Large quantities of recombinantly overproduced [Fe-S] cluster-containing proteins are necessary for their in-depth biochemical characterization. Commercially available E. coli strain BL21(DE3) and its derivatives have a mutation that inactivates the function of one of the two native pathways (Suf pathway) responsible for cluster biogenesis. Correction of the mutation, combined with sequence changes that elevate Suf protein levels, can increase yield and cluster occupancy of [Fe-S] cluster-containing enzymes, facilitating the biochemical analysis of this fascinating group of proteins.
KEYWORDS: Fe-S protein overproduction, E. coli B, iron-sulfur biogenesis, Suf pathway, Fe-S cluster biogenesis, Fe-S protein overexpression, Suf
ABSTRACT
Structural and spectroscopic analysis of iron-sulfur [Fe-S] cluster-containing proteins is often limited by the occupancy and yield of recombinantly produced proteins. Here we report that Escherichia coli BL21(DE3), a strain routinely used to overproduce [Fe-S] cluster-containing proteins, has a nonfunctional Suf pathway, one of two E. coli [Fe-S] cluster biogenesis pathways. We confirmed that BL21(DE3) and commercially available derivatives carry a deletion that results in an in-frame fusion of sufA and sufB genes within the sufABCDSE operon. We show that this fusion protein accumulates in cells but is inactive in [Fe-S] cluster biogenesis. Restoration of an intact Suf pathway combined with enhanced suf operon expression led to a remarkable (∼3-fold) increase in the production of the [4Fe-4S] cluster-containing BchL protein, a key component of the dark-operative protochlorophyllide oxidoreductase complex. These results show that this engineered “SufFeScient” derivative of BL21(DE3) is suitable for enhanced large-scale synthesis of an [Fe-S] cluster-containing protein.
IMPORTANCE Large quantities of recombinantly overproduced [Fe-S] cluster-containing proteins are necessary for their in-depth biochemical characterization. Commercially available E. coli strain BL21(DE3) and its derivatives have a mutation that inactivates the function of one of the two native pathways (Suf pathway) responsible for cluster biogenesis. Correction of the mutation, combined with sequence changes that elevate Suf protein levels, can increase yield and cluster occupancy of [Fe-S] cluster-containing enzymes, facilitating the biochemical analysis of this fascinating group of proteins.
INTRODUCTION
Iron-sulfur [Fe-S] proteins are integral to the activity of numerous biological processes, including respiration, nitrogen fixation, photosynthesis, DNA replication and repair, RNA modification, and gene regulation (1–3). In Escherichia coli K-12, there are two multiprotein systems, Isc and Suf, dedicated to the biosynthesis of various [Fe-S] clusters and their incorporation into 140 known [Fe-S] proteins (4–8). The Isc system is encoded by the isc operon, composed of the iscRSUA-hscBA-fdx-iscX genes (Fig. 1a). The Suf system is encoded by its cognate sufABCDSE (suf) operon (Fig. 1a). E. coli strains carrying defects in both systems are not viable due to a nonfunctional isoprenoid biosynthetic pathway, which relies on two [Fe-S] enzymes (9–11), highlighting the significance of these [Fe-S] cluster biogenesis systems for essential life processes (12). However, the Isc and Suf systems display some functional redundancy, as cells lacking only one system remain viable. Nevertheless, individual enzyme components of the two systems are not interchangeable, reinforcing that the scaffolds for building [Fe-S] clusters are functionally different (13, 14). Further, little is known about the specificity of these biogenesis pathways for particular client proteins, with some cases driven by protein levels and/or environmental conditions (15). Under normal growth conditions, the Isc system is thought to play the major role in [Fe-S] cluster biogenesis, but under conditions of stress, such as oxidative stress or iron-limiting conditions, the Suf system is reported to assume a greater role (4, 6, 16). Interestingly, some bacteria, archaea, and plant plastids contain only the Suf machinery, serving as the sole [Fe-S] cluster biogenesis machinery (4–7, 15).
