have put Lon on the AMR map, and, given its role in the pathogenesis of bacteria such as Salmonella enterica serovar Typhimurium and Pseudomonas aeruginosa, inhibitors of Lon have been suggested as novel antimicrobials (23–27).
In addition to influencing survival in antibiotics, a few studies have shown that Lon modifies the evolutionary dynamics of AMR in E. coli. Nicoloff and Andersson (22) showed that Lon-deficient E. coli evolved multidrug resistance more rapidly that wild-type bacteria. This was attributed to insertion (IS) element-mediated duplication of genes coding for the AcrAB drug efflux pump. Interestingly, the resistance-conferring effects of this duplication were contingent on the absence of Lon (22). In a similar vein, Bershtein et al. (28) have demonstrated that Lon activity has the potential to influence genotype-phenotype correlation in E. coli. Lon, together with bacterial chaperones such as GroEL, modifies the folding environment within bacterial cells, which can have a profound effect on the fitness of mutant bacteria. It was demonstrated that loss of bacterial fitness due to destabilizing mutations in dihydrofolate reductase (DHFR) can be partially compensated by loss of Lon activity (28). Further, Lon activity can also influence the level of resistance conferred by mutations in DHFR to the antifolate antibiotic trimethoprim. This was also attributed to the activity of Lon against partially folded mutants of DHFR and, in particular, was found to impact the phenotypes of DHFR with multiple mutations (29). Thus, the presence or absence of Lon appears to be an important cell-intrinsic factor governing adaptation to drugs in bacteria such as E. coli.
In this study, I found that loss of Lon activity altered the survival and growth of E. coli upon trimethoprim challenge. Trimethoprim is a competitive inhibitor of bacterial DHFR enzymes. Clinical resistance to this antibiotic is mediated primarily by acquisition of drug-resistant, plasmid-encoded DHFR (30). However, genomic resistance to trimethoprim does evolve rapidly in laboratory strains of E. coli and primarily maps to a few mutational hot spots within endogenous DHFR, encoded by the folA gene (31–33). This has made genomic trimethoprim resistance in E. coli an attractive model to investigate the evolution of drug resistance (29, 31–33). I report that the phenotypic effects of Lon deficiency varied qualitatively and quantitatively depending on drug concentration, being beneficial at low drug concentrations but detrimental in the presence of high drug concentrations. Using genetic tools, I provide mechanistic explanations for each of these observations. Further, I show that Lon deficiency can enhance the mutational potential for trimethoprim resistance of DHFR. Through these analyses, I show that perturbation of master regulators such as Lon can produce highly contingent phenotypes owing to their multifaceted influence on bacterial physiology.
RESULTS
Drug concentration-dependent effects of Lon deficiency in E. coli challenged with trimethoprim.
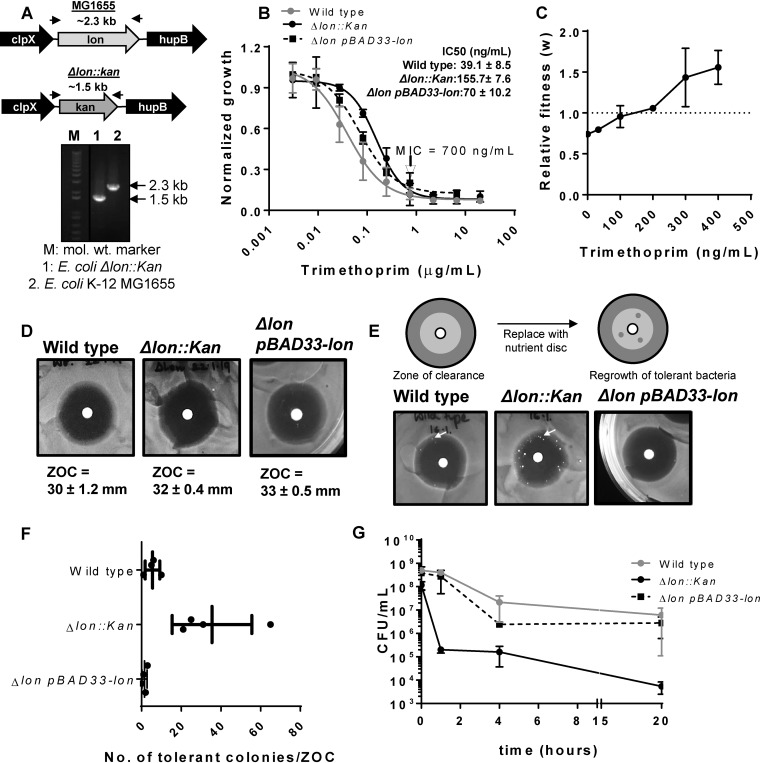
In order to test how the Lon protease impacted the intrinsic susceptibility of E. coli to antibiotics, I created a Lon-deficient strain by replacing the lon gene with a kanamycin resistance cassette (here referred to as E. coli Δlon) (Fig. 1A). I then compared the dose-response characteristics of Lon-deficient and wild-type E. coli strains against 5 antibiotics with different mechanisms of action. Deletion of lon had no detectable impact on susceptibility to ampicillin, chloramphenicol, nalidixic acid, and rifampin (see Fig. S1 in the supplemental material). However, Lon deletion significantly improved the ability of E. coli to resist growth inhibition by trimethoprim. E. coli Δlon had an ∼4-fold-higher IC50 (concentration needed for 50% growth inhibition) for trimethoprim than the wild-type strain (Fig. 1B), though no change in the MIC of the drug was detectable (Fig. 1B). These effects could be partially reversed by expressing Lon protease heterologously in the Δlon strain (Fig. 1B). This observation suggested that loss of Lon may be advantageous at sub-MICs of trimethoprim. To confirm this hypothesis, I allowed wild-type and Δlon E. coli strains to compete in the presence or absence of trimethoprim. Deletion of lon was costly, as it reduced the relative fitness (w) of E. coli in drug-free medium (Fig. 1C). However, the presence of sub-MIC trimethoprim in growth medium enhanced the relative fitness of the Δlon strain in a dose-dependent manner, such that at concentrations higher than 200 ng/ml, Lon-deficient E. coli outcompeted the wild type (Fig. 1C). I next asked if the enhanced growth of E. coli Δlon in sub-MIC trimethoprim could be attributed to greater drug tolerance. E. coli Δlon showed wild-type-like zones of clearance (ZOCs) in a standard disc diffusion assay for trimethoprim sensitivity, in line with the similar MICs of the two strains (Fig. 1D). However, in a TDtest for drug tolerance (34) that assays for the presence of surviving bacteria within the ZOC, E. coli Δlon showed significantly greater trimethoprim-tolerance levels than the wild type (Fig. 1E and F). The enhanced tolerance of E. coli Δlon could be reversed upon expression of Lon protease in this strain, verifying that the observed phenotypes were indeed due to loss of Lon protease (Fig. 1E and F). These data demonstrated that the Lon protease suppressed the intrinsic trimethoprim tolerance of wild-type E. coli and explained why loss of Lon was beneficial at sub-MIC levels of trimethoprim.
FIG 1.
Impact of Lon deficiency on intrinsic trimethoprim tolerance and resistance is contingent on drug concentration. (A) Confirmation of the Δlon::kan strain using genomic PCR. The upper panel shows the genome neighborhood of the lon gene in wild-type (MG1655) or Δlon::kan strains (not drawn to scale). Locations of primers used for PCR are indicated by arrows, and the expected amplicon sizes are also indicated. The lower panel shows the PCR confirmation of replacement of the lon gene with a kanamycin resistance cassette. mol. wt., molecular weight. (B) Trimethoprim dose-response curves for wild-type, Δlon::kan, and pBAD33-Lon-harboring E. coli Δlon::kan strains. Peak optical density after 15 to 18 h of growth at each trimethoprim concentration was normalized to growth in drug-free medium (normalized growth). Means ± standard deviations (SD) of results from 3 independent experiments are plotted. The MICs for the two strains were identical and are indicated in the graph. IC50 values (means ± standard errors of the means [SEM]) estimated from the data are provided as an inset. (C) Relative fitness (w) of E. coli Δlon::kan compared to wild-type E. coli calculated as a function of trimethoprim concentration. Fitness of the wild type is represented by a value of 1 by definition and marked with a dotted line. Means ± SD of results from 3 independent experiments are plotted. (D) Trimethoprim resistance of E. coli Δlon::kan, pBAD33-Lon rescue strain, and the wild type assessed by a disc diffusion assay. Similar diameters of zones of clearance (ZOC) were observed for the wild-type and Δlon::kan strains. Means ± SD of the diameters of the ZOC from 3 independent experiments are shown. (E) TDtest for trimethoprim tolerance for E. coli Δlon::kan, pBAD33-Lon rescue strain, and wild type. The antibiotic disc in a standard disc diffusion assay is replaced with a nutrient disc to allow growth of surviving bacteria within the ZOC (see schematic). Trimethoprim-tolerant bacteria (indicated by arrow) were more numerous for E. coli Δlon::kan than for the wild-type or plasmid rescue strain. Representative data from experiments performed at least thrice are shown. (F) Quantitation of the number of trimethoprim-tolerant colonies obtained in the ZOC from 3 or 4 independent experiments. (G) Survival of wild-type, E. coli Δlon::kan, and pBAD33-Lon rescue strains at 10 μg/ml (∼14× MIC) trimethoprim. Means ± SEM of results from 3 independent experiments are plotted.
Importantly, trimethoprim-tolerant colonies of E. coli Δlon were observed mainly at the periphery of the ZOC and not close to the disc where the concentration of trimethoprim is expected to be the highest (Fig. 1E). This prompted me to ask whether the beneficial effects of Lon deficiency also persisted at high trimethoprim concentrations. Trimethoprim is known to be bacteriostatic for E. coli. In line with this, treatment of wild-type cultures with 10 μg/ml trimethoprim (∼14× MIC) had a modest effect on the viability of E. coli even after 20 h of drug exposure (Fig. 1G). Surprisingly, a sharp decline in the numbers of viable bacteria was observed in E. coli Δlon cultures treated with the same concentration of trimethoprim within the first hour of treatment. This reduction persisted even after a longer duration of treatment, indicating that at concentrations higher than the MIC, Lon deficiency was detrimental for E. coli (Fig. 1G). Once again, plasmid-based expression of Lon protease in the E. coli Δlon strain was able to rescue the observed phenotype (Fig. 1G). Thus, the phenotype of Lon deficiency in E. coli was contingent on the trimethoprim concentration; it was beneficial and conferred a selective advantage at sub-MIC levels, while Lon deficiency was detrimental for the survival of E. coli at concentrations higher than the MIC.
Different molecular mechanisms for the effects of Lon deficiency at sub-MICs and high trimethoprim concentrations.
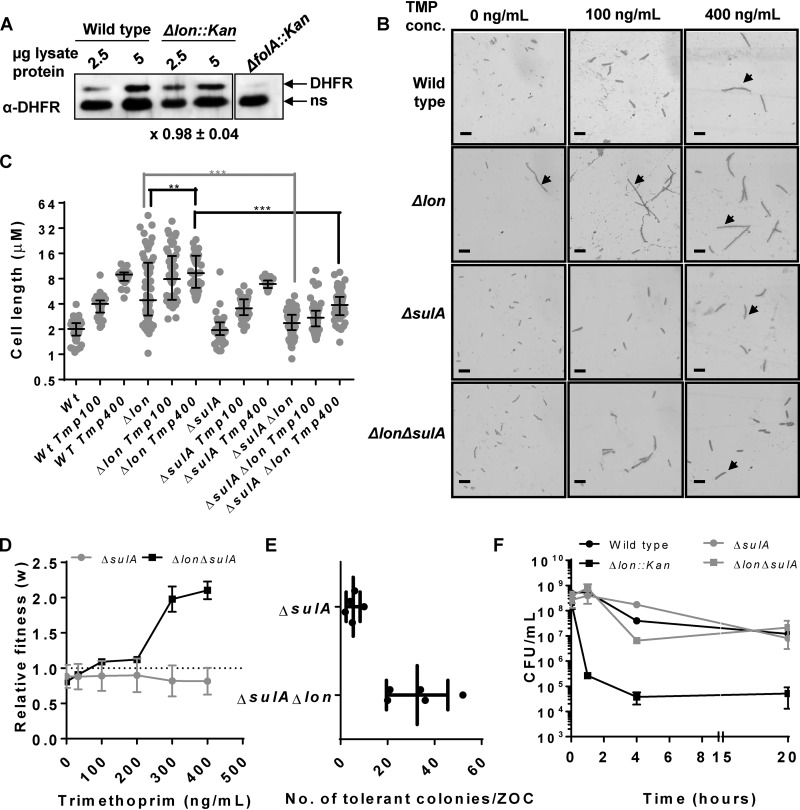
The opposite effects of Lon deficiency at low and high trimethoprim concentrations were curious and warranted investigation of the underlying molecular mechanisms. On the basis of published literature, I surmised that there are 3 molecular pathways that might link Lon with altered trimethoprim tolerance/resistance. First, Lon protease is known to regulate the steady-state levels of bacterial DHFRs expressed in E. coli (35). Since DHFR is the target of trimethoprim, changes in its expression level may directly alter susceptibility to the antibiotic. However, the expression level of endogenous DHFR did not appreciably change upon lon deletion, negating this possibility (Fig. 2A). Second, Lon regulates the levels of SulA, an inhibitor of bacterial cytokinesis. In the absence of Lon, SulA accumulates and leads to arrested cell division and filamentation (13, 14). Trimethoprim treatment also induces filamentation in E. coli; hence, I reasoned that enhanced filamentation of the Δlon strain may alter its response to trimethoprim challenge. In order to test this, I generated a ΔsulA strain of E. coli as well as a Δlon ΔsulA double mutant. As expected, Lon-deficient E. coli formed filaments under standard growth conditions. Treatment with sub-MICs of trimethoprim further stimulated filamentation in Lon-deficient E. coli (Fig. 2B and C). This enhancement was due to a greater proportion of the population showing filamentation rather than to an increase in the length of the filaments seen. E. coli Δlon ΔsulA did not show filamentation in drug-free medium (Fig. 2B and C), reconfirming that the hyperfilamentation phenotype of E. coli Δlon was due to SulA accumulation. However, in the presence of trimethoprim, both the ΔsulA and the Δlon ΔsulA strains showed wild type-like filamentation (Fig. 2B and C). In line with this, E. coli Δlon ΔsulA continued to show trimethoprim-dependent enhancement in competitive fitness (Fig. 2D) and greater trimethoprim tolerance in a TDtest (Fig. 2E). Thus, SulA accumulation could not explain the phenotypes of E. coli Δlon at sub-MIC trimethoprim concentrations. To check whether SulA accumulation contributed to the phenotypes of E. coli Δlon at high trimethoprim concentrations, the survival of E. coli strains Δlon ΔsulA and ΔsulA at ∼14× the MIC of the drug was analyzed. Interestingly, loss of SulA restored the survival of Lon-deficient E. coli in the presence of a high trimethoprim concentration to wild type-like levels (Fig. 2F). These data proved that while SulA-dependent filamentation did not contribute to greater survival of Lon-deficient E. coli at low trimethoprim concentrations, it explained the detrimental effects of Lon deficiency at a high trimethoprim concentration.
FIG 2.
SulA-dependent hyperfilamentation explains the effects of Lon deficiency at high drug concentrations but not low drug concentrations. (A) Levels of endogenous DHFR in lysates of wild-type or Lon-deficient (Δlon::kan) E. coli assayed by immunoblotting. DHFR-specific polyclonal antibody (α-DHFR) was used as the primary antibody. E. coli ΔfolA was used as a control to differentiate between specific (DHFR) and nonspecific (ns) immune-reactive bands. Two different amounts of lysate protein (2.5 μg and 5 μg) were used for each strain to ensure that the Western blot was not saturated. Fold expression of DHFR (Δlon/wild type) was calculated using densitometric analysis of band intensities. Means ± SD of results from 3 independent experiments are indicated below the image. (B) Filamentation of indicated genotype of E. coli growing in drug-free medium or in the presence of sub-MICs of trimethoprim (TMP). Filamentous bacteria are indicated by an arrow. Images shown are representative of results from at least 3 independent experiments. Scale bar = 2 μm. (C) Quantitation of cell lengths shown as a scatter of individual values from 3 independent experiments. Median and interquartile range of data from each of the strains are shown. Statistical significance was tested using a Student's t test (***, P < 0.01; **, P < 0.02). (D) Relative fitness (w) of E. coli ΔsulA or E. coli ΔsulA Δlon strains compared to wild-type E. coli calculated as a function of trimethoprim concentration. Fitness of the wild type is represented by a value of 1 by definition and marked with a dotted line. Means ± SD of results from 3 independent experiments are plotted. (E) Number of trimethoprim-tolerant colonies of E. coli ΔsulA Δlon or E. coli ΔsulA as observed in a TDtest. Results from 4 or 5 independent experiments are plotted. (F) Survival of wild type and indicated mutants of E. coli at 10 μg/ml (∼14× MIC) trimethoprim. Means ± SEM of results from 3 independent experiments are plotted.
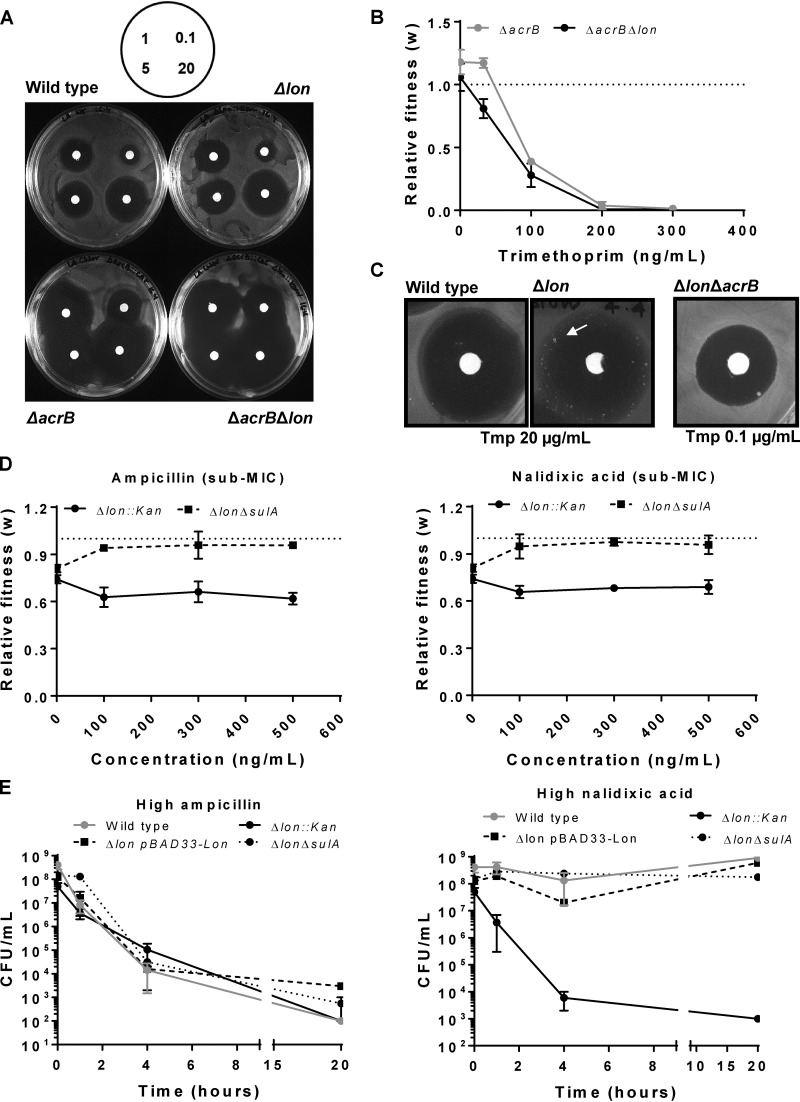
Finally, Lon is known to restrict the transcription of the AcrAB drug efflux pump by regulating the levels of the MarA transcription factor (20). In the absence of Lon, AcrAB expression is enhanced and has been linked to low-level resistance to antibiotics such as ciprofloxacin and amoxicillin (21, 22). To test if the phenotypes of E. coli Δlon at trimethoprim sub-MICs could be attributed to higher levels of expression of the AcrAB efflux pump, I generated ΔacrB and Δlon ΔacrB strains and monitored their survival at low concentrations of trimethoprim. Deletion of acrB conferred hypersusceptibility to trimethoprim in Lon-deficient and Lon-expressing backgrounds, indicating that trimethoprim is indeed subjected to efflux by the AcrAB pump (Fig. 3A). In line with this finding, E. coli strain Δlon ΔacrB did not show enhanced competitive fitness in trimethoprim-supplemented medium (Fig. 3B). In fact, the fitness of this strain was reduced in a dose-dependent manner in the presence of trimethoprim. E. coli Δlon ΔacrB also did not show trimethoprim tolerance in a TDtest (Fig. 3C). These data demonstrated that the greater trimethoprim tolerance of E. coli Δlon at trimethoprim sub-MICs was due to enhanced drug efflux. Taking the results together, loss of Lon had opposite effects at trimethoprim sub-MICs and high concentrations due to two different molecular mechanisms. While SulA-mediated filamentation was responsible for trimethoprim hypersensitivity at high drug concentrations, AcrAB-mediated drug efflux explained trimethoprim tolerance of Lon-deficient E. coli at low drug concentrations.