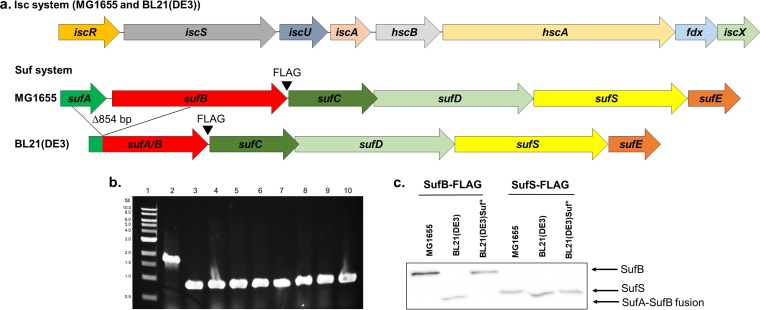
FIG 1.
An in-frame deletion between sufA and sufB renders the suf operon inactive in BL21(DE3). (a) Diagram of the isc and suf operons present in MG1655 and BL21(DE3); BL21(DE3) has an 854-bp deletion in the suf operon resulting in an in-frame fusion of the sufA and sufB genes. (b) The presence of the 854-bp deletion was tested in commercial lineages of BL21(DE3) using PCR analysis. Lane 2 shows the expected 1,641-bp product from E. coli K-12 MG1655. Lanes 3 through 10 show the 787-bp PCR product predicted from the 854-bp deletion present in strains BL21(DE3), NiCo21(DE3), Lemo21(DE3), C41(DE3), Rosetta2(DE3)pLysS, BLR(DE3)pLysS, BL21(DE3)Ai, and BL21(DE3)codon plus. (c) Western blot analysis using an anti-FLAG antibody revealed the production of full-length SufB protein in MG1655 and in strain BL21(DE3)Suf+, in which the sufA and sufB genes were properly restored. Full-length SufS protein was present in all strains.
To accelerate biochemical studies of [Fe-S] proteins, genes encoding proteins of interest are often heterologously expressed in engineered E. coli strains designed for overproduction of proteins. A major challenge in the field is to obtain large enough quantities of proteins at high concentrations that are also maximally occupied with [Fe-S] clusters (17) to enable spectroscopic and structural studies. Increasing the level of the housekeeping Isc pathway imparts variable improvement in [Fe-S] cluster protein yields (17–20). However, to our knowledge, a similar approach has not been examined for the Suf pathway despite it being the sole pathway for [Fe-S] biogenesis in many organisms.
A commonly used strain for [Fe-S] protein overproduction is E. coli BL21(DE3) or its derivatives. The ancestry of the parent strain for the modern-day BL21(DE3) can be traced back to E. coli B strains established by Delbrück and Luria in the 1920s (21). The sequence of the BL21(DE3) genome, published in 2009, revealed many sequence changes compared to the sequence of another E. coli B strain, REL606 (21–23). Among these differences was an in-frame deletion between sufA and sufB within the suf operon, encoding the Suf [Fe-S] biogenesis pathway. Here we show that BL21(DE3) is defective for Suf-dependent [Fe-S] biogenesis, and we corrected the deletion with sequences found in E. coli K-12. We also tested commercially available BL21(DE3) derivatives to determine if they carry the same deletion of sufAB, which was suggested to arise from UV treatment early in the lineage of BL21 (22). By altering the promoter sequences of the corrected allele in BL21(DE3), we developed a strain with increased levels of the Suf pathway and tested whether it improved the yield of [Fe-S] proteins. This strain may be of general use in applications that require overproduction of [Fe-S] proteins for structural and spectroscopic studies in which large quantities of protein are required.
RESULTS
Confirming the in-frame partial deletion of sufA and sufB in BL21(DE3) and in commercial derivatives.