FIG 3.
Trimethoprim tolerance of Lon-deficient E. coli at sub-MICs is dependent of AcrAB-mediated drug efflux. (A) Trimethoprim resistance of indicated genotypes tested by a disc diffusion assay. Concentrations of trimethoprim used in the antibiotic discs are indicated in micrograms per milliliter in the schematic. Representative data from experiments performed twice are shown. (B) Relative fitness (w) of E. coli ΔacrB strain or E. coli ΔacrB Δlon strain compared to wild-type E. coli calculated as a function of trimethoprim concentration. Fitness of the wild type is represented by a value of 1 by definition and marked with a dotted line. Means ± SD of results from 3 independent experiments are plotted. (C) TDtest showing trimethoprim-tolerant bacteria (arrow) in E. coli Δlon but not in wild-type or ΔacrB Δlon strains. The concentration of trimethoprim used for E. coli ΔacrB Δlon was lower than wild type and Δlon to obtain comparable ZOC diameters. Representative data from experiments performed at least thrice are shown. (D) Relative fitness (w) of E. coli Δlon or E. coli ΔsulA Δlon strains compared to wild-type E. coli calculated as a function of ampicillin or nalidixic acid concentration. Fitness of the wild type is represented by a value of 1 by definition and marked with a dotted line. Means ± SD of results from 3 independent experiments are plotted. (E) Survival of wild type and indicated mutants of E. coli at a high concentration of ampicillin (20 μg/ml) or nalidixic acid (50 μg/ml). Means ± SEM of results from 3 independent experiments are plotted.
Since neither of the molecular mechanisms described above involved DHFR, it was possible that these phenotypes of Lon deficiency also extend to other antibiotics. I chose ampicillin and nalidixic acid as representative bactericidal and bacteriostatic antibiotics, respectively, for these studies. The fitness of E. coli Δlon was not potentiated at sub-MICs of both these antibiotics, in stark contrast to the results obtained for trimethoprim. In fact, a small but significant decrease in fitness was observed at low ampicillin and nalidixic acid levels compared to drug-free medium (Fig. 3D). These results were in line with the drug dose-response curves shown in Fig. S1 and indicated that the AcrAB upregulation in the Lon-deficient bacteria was likely insufficient in the context of these antibiotics. At a high ampicillin concentration, no measurable difference in survival rate of E. coli Δlon was observed compared to the wild type (Fig. 3E). However, when exposed to a high concentration of nalidixic acid, E. coli Δlon had severely compromised survival. This was rescued by heterologous expression of Lon, confirming that, as seen with trimethoprim, survival in the presence of a high concentration of nalidixic acid required the Lon protease (Fig. 3E). Further, deletion of sulA from E. coli Δlon also rescued survival in the presence of a high concentration of nalidixic acid (Fig. 3E). Thus, Lon deficiency produced very similar phenotypic effects at high concentrations of trimethoprim and nalidixic acid but not ampicillin. Interestingly, deletion of sulA from E. coli Δlon also allowed a small increase in fitness at low ampicillin and nalidixic acid concentrations (Fig. 3D), though this enhancement was not as dramatic as that observed in at low trimethoprim concentrations (Fig. 1C and 2D). These data indicated that Lon deficiency also altered responses to other antibiotics and that these effects were antibiotic specific. Further, SulA-dependent filamentation was more relevant in the case of ampicillin and nalidixic acid even at low drug concentrations, once again pointing to the high level of contingency with respect to the phenotypes of Lon-deficient E. coli.
Lon deficiency enhances trimethoprim resistance conferred by a mutant DHFR allele.
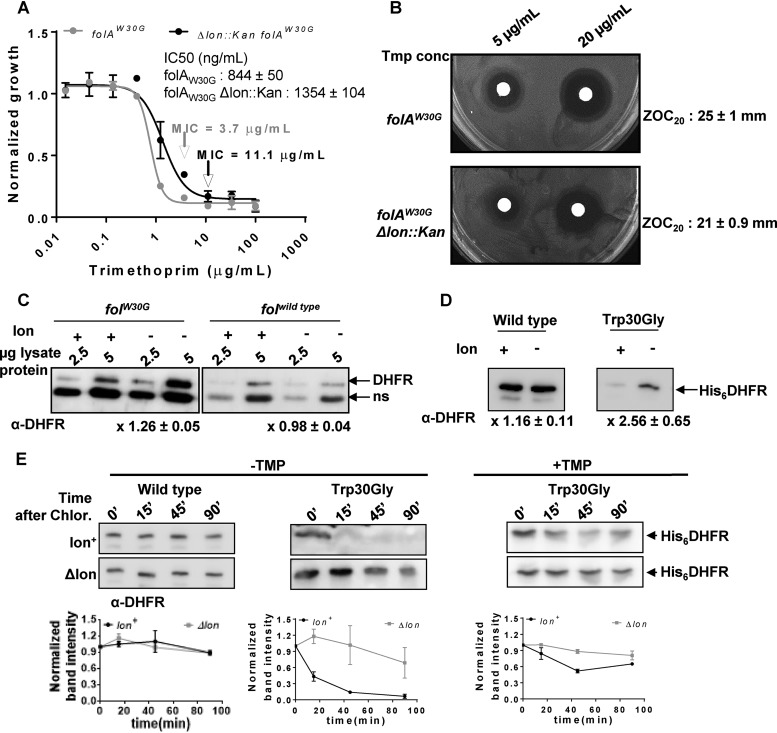
Having established a role for the Lon protease in modulating intrinsic trimethoprim tolerance of wild type E. coli, I next asked how Lon influenced mutationally acquired trimethoprim resistance. I deleted the lon gene from a previously isolated spontaneous trimethoprim-resistant strain harboring the Trp30Gly mutation in its endogenous copy of DHFR (folAW30G) (31). Compared to the results seen in a trimethoprim-sensitive background, deletion of lon in a trimethoprim-resistant background led to a 3-fold increase in drug MIC, in addition to enhancement in the IC50 (Fig. 4A). This change in MIC was also observed as a reduction in ZOC diameter in a disc diffusion assay (Fig. 4B). Interestingly, lon deletion led to significant enhancement in the expression levels of Trp30Gly DHFR in comparison to wild-type DHFR (Fig. 4C). This observation also held true for heterologously expressed wild-type and Trp30Gly DHFRs, negating the possibility that the observed differences in expression levels were due to feedback or transcriptional regulation (Fig. 4D). Previous work demonstrated that the Trp30Gly mutation destabilizes DHFR (31). I reasoned, therefore, that the Trp30Gly mutation may render DHFR more prone to proteolysis in Lon-expressing bacteria. In order to test this, I assayed the in vivo stabilities of plasmid-expressed wild-type and Trp30Gly DHFRs using a chloramphenicol chase (“shutoff”) assay. I found that the stability of wild-type DHFR was unaffected by the presence of Lon over the duration of the assay (90 min after addition of chloramphenicol) (Fig. 4E). However, in the presence of Lon, Trp30Gly DHFR was rendered very unstable in E. coli. The difference in in vivo stability of Trp30Gly DHFR was partially rescued by the presence of trimethoprim in the growth medium, in line with earlier studies that showed the stabilizing effects of inhibitors on DHFR (36, 37). However, even in the presence of the antibiotic, the lifetime of Trp30Gly DHFR was longer in Lon-deficient E. coli than in Lon-expressing bacteria (Fig. 4E). Thus, the diminished expression level of Trp30Gly DHFR was attributable to its lower stability in Lon-expressing E. coli, which in turn explained why the trimethoprim resistance of this mutant was enhanced by lon deletion (Fig. 4E).
FIG 4.
Lon deficiency potentiates the trimethoprim resistance level of a spontaneous drug-resistant isolate by increasing the in vivo stability of mutant DHFR. (A) Dose-response curves for a trimethoprim-resistant isolate of E. coli harboring the folAW30G allele and its Δlon::kan derivative. Peak optical density after 15 to 18 h of growth at each trimethoprim concentration was normalized to growth in drug-free medium (normalized growth). Means ± SD of results from 3 independent experiments are plotted. The drug MIC values for each of the strains are indicated in the graph with an arrow. IC50 values (means ± SEM) estimated from the data are provided as an inset. (B) Trimethoprim resistance of E. coli folAW30G and its Δlon::kan derivative assessed by a disc diffusion assay at the indicated concentrations of trimethoprim. Diameters of zone of clearance were estimated at 20 μg/ml of the antibiotic (ZOC20). Means ± SD of the diameters of the ZOC from 3 independent experiments are shown. (C) Levels of endogenous DHFR in lysates of drug-sensitive (folAwild type) or drug-resistant (folAW30G) E. coli strains and their Δlon::kan derivatives assayed by immunoblotting. DHFR polyclonal antibody (α-DHFR) was used as the primary antibody. Specific (DHFR) and nonspecific (ns) immune-reactive bands are indicated. Two different amounts of lysate protein (2.5 μg and 5 μg) were used for each strain to ensure that the Western blot was not saturated. Fold expression of DHFR (Δlon/wild type) was calculated using densitometric analysis of band intensities. Means ± SD of results from 3 independent experiments are indicated below the image. (D) Levels of plasmid-encoded wild type or Trp30Gly mutant DHFR (His6DHFR) in Lon-deficient Lon-expressing E. coli assayed by immunoblotting using DHFR polyclonal antibody (α-DHFR). Fold expression of DHFR (Δlon/wild type) was calculated using densitometric analysis of band intensities. Means ± SD of results from 3 independent experiments are indicated below the image. (E) In vivo stabilities of plasmid-encoded wild-type or Trp30Gly mutant DHFR (His6DHFR) in Lon-deficient or Lon-expressing E. coli assayed using a chloramphenicol (Chlor.) chase shutoff assay. After inhibition of protein synthesis by addition of chloramphenicol, levels of DHFR were assessed using immunoblotting with DHFR-specific polyclonal antibody (α-DHFR) at the indicated times. For Trp30Gly DHFR, the assay was performed in the presence or absence of trimethoprim in the growth medium as indicated. Representative immunoblot data from experiments performed at least thrice are shown. Below each blot, quantitation of band intensities normalized to the intensity measured at 0 min is plotted as means ± SD of results from 3 independent experiments.
Lon suppresses the mutational potential for trimethoprim resistance at 3 hot spots in DHFR.
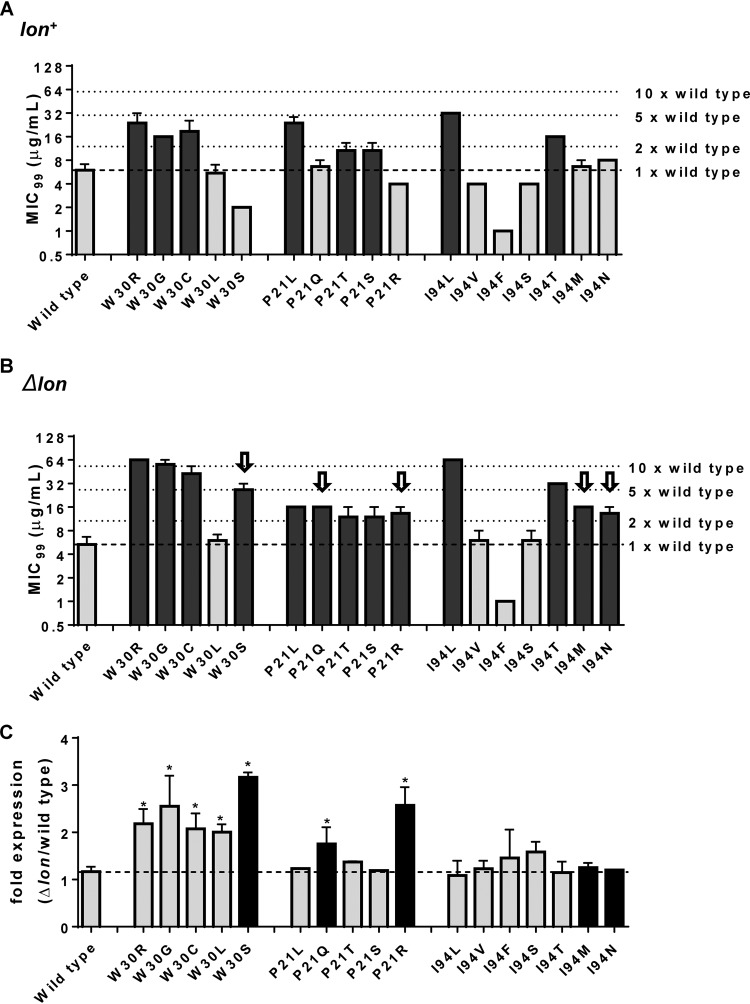
Next, I asked whether the phenotypes of mutations at different hot spots within DHFR were also influenced by lon deletion, similarly to what was observed for Trp30Gly. I designed a mutant library in which all possible unique missense mutations accessible through single-base-pair substitution at three hot spots (Pro21, Trp30, and Ile94) were engineered in a plasmid-encoded copy of DHFR (Fig. S2). These three hot spots were selected for this investigation since they are present in structurally distinct parts of the DHFR enzyme. Pro21 is part of the Met20 catalytic loop, Trp30 is an important stabilizing residue needed for folding, and Ile94 is an active-site residue (Fig. S3). All mutations could be successfully engineered in DHFR, except Pro21Ala, which, even after repeated attempts, accumulated unwanted second-site mutations. Pro21Ala was therefore excluded from the analysis. This resulted in 17 different DHFR mutants. In order to test whether a mutation conferred trimethoprim resistance, the MIC99 of each DHFR mutant expressed in Lon-replete and Lon-deficient backgrounds was calculated and compared with that of wild-type DHFR. An increase in the MIC99 by at least 2-fold compared to wild-type DHFR was considered representative of drug resistance. The number of mutations at each of the hot spots that conferred trimethoprim resistance compared to the total number of unique, nonsynonymous substitutions possible was referred to as the “potential for resistance” at that hot spot. Using these criteria, the potential for resistance at Pro21 and Trp30 was found to be 3/5 whereas that at Ile94 was 2/7 in a Lon-expressing background. Interestingly, the potential for resistance at Pro21 and Trp30 in a Lon-deficient genetic background increased to 4/5 each, while that at Ile94 increased to 4/7 (Fig. 5A and B). This increase occurred because 5 mutations from the library, namely, Pro21Gln, Pro21Arg, Trp30Ser, Ile94Asn, and Ile94Met, conferred resistance to trimethoprim only in Lon-deficient bacteria. Three of these, namely, Pro21Gln, Ile94Asn, and Ile94Met, had a wild-type-like phenotype in Lon-expressing bacteria, while Pro21Arg and Trp30Ser were hypersensitive to trimethoprim in bacteria expressing the Lon protease (Fig. 5A and B). These analyses showed that deletion of lon expanded the repertoire of DHFR mutants able to confer trimethoprim resistance to E. coli.
FIG 5.
Lon deficiency expands the potential for trimethoprim-resistant mutations at 3 hot spots of resistance in DHFR. (A and B) MIC99 values of wild-type (A) and Δlon (B) E. coli strains harboring plasmids expressing wild-type DHFR or the indicated mutants at 3 resistance hot spots. Mutants that were at least 2-fold more resistant than the wild type are shown as black bars, while those that showed a wild type-like or hypersensitive phenotype are shown as gray bars. Mutants that conferred resistance only in Δlon E. coli are pointed out with arrows in panel B. Means ± SD of results from at least 3 measurements are plotted. (C) Mean fold differences in expression levels of plasmid-encoded wild-type or mutant DHFR (Δlon/wild type) are plotted along with SEM. Wild-type DHFR showed similar levels of expression on lon+ and Δlon backgrounds. Mutants that conferred resistance only in E. coli Δlon are shown as black bars. Statistical significance was tested using a Student's t test. *, P < 0.05.
Expanded mutation potential for trimethoprim resistance in Lon-deficient bacteria is explained by proteolytic stability of mutants in the absence of Lon.
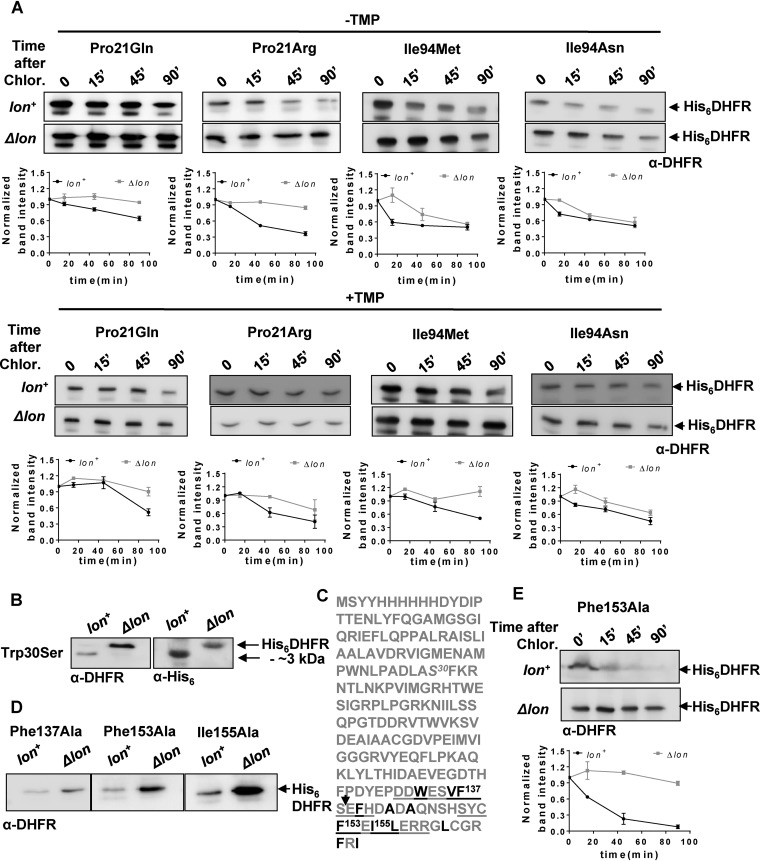
Could the expanded repertoire of trimethoprim-resistant DHFR mutations in Δlon bacteria be explained by enhanced in vivo stabilities? In order to answer this, I initially estimated the expression level of wild-type DHFR and of each of the mutants from the library described above in Lon-deficient and Lon-expressing E. coli. As with Trp30Gly, all of the mutations at Trp30, except Trp30Leu, showed a dramatic enhancement of expression level as well as potentiation of trimethoprim resistance in Lon-deficient E. coli (Fig. 5C). Pro21Gln and Pro21Arg, both of which conferred trimethoprim resistance only in a Δlon background, were also expressed to higher levels in the absence of Lon. In line with this result, both Pro21Arg and Pro21Gln showed lowered stability in Lon-expressing E. coli, though the magnitude of this effect was far greater for Pro21Arg than for Pro21Gln (Fig. 6A). The expression levels of other Pro21 mutants were unaffected by lon deletion (Fig. 5C). All mutants at Ile94 had similar expression levels in Lon-expressing and Lon-deficient E. coli (Fig. 5C). Particularly surprising was the fact that Ile94Asn and Ile94Met, both of which conferred trimethoprim resistance only in a Δlon background, did not show a significant enhancement in expression level upon lon deletion (Fig. 5). Stability measurements of these two mutants, however, verified that both mutations reduced stability of DHFR more dramatically in Lon-expressing bacteria than in Δlon E. coli (Fig. 6A). Even in the presence of trimethoprim in the growth medium, these mutants were less stable in Lon-expressing bacteria than in Lon-deficient E. coli (Fig. 6A). Thus, Lon-dependent in vivo stability could explain the different phenotypes of DHFR mutants in the 2 genetic backgrounds tested.
FIG 6.