Previous sequencing of the BL21(DE3) genome indicated that it contains a genomic deletion encompassing portions of sufA, which functions in delivering [Fe-S] clusters to apoproteins, and sufB, which acts as a scaffold for building [Fe-S] clusters (22). PCR amplification of the suf operon from our laboratory stock of BL21(DE3) using primers flanking the two genes generated a DNA fragment that is 854 bp smaller than the size expected for intact sufA-sufB observed for the reference strain E. coli K-12 MG1655 (Fig. 1b), indicating the presence of the deletion. We also tested whether other commercially available strain derivatives of BL21(DE3) have a similar deletion. PCR amplification of the sufA-sufB genes from 7 different commercial strains [Ni-Co21(DE3), Lemo21(DE3), C41(DE3), Rosetta2(DE3)pLysS, BLR(DE3)pLysS, BL21(DE3)Ai, and BL21(DE3)codon plus] revealed the same 854-bp deletion in the sufA-sufB genes as observed for the parent BL21(DE3) (Fig. 1b).
The 854-bp deletion within the suf operon renders the Suf pathway nonfunctional.
DNA sequencing of sufA and sufB of BL21(DE3) also confirmed the same nucleotide deletion boundaries within sufA and sufB as reported previously (22). Comparison of the sufABCDSE operon DNA sequences from E. coli K-12 strain MG1655 and E. coli B strain BL21(DE3) indicated that the 854-bp in-frame deletion within the BL21(DE3) sufABCDSE operon encompassed the last 79 codons and the first 202 codons of the sufA and sufB coding sequences, respectively, generating a predicted SufA/B fusion protein of ∼37 kDa (Fig. 1a). A FLAG epitope was engineered at the C-terminal end of SufB to test whether this fusion protein accumulated in cells by Western blotting using an anti-FLAG antibody. In the BL21(DE3) strain background, a protein that was smaller than that present in the reference strain MG1655 was detected, consistent with the size of the predicted SufA/B fusion protein (Fig. 1c). This deletion had no effect on downstream sufS expression since similar levels of FLAG-tagged SufS were observed for MG1655 and BL21(DE3) (Fig. 1c).
Activity of this mutant fusion protein was tested using P1 vir to transduce into BL21(DE3) a ΔiscSUA-hscBA-fdx::kan allele which requires a functional Suf pathway for growth (24). No BL21(DE3) derivatives were recovered, suggesting that this fusion protein, and consequently the Suf pathway, is nonfunctional in this strain. To quantitatively demonstrate this, we genetically linked the ΔiscSUA-hscBA-fdx::kan allele to a gua-26::Tn10 (tetracycline-resistant [Tetr]) allele and established the cotransduction linkage between these two markers upon their subsequent transduction into MG1655 and BL21(DE3). After initially selecting for transductants on tetracycline-containing medium and then screening ≥50 of these colonies for kanamycin resistance (Kanr), we observed that 38% of the MG1655 transductants exhibited Kanr. In contrast, none of the BL21(DE3) transductants were Kanr, thus providing further evidence of a nonfunctional Suf pathway in this strain.
A derivative of BL21(DE3) was then constructed in which the mutant suf operon was replaced with an intact suf operon from MG1655 using P1 vir transduction. Instead of the fusion protein observed in BL21(DE3), this genetically restored BL21(DE3)Suf+ strain (PK13235) generates full-length SufB, as confirmed by Western blotting (Fig. 1c).
Designing a functional Suf-containing strain for [Fe-S] protein production.
Since our and other laboratories routinely utilize BL21(DE3) to overproduce recombinant [Fe-S] cluster-containing proteins, we tested whether restoring the Suf system would improve the yield of recombinant [Fe-S] cluster-containing proteins. Because the suf operon is repressed by the transcriptional regulator Fur under standard growth conditions (6, 14), we constructed a variant of BL21(DE3)Suf+ that also contains a mutated Fur binding site (here designated Suf++) within the sufA promoter region. This mutation increases suf expression at least 4-fold under aerobic conditions (24). Thus, we expect this BL21(DE3)Suf++ strain (PK11466) to have enhanced levels of the Suf machinery, in addition to functional SufA and SufB proteins. In contrast to the case with BL21(DE3), viable colonies of BL21(DE3)Suf+ and BL21(DE3)Suf++ were recovered upon deletion of iscSUA-hscBA-fdx, indicating that the Suf pathway is indeed functional in these two strains.