In vivo proteolytic stabilities explain disparities between trimethoprim-resistant phenotypes of DHFR mutants in lon+ and Δlon backgrounds. (A) In vivo stabilities of plasmid-encoded mutants of DHFR (His6DHFR) in Lon-deficient or Lon-expressing E. coli assayed using a chloramphenicol chase shutoff assay. After inhibition of protein synthesis by addition of chloramphenicol, levels of DHFR were assessed using immunoblotting with DHFR-specific polyclonal antibody (α-DHFR) at the indicated times in the presence or absence of trimethoprim in the growth medium. Representative immunoblot data from experiments performed at least thrice are shown. Below each blot, quantitation of band intensities normalized to intensity at 0 min is plotted as means ± SD of results from 3 independent experiments. (B) Clipping of Trp30Ser DHFR in lon+ E. coli. DHFR expression in lysates of lon+ or Δlon E. coli harboring plasmid coding for His6-DHFR Trp30Ser was analyzed by immunblotting. The clipped fragment of Trp30Ser DHFR was roughly 3 kDa smaller than full-length Trp30Ser DHFR and showed immunoreactivity with anti-DHFR polyclonal and anti-His monoclonal antibodies, indicating clipping at the C terminus of the protein. Due to the relatively lower level of immunoreactivity with anti-His antibody, lysates in which DHFR was overproduced by induction with IPTG were used for immunoblots performed with this antibody. (C) Sequence of plasmid-encoded Trp30Ser DHFR. The Trp30Ser mutation is indicated in italics. The 2 buried β-sheets at the C terminus of DHFR are underlined, and hydrophobic residues are shown in black. Phe137, Ph153, and Ile155, all of which are important for folding of DHFR, are indicated. The expected proteolytic site that would result in an approximately 3-kDa-smaller fragment is indicated by an arrow. (D) Levels of plasmid-encoded Phe137Ala, Phe153Ala, and Ile155Ala mutant DHFR (His6DHFR) in Lon-deficient or Lon-expressing E. coli assayed by immunoblotting using DHFR polyclonal antibody (α-DHFR). Representative data from 3 independent experiments are shown. (E) In vivo stabilities of plasmid-encoded Phe153Ala DHFR (His6DHFR) in Lon-deficient or Lon-expressing E. coli assayed using a chloramphenicol chase assay. After inhibition of protein synthesis by addition of chloramphenicol, levels of DHFR were assessed using immunoblotting with DHFR-specific polyclonal antibody (α-DHFR) at the indicated times. Representative data from experiments performed thrice are shown.
Interestingly, Trp30Ser, when expressed in wild-type E. coli, was naturally clipped to a protein that was approximately 3 kDa smaller than the full-length enzyme (based on electrophoretic mobility) (Fig. S4). In Δlon bacteria, however, Trp30Ser was unclipped and showed electrophoretic mobility similar to that seen with full-length DHFR (Fig. 6B). Arguing that clipped Trp30Ser was a proteolytic intermediate, I exploited the fact that the plasmid-encoded DHFR used in this study had a hexahistidine tag at the N terminus to ask where the determinants of proteolysis of DHFR might lie. Clipped Trp30Ser in Lon-expressing bacteria showed immunoreactivity with an antihexahistidine antibody, indicating that this mutant was cleaved at the C terminus (Fig. 6B). The C terminus of DHFR is primarily made up of buried β-sheets containing several hydrophobic residues. Three of these, namely, Phe137, Phe153, and Ile155 (Fig. 6C), interact with Trp30 in a hydrophobic tetrad and play a role in stabilizing the DHFR fold (31). Since several proteases, including Lon, recognize exposed hydrophobic residues as signals for proteolysis (38), I asked whether mutations at Phe137, Phe153, and Ile155 would also destabilize DHFR in Lon-expressing bacteria. This was indeed the case, as evidenced by enhanced expression levels of Phe137Ala, Phe153Ala, and Ile155Ala mutants of DHFR in Lon-deficient E. coli (Fig. 6D). Further, for the Phe153Ala mutant that closely phenocopies Trp30 mutants (31), a significant decrease in stability was observed in a Lon-expressing background (Fig. 6E). These results substantiated that mutation-induced perturbation of a C-terminal hydrophobic patch in DHFR could make the protein prone to degradation in E. coli.
DISCUSSION
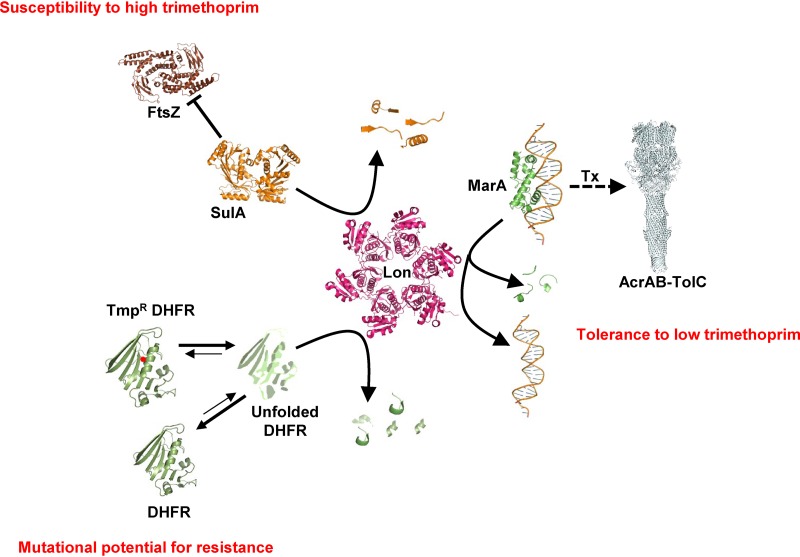
During the evolution of AMR, bacteria accumulate mutations that allow survival in the presence of antibiotics. Drug-resistant mutations have pleiotropic effects on bacterial physiology that are, in part, results of interactions with other genes (39). Therefore, genetic context is a key factor that determines the phenotypes and the fitness landscape of drug-resistant bacteria. A second layer of complexity is introduced by environmental variables, such as drug concentration, that further alter the phenotypes of resistant mutants. Ultimately, these factors together impact the evolutionary dynamics of AMR. The present study investigated how Lon, a master regulator protease, regulates AMR in E. coli. The key finding of this study is that Lon deficiency impacted the ability of E. coli to resist trimethoprim challenge in a highly contingent manner, i.e., in a manner dependent on drug concentration as well as on genetic background. At sub-MIC drug pressure levels, Lon deficiency potentiated the intrinsic trimethoprim tolerance of E. coli. On the other hand, at higher concentrations, absence of Lon conferred hypersensitivity to trimethoprim. Further, Lon deficiency was epistatic with respect to DHFR mutations, expanding the repertoire of mutations conferring trimethoprim resistance as well as potentiating the levels of resistance conferred by them. Importantly, each of these phenomena was explained by unique molecular mechanisms. Trimethoprim tolerance conferred by Lon deficiency could be attributed to exaggerated drug efflux. Hyperfilamentation explained the phenotypic effects of Lon deficiency at high drug levels, while altered drug target stability and expression levels explained the effects of Lon protease on the phenotypes of trimethoprim-resistant alleles of DHFR. Dissection of these phenomena at the molecular level demonstrates the multipronged influence that master regulators of bacterial physiology have on AMR and underlines the importance of the genetic and environmental contexts in understanding the phenotypes of drug-resistant bacteria (represented schematically in Fig. 7).
FIG 7.
Schematic representation of 3 different molecular mechanisms that explain the highly contingent phenotypes of Lon deficiency in E. coli challenged with trimethoprim. X-ray crystal structures or electron micrographic structures of E. coli proteins, or of homologs from other bacteria, are used to represent the various molecular players involved (Lon PDB, 1RR9; SulA PDB, 1OFT; MarA-DNA PDB, 1BL0; FtsZ PDB, 5IMJ; AcrABZ-TolC PDB, 5O66; DHFR PDB, 7DFR). Lon contributes to low trimethoprim tolerance by regulating the levels of MarA and, in turn, of the AcrAB-TolC efflux system. Lon regulates susceptibility to high levels of trimethoprim by modulating the levels of SulA, an inhibitor of the FtsZ cytokinesis protein. Lon also degrades misfolded DHFR, thus altering the steady-state expression of the protein. Trimethoprim-resistant mutants of DHFR (Tmpr DHFR; indicated with a red dot) are more likely to misfold than the wild type and hence more likely to be degraded by Lon. This activity of Lon determines the mutational potential of DHFR for evolving trimethoprim resistance. The schematic is not drawn to scale.
The information detailing the impact of Lon on intrinsic trimethoprim tolerance at sub-MIC drug levels represents the first report of its kind and identifies Lon as a novel mediator of drug tolerance in bacteria. Drug-tolerant and persistent bacteria, though not resistant to antibiotics themselves, can act as precursors to drug-resistant strains (40). The contribution of these phenotypes to AMR evolution is being increasingly appreciated in laboratory and clinical settings. Both tolerance and persistence ensure survival of a few bacteria under conditions of lethal drug pressure. Surviving bacteria can subsequently accumulate resistance-conferring mutations, thus preventing extinction. Persistence stems from within-population heterogeneities in the metabolic state or from the growth rate of bacteria (40). Mutations that enhance the proportion of persistent bacteria in a population typically map to bacterial growth modulators such as toxin-antitoxin systems (41). Tolerance, though closely related to persistence, has proven to be more complex to characterize, and a series of recent studies have defined drug-tolerant bacteria in terms of “time-to-kill” analyses (5, 40). In the present report, I have shown that Lon deficiency enhances survival upon trimethoprim challenge by the use of a TDtest, which is unable to distinguish between persisters and drug-tolerant bacteria. However, the effects of Lon deficiency described here are more akin to drug tolerance than persistence, even though Lon was implicated in persistence in responses to other antibiotic in earlier studies (19, 42). I have drawn this conclusion on the basis of the following considerations. First, persistence phenotypes are manifested at high drug doses, i.e., in antibiotic regimes that kill actively dividing bacteria but do not affect metabolically dormant bacteria (40). This would imply that if Lon deficiency represented a “high-persistence” mutation, it would be beneficial only at high drug doses (i.e., doses higher than the MIC). In the present study, however, Lon deficiency was found to be beneficial only at sub-MIC trimethoprim levels. Second, earlier studies attributed Lon-dependent persistence, in large part, to SulA levels (42). In the present study, enhanced trimethoprim tolerance of Lon-deficient E. coli was found to be due to drug efflux and independent of SulA.
The identification of Lon as a suppressor of drug tolerance adds a new dimension to the roles of this molecule in modulating responses to antibiotic insult. Loss of Lon activity has been selected repeatedly under laboratory conditions. For instance, E. coli bacteria selected on many antibiotics yielded strains that did not produce Lon due to insertion of the IS186 transposable element in the lon gene promoter (15, 22). In line with the data presented in the present study, earlier reports of selection of Lon-deficient E. coli under conditions of antibiotic stress indicated that the effects were also related to greater levels of drug efflux in Lon-deficient bacteria (15, 22). Thus, loss of Lon seems to be associated with a frequently encountered “first-step” mutation that allows rapid adaptation to several antibiotics, at least under laboratory conditions. Further, the fitness advantage conferred by Lon deficiency at sub-MICs of trimethoprim is relevant in the context of selection under conditions of low drug pressure. Sub-MICs of antibiotics are likely to be encountered by bacteria in natural habitats such as soil and water and are known to select for clinically relevant drug resistance (2, 3, 43). Mounting evidence now suggests that the evolutionary trajectories that sub-MIC selection can follow are different from those seen under conditions of high drug concentrations. In at least two different cases, sub-MIC drug pressures were also shown to select for very high-level drug resistance (2, 3). The results obtained in the present study mirror these results conceptually, as Lon deficiency enhanced the level of trimethoprim resistance (i.e., the drug MIC) for several of the mutants tested. This would imply that Lon deficiency, once selected for, can facilitate the evolution of higher levels of drug resistance without the need for multiple resistance-conferring mutations. More importantly, the results of the present study show that in addition to modifying the phenotypes of mutants, Lon deficiency can also expand the spectrum of mutations that confer resistance. In conjunction with other effects of Lon deficiency reported earlier, such as genomic instability (15), the expanded repertoire of resistant mutants is likely to potentiate resistance evolution. Though this study was restricted to examination of DHFR and trimethoprim, the idea of the potential of a gene for resistance acquisition has been explored for other systems as well. For instance, the mutational potential for resistance of β-lactamase enzymes has been defined in the past using directed evolution (44, 45). In those studies, the genetic background of bacteria was not taken into consideration. The results obtained in the present study point out the importance of genetic background in truly being able to define the potential for drug resistance of a gene or an organism.
The interaction between Lon and unstable mutants of DHFR was demonstrated earlier (28). Bershtein et al. (28) found that destabilizing mutations can trap DHFR in the molten globule state, which may then be either folded into a near-native conformation by the action of chaperones or degraded by proteases such as Lon. This tussle between chaperones and proteases was found to influence the availability of DHFR in bacterial cells and hence to modulate organismal fitness (28). The findings of the present study relate that idea to stability-function trade-offs occurring during the evolution of drug resistance in bacteria, now an established phenomenon (29, 31, 46–48). I propose that destabilization of DHFR due to mutations that confer trimethoprim resistance results in its degradation, presumably by Lon itself, which provides a simple mechanism explaining why certain mutations are able to confer trimethoprim resistance in only Lon-deficient E. coli. However, it must be noted that wild-type DHFR is immune to Lon activity. Indeed, data from this study and from proteomics studies on Lon-deficient E. coli (17) have shown that Lon does not use wild-type DHFR as a substrate in vivo. Taken together, these results indicate that adaptive mutations can convert proteins like DHFR into substrates of quality control proteases such as Lon. It is not unreasonable to envisage, therefore, that Lon may impose similar constraints on the fitness of drug-resistant mutations in other drug targets as well. More generally, this observation presents a relevant system to investigate the adaptive costs of quality control mechanisms in living systems. Quality control mechanisms function to minimize errors in information transfer. Mutations, for instance, are prevented during DNA replication by repair enzymes, and mistranslation of proteins is prevented by a number of checks and balances imposed by the ribosome itself. However, both these processes are expected to retard adaptation, as they strive to minimize variation, which is the substrate for evolution. In the context of AMR evolution, loss of DNA repair machinery (8, 49, 50) and error-prone translation (51–53) facilitate survival in the face of antibiotics, eventually allowing the emergence of drug resistance. Lon plays the role of posttranslational quality control in bacterial cells, clearing misfolded proteins. The finding that Lon deficiency widens the mutational scope for resistance is therefore analogous to observations from studies of other quality control systems. Other components of posttranslational quality control have already been implicated in shaping the evolutionary dynamics of living organisms. Molecules that are part of the cell’s protein quality control machinery, in particular, chaperones like Hsp90, are thought of as capacitors of evolution (54). Hsp90 is known to buffer the phenotypes of mutations, and its loss alters adaptation to novel environments in many different systems by allowing new phenotypes to be expressed (54). Though Lon may not itself be a capacitor of evolution, it may act in concert with cellular chaperones to similarly modulate evolution to novel environments. It will be instructive to look at how Lon and other quality control molecules participate in drug resistance evolution in the future.
In summary, antibiotic resistance evolution, though highly directional, can be influenced significantly by the environmental and genetic context. Master regulators such as Lon modify the evolutionary outcomes and propensity for AMR evolution. One of the important findings that emerged from this study is that the effects of Lon deficiency in trimethoprim-sensitive and -resistant backgrounds, though phenomenologically similar, were mechanistically different. Understanding the effects of master regulator molecules on the outcomes of resistance evolution hence must take into account their diverse activities in bacterial cells. It remains to be seen how general these observations are in the context of resistance and tolerance to other antibiotics and whether other master regulators in bacteria impinge similarly on the evolutionary trajectories of bacteria under drug pressure.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
Strains used and generated in this study are listed in Table 1. All single-gene knockouts were generated by gene replacement using the lambda red recombination system (55) and the oligonucleotide primers listed in Table 2. Double knockouts were generated using P1 transduction. Strains were routinely cultured in Luria-Bertani (LB) broth or on LB agar at 37°C. Liquid cultures were grown with shaking at 180 to 200 rpm. Kanamycin (25 μg/ml), chloramphenicol (25 μg/ml), or ampicillin (100 μg/ml) was added as needed. For E. coli ΔfolA, thymidine was added to the growth medium at 100 μg/ml. The growth medium was supplemented with trimethoprim (Sigma-Aldrich-Merck) as needed at the appropriate concentration from a stock solution (usually 10 to 20 mg/ml) made in dimethyl sulfoxide (DMSO).
TABLE 1.
List of strains used in this study
| Strain | Details | Source or reference |
|---|---|---|
| E. coli K-12 MG1655 | Wild type | Kind gift from Sutirth Dey, IISER (Pune, India)a |
| E. coli Δlon::kan | Lon-deficient derivative of wild type | This study |
| E. coli folAW30G | Spontaneous trimethoprim-resistant isolate derived from wild type harboring the Trp30Gly mutations in its genomic copy of folA | 31 |
| E. coli folAW30G Δlon::kan | Lon-deficient derivative of E. coli folAW30G | This study |
| E. coli ΔsulA::cat | SulA-deficient derivative of wild type | This study |
| E. coli ΔsulA::cat Δlon::kan | Lon-deficient derivative of E. coli ΔsulA::cat | This study |
| E. coli ΔacrB::cat | AcrB-deficient derivative of wild type | This study |
| E. coli ΔacrB::cat Δlon::kan | Lon-deficient derivative of E. coli ΔacrB::cat | This study |
| E. coli ΔfolA::kan | DHFR-deficient derivative of wild type | Kind gift from Peter Wright, Scripps Research Institute (57) |
IISER, Indian Institute of Science Education and Research.
TABLE 2.
List of oligonucleotide primers used in this study
| Name | Sequence (5′–3′) |
|---|---|
| Oligonucleotides for gene knockouts | |
| Lon_KO_pKD4_fwd | GTGAAGCACAGTCGTGTCATCTGATTACCTGGCGGAAATTAAACTAAGAGTGTGTAGGCTGGAGCTGCTTC |
| Lon_KO_pKD4_rev | GATCGGCAATTACGTTGTCAGGAATCTCTTCCAGATCGCGTTTATTTTCGCATATGAATATCCTCCTTA |
| SulA_up_pKD3_fwd | AAAAGTTCCAGGATTAATCCTAAATTTACTTAATGATACAAATTAGAGTGTGTGTAGGCTGGAGCTGCTTC |
| SulA_down_pKD3_rev | GGATGTACTGTACATCCATACAGTAACTCACAGGGGCTGGATTGATTATGCATATGAATATCCTCCTTA |
| AcrB_up_pKD3_fwd | TTACGCGGCCTTAGTGATTACACGTTGTATCAATGATGATCGACAGTATGCATATGAATATCCTCCTTA |
| AcrB_down_pKD3_rev | TCAGCCTGAACAGTCCAAGTCTTAACTTAAACAGGAGCCGTTAAGACATGTGTGTAGGCTGGAGCTGCTTC |
| Oligonucleotides for site-directed mutagenesis | |
| EcfolA_P21T_BglII_f | CGCCATGACGTGGAACCTGCCTGCAGATCTCGCCTGGTTTAAACGC |
| EcfolA_P21A_NaeI_f | CGCCATGGCGTGGAACCTGCCGGCCGATCTCGCCTGGTTTAAACGC |
| EcfolA_P21S_PvuII_f | CGCCATGTCGTGGAACCTGCCAGCTGATCTCGCCTGGTTTAAACGC |
| EcfolA_P21R_BglII_f | CGCCATGCGGTGGAACCTGCCTGCAGATCTCGCCTGGTTTAAACGC |
| folA_W30S_NheI_Rev | GGTGTTGCGTTTAAAGCTAGCGAGATCGGCAGGCAGG |
| folA_W30S_NheI_fwd | CCTGCCTGCCGATCTCGCTAGCTTTAAACGCAACACC |
| folA I94V AvaI F | GTGACGTACCCGAGATCATGGTGGTTGGCGGCGGTCGCG |
| folA I94F AvaI F | GTGACGTACCCGAGATCATGGTGTTTGGCGGCGGTCGCG |
| folA I94S AvaI F | GTGACGTACCCGAGATCATGGTGAGTGGCGGCGGTCGCG |
| folA I94N HpaI F | CCAGAAATCATGGTTAACGGCGGCGGTCGCGTTTATG |
| folA I94T AgeI F | CCAGAAATCATGGTGACCGGTGGCGGTCGCGTTTATG |
| folA I94M AvaI F | GTGACGTACCCGAGATCATGGTGATGGGCGGCGGTCGCG |
Plasmids used in this study are listed in Table 3. For ectopic plasmid-based expression of DHFR, previously generated plasmid pPRO-folA (31) or its mutant derivatives were used. DHFR and its mutants were expressed from this plasmid at levels that were detectable (by immunoblotting) without the addition of IPTG (isopropyl-β-d-thiogalactopyranoside); hence, all analyses were conducted without the addition of inducer in the growth medium. Site-directed mutagenesis was carried out as described previously (31) using primers listed in Table 2. All generated mutants were confirmed by restriction digestion and sequencing. Plasmid pBAD33-lon (38) was a gift from Robert Sauer (Addgene plasmid catalog no. 22145; http://n2t.net/addgene:22145; RRID: Addgene_22145).
TABLE 3.