To establish that [Fe-S] cluster biogenesis by the Isc pathway was not perturbed in BL21(DE3)Suf++ due to elevated levels of the Suf machinery, we monitored activity of ectopically expressed FNR, a [4Fe-4S] cluster-containing transcription factor that was previously shown to acquire its cluster primarily from the Isc pathway (25). Under anaerobic growth conditions, FNR activated transcription of a chromosomal PydfZ-lacZ reporter to similar levels (∼28,000 Miller units) in strains BL21(DE3) and BL21(DE3)Suf++, indicating that upregulation of a functional Suf pathway did not further increase FNR activity. Deletion of iscSUA-hscBA-fdx in BL21(DE3)Suf++ decreased FNR activity to 25% of that of the parent strain (∼7,000 Miller units), indicating that in BL21(DE3)Suf++, the Isc pathway plays the major role in FNR [4Fe-4S] cluster biogenesis, as found with MG1655 (25).
Protein yields and [Fe-S] cluster occupancy of BchL-overproducing cultures are improved with a restored and upregulated Suf pathway.
Production and yield of recombinant [Fe-S] cluster-containing proteins were assessed in BL21(DE3)Suf+ and BL21(DE3)Suf++ to determine if the restoration and/or upregulation of the suf operon had any beneficial effects on [Fe-S] cluster biogenesis. We chose two protein complexes from the dark-operative protochlorophyllide (Pchlide) oxidoreductase (DPOR) enzyme from Rhodobacter sphaeroides, which has only the Suf system for [Fe-S] biogenesis, as a test system. DPOR catalyzes the ATP-dependent reduction of Pchlide to chlorophyllide (Chlide) in plant and photosynthetic bacterial systems under low-light or dark conditions (26, 27). DPOR is comprised of two components (Fig. 2a): an electron donor (BchL) and an electron acceptor enzyme (BchN-BchB) (28, 29). BchL exists as a homodimer stabilized by a bridging [4Fe-4S] cluster ligated by 2 cysteine residues from each monomer (29, 30). BchN and BchB exist as an α2β2 heterotetramer with two symmetric halves (Fig. 2a). Each half contains one [4Fe-4S] cluster that accepts an electron from reduced BchL and an active site for substrate (Pchlide) binding and reduction (31). Two rounds of electron transfer from BchL to BchN-BchB are required to reduce the C-17=C-18 double bond in Pchlide to form Chlide (32).
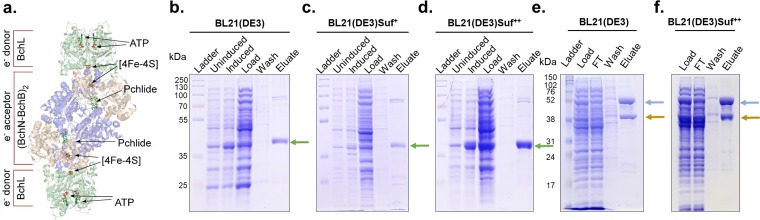
FIG 2.
Removing transcriptional repression and restoring the function of the suf operon in BL21(DE3) result in enhanced production of the [4Fe-4S] cluster BchL protein. (a) Crystal structure of the DPOR complex (PDB code 2YNM) consisting of BchL (green) and BchB(blue)-BchN(brown) proteins. Locations of [4Fe-4S] clusters and the binding sites for Pchlide and ATP are indicated. (b to d) Synthesis and affinity purification of BchL from BL21(DE3) (b), BL21(DE3)Suf+ (c), and BL21(DE3)Suf++ (d). The green arrow indicates the position of BchL following SDS-PAGE. (e and f) Analogous purification of the BchN-BchB complex from BL21(DE3) (e) and BL21(DE3)Suf++ (f). Brown and blue arrows indicate the positions of BchN and BchB, respectively. Gels are representative of results from three independent experiments.
We compared BchL and BchN-BchB syntheses, protein yields, iron contents, and protein activities from three strains: (i) BL21(DE3), (ii) BL21(DE3)Suf+, and (iii) BL21(DE3)Suf++. Strains containing plasmids carrying open reading frames for BchL or BchN-BchB under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible T7 promoter were grown and used for synthesis and purification of the proteins under identical conditions. For BchL, protein accumulated to similar cellular levels after induction for BL21(DE3) and BL21(DE3)Suf+ (Fig. 2b and c) but was markedly higher for BL21(DE3)Suf++ (Fig. 2d). BchL protein was then purified from each strain using affinity Ni2+-nitrilotriacetic acid (NTA) chromatography. The elevated protein levels in BL21(DE3)Suf++ resulted in a reproducible ∼3-fold increase in the overall yield of purified BchL protein obtained from 1 liter of cells (3.62 ± 0.54 mg/liter) compared to that for BL21(DE3) (1.32 ± 0.63 mg/liter) or BL21(DE3)Suf+ (0.81 ± 0.23 mg/liter) (Fig. 3a). The increased yield from BL21(DE3)Suf++ may reflect increased stability of BchL if loading of the protein with [4Fe-4S] clusters protects the protein from proteolysis. If this is the case, then the levels of the Fe-S cluster biogenesis machinery but not BchL protein are limiting for formation of [4Fe-4S]-BchL in this strain background.
FIG 3.
BL21(DE3) Suf++ increases yield and [Fe-S] cluster formation/load ratio and maintains enzyme activity. (a) Quantification of the final protein yield of BchL and BchN-BchB is plotted and shows enhanced yield of BchL in BL21(DE3)Suf++. No difference in the yields of the BchN-BchB complex was observed between strains. The left axis refers to dark gray bars, and the right axis refers to light gray bars. (b) The total iron content in the purified BchL protein was measured and plotted as a percentage of total occupancy. BL21(DE3) and BL21(DE3)Suf+ produced similar (2-tailed Student’s t test assuming unequal variance, n = 3) iron contents (P = 0.72). However, BL21(DE3)Suf++ had improved iron content compared to those of BL21(DE3) and BL21(DE3)Suf+ (P = 0.0053 and 0.0226, respectively). (c) The ratio of [4Fe-4S] cluster per gram of wet cell paste is plotted for BchL and BchN-BchB. (d) Growth curves for BL21(DE3) and BL21(DE3)Suf++ that carry the bchL overexpression plasmid are plotted. Strains showed similar doubling times [0.43 ± 0.10 and 0.41 ± 0.058 h for BL21(DE3) and BL21(DE3)Suf++, respectively; P = 0.61 from 2-tailed Student’s t test assuming unequal variance, n = 9] but reached an OD600 of 0.2 at different times [5.91 ± 0.12 and 5.19 ± 0.17 h for BL21(DE3) and BL21(DE3)Suf++, respectively; P = 4.92E−08 from 2-tailed Student’s t test assuming unequal variance, n = 9]. (e) BchL and BchN-BchB proteins purified from all three strains reduced Pchlide to Chlide. The lack of reduction in the absence of ATP is shown as a control. The y axis displays absorbance corresponding to the no-ATP data set. The other three experimental traces have been numerically shifted to better visualize the traces. Data in all panels show averaged results from three independent experiments, and the standard deviations are plotted.