List of plasmids used in this study
| Name | Description | Source or reference |
|---|---|---|
| pPRO-folA | E. coli folA gene cloned into pPRO-Ex-Htb plasmid for heterologous expression (adds a hexa-His tag at the N terminus) | 31 |
| pPRO-folA P21L/P21Q/P21T/P21R/P21S | Derivative of pPRO-folA expressing Pro21 mutants of DHFR | 31, this study |
| pPRO-folA W30R/W30G/W30L/W30S/W30C | Derivative of pPRO-folA expressing Trp30 mutants of DHFR | 31, this study |
| pPRO-folA I94V/I94F/I94L/I94S/I94N/I94T/I94M | Derivative of pPRO-folA expressing Ile94 mutants of DHFR | 31, this study |
| pBAD33-Lon | Plasmid for expression of Lon protease from an arabinose inducible promoter | 38 |
| pKD46 | Plasmid for expression of lambda red recombination proteins | 55 |
| pKD4 | Plasmid used as the template for amplification of kanamycin resistance cassette | 55 |
| pKD3 | Plasmid used as the template for amplification of chloramphenicol resistance cassette | 55 |
Measuring trimethoprim resistance and tolerance.
For wild-type E. coli and E. coli folAW30G and their Δlon derivatives, MIC and IC50 (concentration needed for 50% growth inhibition) values were determined in liquid cultures using the following protocol. In the wells of a 96-well plate, a 3-fold-dilution series of trimethoprim (100 μg/ml to 7.5 ng/ml) was set up using serial dilution with LB as the dilutant. Saturated cultures of appropriate strains were diluted 100-fold for each trimethoprim concentration such that the final volume in each well was 100 μl. Wells at the periphery of the plate were filled with deionized water to prevent evaporative loss. After 15 to 18 h of incubation at 37°C, the optical density (at 600 nm) was measured in a microplate reader (Varioskan LUX multimode reader; Thermo Scientific). Growth at each trimethoprim concentration was normalized to growth in the absence of antibiotic for each strain. Normalized values were plotted against the log of trimethoprim concentration. The lowest concentration of trimethoprim at which growth was ≤10% of that seen with the control was defined as the MIC. IC50 values were determined by fitting a variable slope inhibition curve to the data presented above using GraphPad Prism software (version 6.07). All measurements were conducted at least thrice.
For determinations of drug resistance on solid medium, a standard disc diffusion assay was used. Briefly, ∼5 × 107 bacteria of the appropriate strain were spread on an LB agar plate. A sterile disc punched out of Whatman No. 3 paper was placed on the plate, and 8 μl of a trimethoprim solution of the appropriate concentration was dropped onto the disc. Once dry, the plate was closed and incubated at 37°C for 15 to 18 h. The diameter of the zone of clearance (ZOC) was measured, and the plates were photographed in a gel documentation system. For measuring drug tolerance, a TDtest was used (34) with slight modification. After obtaining a ZOC with trimethoprim, the antibiotic disc was replaced with a sterile disc and 8 μl of sterile glucose solution (20%) was dropped onto the disc. Once dry, the plate was closed and incubated at 37°C for ∼20 h and photographed.
Trimethoprim resistance conferred by plasmid-expressed DHFR or its mutants was measured as described earlier (31). Briefly, 5-μl volumes of a 10-fold-dilution series of saturated cultures of E. coli were spotted onto LB agar supplemented with various concentrations of trimethoprim (0 μg/ml to 128 μg/ml). Growth was visualized after ∼20 h of incubation at 37°C, and the maximum dilution of culture allowing visible growth was noted at each concentration of trimethoprim. MIC99 was defined as the lowest concentration of trimethoprim not allowing growth at a 100-fold dilution of culture.
To monitor the impact of high trimethoprim concentrations, mid-log-phase cultures of the appropriate strains were treated with trimethoprim (10 mg/ml; ∼14× MIC). The levels of viability of the treated cultures were monitored by serial dilution and plating at 0, 1, 4, and 20 h after addition of trimethoprim. To investigate the effects on E. coli of other antibiotics at high concentrations, 25 μg/ml ampicillin or 50 μg/ml nalidixic acid was used in a similar assay.
Calculating relative fitness.
The relative fitness levels of various strains were calculated as described previously by Lenski (56). Saturated cultures of appropriate mutant and wild-type E. coli strains were mixed 1:1 volumetrically and inoculated 1:1,000 in 3 ml of LB. Trimethoprim was added at the required concentration. Strains were allowed to compete for 24 h at 37°C with shaking at 180 to 200 rpm. Initial and final densities of competing strains was determined by plating serially diluted cultures onto LB agar (total count) and onto LB agar supplemented with kanamycin or chloramphenicol as needed. Relative fitness (w) was calculated using the following formula:
where Tf and Ti are the final and initial densities in CFU per milliliter of the test strain and Wf and Wi are the final and initial densities (in CFU per milliliter) of the wild type.
Filamentation assay.
For analyzing trimethoprim-induced filamentation, saturated cultures of appropriate strains were diluted 1:100 into fresh medium supplemented with 100 ng/ml or 400 ng/ml trimethoprim and allowed to grow at 37°C with shaking at 180 to 200 rpm. Bacteria from each of the cultures were then streaked onto glass slides using a Nichrome loop, stained with 0.1% safranin (HiMedia), and visualized using a ×100 oil immersion lens on a compound microscope (Zeiss). Multiple fields were imaged for each strain. Bacterial cell lengths were then estimated using ImageJ for at least 30 bacteria per culture. Two or three cultures were used for each strain.
Estimating expression level of DHFR.
For estimating the steady-state expression level of DHFR, 3 to 5 ml of mid-log-phase cultures of the appropriate strain were harvested by centrifugation. Bacteria were lysed in 500 μl of lysis buffer (50 mM Tris–HCl [pH 7.5]), 100 mM NaCl, 5 mM β-mercaptoethanol, 10% glycerol) in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF) by sonication. Total protein concentrations in lysates were estimated using the Bradford assay. For determination of endogenous DHFR levels, 2.5 and 5 μg lysate protein was subjected to SDS-PAGE and electroblotted onto polyvinylidene difluoride (PVDF) membranes. DHFR was visualized using immunoblotting with polyclonal anti-DHFR IgG as described earlier (31). For plasmid-based expression analyses, 1 to 5 μg lysate protein was used depending on the expression level of DHFR mutants. Western blots were quantitated using band intensities in ImageJ. Band intensities were corrected for background and then normalized as follows. For analysis of the expression level of chromosomally or plasmid-expressed wild-type or mutant DHFR, the level of band intensity corresponding to Lon-expressing E. coli was normalized to a value of 1 and the levels of expression in Lon-deficient E. coli bacteria were scaled accordingly. At least 3 independent blots were used for quantitation. For checking if DHFR Trp30Ser had an intact N terminus, lysates were prepared after growth in the presence of 500 μM IPTG. Immunoblotting was performed using antihexahistidine monoclonal antibody (Santa Cruz, catalog no. 8036) at a dilution of 1:5,000 and horseradish peroxidase-conjugated anti-mouse IgG at a dilution of 1:5,000.
In vivo DHFR stability.
The in vivo stability of DHFR was measured using a chloramphenicol chase assay. Saturated cultures of appropriate strain were diluted 1:50 in 5 ml LB, and trimethoprim was added at 1 μg/ml as needed. After 2.5 h of growth, 1 ml was harvested by centrifugation. The cell pellet was resuspended in a suitable volume (usually 100 to 200 μl) of 1× Laemmli sample loading buffer and heated at 95°C for 5 to 10 min. Chloramphenicol was added to the remaining culture at a final concentration of 12.5 μg/ml. After 15, 45, and 90 min of incubation at 37°C, 1-ml aliquots were harvested and prepared for SDS-PAGE as described above. Appropriate volumes of samples were then subjected to SDS-PAGE and immunoblotting for DHFR as described previously. Sample volumes were adjusted such that similar band intensities were obtained in lon+ and Δlon bacteria before the addition of chloramphenicol.
Supplementary Material
ACKNOWLEDGMENTS
I acknowledge the Department of Science and Technology, Government of India, INSPIRE Fellowship, for financial support.
I thank the anonymous reviewers of the manuscript for valuable comments and insights.
I declare that I have no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Harmand N, Gallet R, Jabbour-Zahab R, Martin G, Lenormand T. 2017. Fisher’s geometrical model and the mutational patterns of antibiotic resistance across dose gradients. Evolution 71:23–37. doi: 10.1111/evo.13111. [DOI] [PubMed] [Google Scholar]
- 2.Matange N, Hegde S, Bodkhe S. 2019. Adaptation through lifestyle switching sculpts the fitness landscape of evolving populations: implications for the selection of drug-resistant bacteria at low drug pressures. Genetics 211:1029–1044. doi: 10.1534/genetics.119.301834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wistrand-Yuen E, Knopp M, Hjort K, Koskiniemi S, Berg OG, Andersson DI. 2018. Evolution of high-level resistance during low-level antibiotic exposure. Nat Commun 9:1599. doi: 10.1038/s41467-018-04059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J, Dong Y, Zhao X, Lee S, Amin A, Ramaswamy S, Domagala J, Musser JM, Drlica K. 2000. Selection of antibiotic-resistant bacterial mutants: allelic diversity among fluoroquinolone-resistant mutations. J Infect Dis 182:517–525. doi: 10.1086/315708. [DOI] [PubMed] [Google Scholar]
- 5.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 6.Chait R, Palmer AC, Yelin I, Kishony R. 2016. Pervasive selection for and against antibiotic resistance in inhomogeneous multistress environments. Nat Commun 7:10333. doi: 10.1038/ncomms10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michel JB, Yeh PJ, Chait R, Moellering RC Jr, Kishony R. 2008. Drug interactions modulate the potential for evolution of resistance. Proc Natl Acad Sci U S A 105:14918–14923. doi: 10.1073/pnas.0800944105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garibyan L, Huang T, Kim M, Wolff E, Nguyen A, Nguyen T, Diep A, Hu K, Iverson A, Yang H, Miller JH. 2003. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair (Amst) 2:593–608. doi: 10.1016/s1568-7864(03)00024-7. [DOI] [PubMed] [Google Scholar]
- 9.Weigand MR, Sundin GW. 2012. General and inducible hypermutation facilitate parallel adaptation in Pseudomonas aeruginosa despite divergent mutation spectra. Proc Natl Acad Sci U S A 109:13680–13685. doi: 10.1073/pnas.1205357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barroso KCM, Previato-Mello M, Batista BB, Batista JH, da Silva Neto JF. 2018. EmrR-dependent upregulation of the efflux pump EmrCAB contributes to antibiotic resistance in Chromobacterium violaceum. Front Microbiol 9:2756. doi: 10.3389/fmicb.2018.02756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frimodt-Moller J, Lobner-Olesen A. 2019. Efflux-pump upregulation: from tolerance to high-level antibiotic resistance? Trends Microbiol 27:291–293. doi: 10.1016/j.tim.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Chung CH, Goldberg AL. 1981. The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc Natl Acad Sci U S A 78:4931–4935. doi: 10.1073/pnas.78.8.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottesman S, Halpern E, Trisler P. 1981. Role of sulA and sulB in filamentation by lon mutants of Escherichia coli K-12. J Bacteriol 148:265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizusawa S, Gottesman S. 1983. Protein degradation in Escherichia coli: the lon gene controls the stability of sulA protein. Proc Natl Acad Sci U S A 80:358–362. doi: 10.1073/pnas.80.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicoloff H, Perreten V, Levy SB. 2007. Increased genome instability in Escherichia coli lon mutants: relation to emergence of multiple-antibiotic-resistant (Mar) mutants caused by insertion sequence elements and large tandem genomic amplifications. Antimicrob Agents Chemother 51:1293–1303. doi: 10.1128/AAC.01128-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winther KS, Gerdes K. 2009. Ectopic production of VapCs from Enterobacteria inhibits translation and trans-activates YoeB mRNA interferase. Mol Microbiol 72:918–930. doi: 10.1111/j.1365-2958.2009.06694.x. [DOI] [PubMed] [Google Scholar]
- 17.Arends J, Griego M, Thomanek N, Lindemann C, Kutscher B, Meyer HE, Narberhaus F. 2018. An integrated proteomic approach uncovers novel substrates and functions of the Lon protease in Escherichia coli. Proteomics 18:e1800080. doi: 10.1002/pmic.201800080. [DOI] [PubMed] [Google Scholar]
- 18.Chowdhury N, Kwan BW, Wood TK. 2016. Persistence increases in the absence of the alarmone guanosine tetraphosphate by reducing cell growth. Sci Rep 6:20519. doi: 10.1038/srep20519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harms A, Fino C, Sorensen MA, Semsey S, Gerdes K. 2017. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. mBio 8:e01964-17. doi: 10.1128/mBio.01964-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffith KL, Shah IM, Wolf RE Jr. 2004. Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol Microbiol 51:1801–1816. doi: 10.1046/j.1365-2958.2003.03952.x. [DOI] [PubMed] [Google Scholar]
- 21.Bhaskarla C, Das M, Verma T, Kumar A, Mahadevan S, Nandi D. 2016. Roles of Lon protease and its substrate MarA during sodium salicylate-mediated growth reduction and antibiotic resistance in Escherichia coli. Microbiology 162:764–776. doi: 10.1099/mic.0.000271. [DOI] [PubMed] [Google Scholar]
- 22.Nicoloff H, Andersson DI. 2013. Lon protease inactivation, or translocation of the lon gene, potentiate bacterial evolution to antibiotic resistance. Mol Microbiol 90:1233–1248. doi: 10.1111/mmi.12429. [DOI] [PubMed] [Google Scholar]
- 23.Breidenstein EB, Janot L, Strehmel J, Fernandez L, Taylor PK, Kukavica-Ibrulj I, Gellatly SL, Levesque RC, Overhage J, Hancock RE. 2012. The Lon protease is essential for full virulence in Pseudomonas aeruginosa. PLoS One 7:e49123. doi: 10.1371/journal.pone.0049123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Culp E, Wright GD. 2017. Bacterial proteases, untapped antimicrobial drug targets. J Antibiot (Tokyo) 70:366–377. doi: 10.1038/ja.2016.138. [DOI] [PubMed] [Google Scholar]
- 25.Frase H, Hudak J, Lee I. 2006. Identification of the proteasome inhibitor MG262 as a potent ATP-dependent inhibitor of the Salmonella enterica serovar Typhimurium Lon protease. Biochemistry 45:8264–8274. doi: 10.1021/bi060542e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takaya A, Suzuki M, Matsui H, Tomoyasu T, Sashinami H, Nakane A, Yamamoto T. 2003. Lon, a stress-induced ATP-dependent protease, is critically important for systemic Salmonella enterica serovar typhimurium infection of mice. Infect Immun 71:690–696. doi: 10.1128/iai.71.2.690-696.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takaya A, Tomoyasu T, Tokumitsu A, Morioka M, Yamamoto T. 2002. The ATP-dependent lon protease of Salmonella enterica serovar Typhimurium regulates invasion and expression of genes carried on Salmonella pathogenicity island 1. J Bacteriol 184:224–232. doi: 10.1128/jb.184.1.224-232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bershtein S, Mu W, Serohijos AW, Zhou J, Shakhnovich EI. 2013. Protein quality control acts on folding intermediates to shape the effects of mutations on organismal fitness. Mol Cell 49:133–144. doi: 10.1016/j.molcel.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodrigues JV, Bershtein S, Li A, Lozovsky ER, Hartl DL, Shakhnovich EI. 2016. Biophysical principles predict fitness landscapes of drug resistance. Proc Natl Acad Sci U S A 113:E1470–E1478. doi: 10.1073/pnas.1601441113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brolund A, Sundqvist M, Kahlmeter G, Grape M. 2010. Molecular characterisation of trimethoprim resistance in Escherichia coli and Klebsiella pneumoniae during a two year intervention on trimethoprim use. PLoS One 5:e9233. doi: 10.1371/journal.pone.0009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matange N, Bodkhe S, Patel M, Shah P. 2018. Trade-offs with stability modulate innate and mutationally acquired drug resistance in bacterial dihydrofolate reductase enzymes. Biochem J 475:2107–2125. doi: 10.1042/BCJ20180249. [DOI] [PubMed] [Google Scholar]
- 32.Palmer AC, Toprak E, Baym M, Kim S, Veres A, Bershtein S, Kishony R. 2015. Delayed commitment to evolutionary fate in antibiotic resistance fitness landscapes. Nat Commun 6:7385. doi: 10.1038/ncomms8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toprak E, Veres A, Michel JB, Chait R, Hartl DL, Kishony R. 2011. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet 44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gefen O, Chekol B, Strahilevitz J, Balaban NQ. 2017. TDtest: easy detection of bacterial tolerance and persistence in clinical isolates by a modified disk-diffusion assay. Sci Rep 7:41284. doi: 10.1038/srep41284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bershtein S, Serohijos AW, Bhattacharyya S, Manhart M, Choi JM, Mu W, Zhou J, Shakhnovich EI. 2015. Protein homeostasis imposes a barrier on functional integration of horizontally transferred genes in bacteria. PLoS Genet 11:e1005612. doi: 10.1371/journal.pgen.1005612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwamoto M, Bjorklund T, Lundberg C, Kirik D, Wandless TJ. 2010. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem Biol 17:981–988. doi: 10.1016/j.chembiol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. 2001. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell 7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 38.Gur E, Sauer RT. 2008. Recognition of misfolded proteins by Lon, a AAA(+) protease. Genes Dev 22:2267–2277. doi: 10.1101/gad.1670908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schenk MF, de Visser JA. 2013. Predicting the evolution of antibiotic resistance. BMC Biol 11:14. doi: 10.1186/1741-7007-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 41.Germain E, Castro-Roa D, Zenkin N, Gerdes K. 2013. Molecular mechanism of bacterial persistence by HipA. Mol Cell 52:248–254. doi: 10.1016/j.molcel.2013.08.045. [DOI] [PubMed] [Google Scholar]
- 42.Theodore A, Lewis K, Vulic M. 2013. Tolerance of Escherichia coli to fluoroquinolone antibiotics depends on specific components of the SOS response pathway. Genetics 195:1265–1276. doi: 10.1534/genetics.113.152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gullberg E, Cao S, Berg OG, Ilback C, Sandegren L, Hughes D, Andersson DI. 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 7:e1002158. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barlow M, Hall BG. 2002. Predicting evolutionary potential: in vitro evolution accurately reproduces natural evolution of the tem beta-lactamase. Genetics 160:823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salipante SJ, Hall BG. 2003. Determining the limits of the evolutionary potential of an antibiotic resistance gene. Mol Biol Evol 20:653–659. doi: 10.1093/molbev/msg074. [DOI] [PubMed] [Google Scholar]
- 46.Abriata LA, Salverda ML, Tomatis PE. 2012. Sequence-function-stability relationships in proteins from datasets of functionally annotated variants: the case of TEM beta-lactamases. FEBS Lett 586:3330–3335. doi: 10.1016/j.febslet.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 47.Olabode AS, Kandathil SM, Lovell SC, Robertson DL. 2017. Adaptive HIV-1 evolutionary trajectories are constrained by protein stability. Virus Evol 3:vex019. doi: 10.1093/ve/vex019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas VL, McReynolds AC, Shoichet BK. 2010. Structural bases for stability-function tradeoffs in antibiotic resistance. J Mol Biol 396:47–59. doi: 10.1016/j.jmb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Norton MD, Spilkia AJ, Godoy VG. 2013. Antibiotic resistance acquired through a DNA damage-inducible response in Acinetobacter baumannii. J Bacteriol 195:1335–1345. doi: 10.1128/JB.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ragheb MN, Thomason MK, Hsu C, Nugent P, Gage J, Samadpour AN, Kariisa A, Merrikh CN, Miller SI, Sherman DR, Merrikh H. 2019. Inhibiting the evolution of antibiotic resistance. Mol Cell 73:157–165.e5. doi: 10.1016/j.molcel.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bratulic S, Toll-Riera M, Wagner A. 2017. Mistranslation can enhance fitness through purging of deleterious mutations. Nat Commun 8:15410. doi: 10.1038/ncomms15410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaudhuri S, Li L, Zimmerman M, Chen Y, Chen YX, Toosky MN, Gardner M, Pan M, Li YY, Kawaji Q, Zhu JH, Su HW, Martinot AJ, Rubin EJ, Dartois VA, Javid B. 28 August 2018, posting date Kasugamycin potentiates rifampicin and limits emergence of resistance in Mycobacterium tuberculosis by specifically decreasing mycobacterial mistranslation. Elife doi: 10.7554/eLife.36782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Javid B, Sorrentino F, Toosky M, Zheng W, Pinkham JT, Jain N, Pan M, Deighan P, Rubin EJ. 2014. Mycobacterial mistranslation is necessary and sufficient for rifampicin phenotypic resistance. Proc Natl Acad Sci U S A 111:1132–1137. doi: 10.1073/pnas.1317580111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Queitsch C, Sangster TA, Lindquist S. 2002. Hsp90 as a capacitor of phenotypic variation. Nature 417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 55.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiser MJ, Lenski RE. 2015. A comparison of methods to measure fitness in Escherichia coli. PLoS One 10:e0126210. doi: 10.1371/journal.pone.0126210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhabha G, Ekiert DC, Jennewein M, Zmasek CM, Tuttle LM, Kroon G, Dyson HJ, Godzik A, Wilson IA, Wright PE. 2013. Divergent evolution of protein conformational dynamics in dihydrofolate reductase. Nat Struct Mol Biol 20:1243–1249. doi: 10.1038/nsmb.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.