We next tested whether the strain modifications influenced the production or yield of the larger BchN-BchB heterotetramer, containing two [4Fe-4S] clusters. Unlike BchL, BchN and BchB accumulated to similar cellular levels in BL21(DE3) and BL21(DE3) Suf++ (Fig. 2e and f). Consequently, we did not obtain enhanced yield of the BchN-BchB complex with BL21(DE3)Suf++ [4.94 ± 0.24 and 4.94 ± 0.13 mg/liter BchN-BchB from BL21(DE3) and BL21(DE3)Suf++, respectively (Fig. 3a)].
We also compared the total iron occupancies within the purified BchL proteins from the three strains. Iron occupancy was determined as follows: (molar [Fe] assayed from isolated protein)/(theoretical molar [Fe] assuming 4 Fe molecules per BchL dimer) × 100. Approximately 70 to 80% iron occupancy for the BchL dimer was observed when purified from BL21(DE3) and BL21(DE3)Suf+, compared to 100% occupancy when isolated from BL21(DE3)Suf++ (Fig. 3b). These data suggest that in addition to enhancing overall protein accumulation and increasing isolated protein yields, the amount of protein carrying an intact [4Fe-4S] cluster is also higher in BL21(DE3)Suf++. In contrast, BchN-BchB showed full [4Fe-4S] cluster occupancy when purified from any of the three strains (data not shown).
To explain why an ∼3-fold increase in BchL production was observed in BL21(DE3)Suf++, we compared the [4Fe-4S] cluster formation/load ratio, which we define as the moles of [4Fe-4S] cluster recovered from purified enzyme per gram of wet cell mass (Fig. 3c). For BchL, BL21(DE3)Suf++ generates and incorporates roughly 3-fold more (5 μmol) clusters per gram of wet cell mass than the other strains. This ratio does not scale for the BchN-BchB protein complex, even though the level of BchN-BchB polypeptide synthesis is lower than the level of BchL. The structural complexity of BchN-BchB and the mechanism of [Fe-S] cluster incorporation might limit protein folding, possibly explaining the lack of increased yields for the BchN-BchB complex. The doubling times for BL21(DE3) and BL21(DE3)Suf++ containing the BchL expression plasmid were also similar (Fig. 3d), although we consistently observed BL21(DE3)Suf++ exiting lag phase faster than BL21(DE3).
To ensure that the protein complexes purified from all the strains were active, we compared substrate reduction by their respective BchL and BchN-BchB proteins in vitro. Protochlorophyllide (substrate) and Chlide (reduced product) have unique spectral characteristics that are monitored through absorbance changes (33). Protochlorophyllide was monitored at its characteristic absorbance peak at 625 nm and formation of Chlide was captured at its absorbance peak at 668 nm (Fig. 3e). In the absence of ATP, no formation of Chlide was observed (black trace in Fig. 3e), and addition of ATP triggered formation of a Chlide peak (red trace in Fig. 3e). The kinetics of substrate reduction were similar between the preparations when purified protein concentrations were normalized for reactions [0.24 ± 0.02, 0.27 ± 0.03, and 0.24 ± 0.001 μM/min for proteins produced from BL21(DE3), BL21(DE3)Suf+, and BL21(DE3)Suf++, respectively].
DISCUSSION
Restoring the Suf [Fe-S] cluster biogenesis pathway and removing its transcriptional repression led to a marked enhancement in the production and purification of a [4Fe-4S] cluster protein. We call this BL21(DE3)Suf++ strain “SufFeScient” for its potential application in improving production and yields of other [Fe-S] cluster-containing proteins. The SufFeScient strain can improve yields in at least the case shown here; however, its utility must be experimentally determined for given target proteins since we do not have a sophisticated understanding of how subunit complexity, protein structure, and local cluster environment impact biogenesis of target enzymes. For the BchL homodimer, the [4Fe-4S] cluster is exposed (Fig. 2a), whereas the two [4Fe-4S] clusters in the BchN-BchB complex are buried deep within the dimer interface in the context of a heterotetramer (Fig. 2a). Additionally, the relative efficiencies and preferences of the Isc versus Suf systems in incorporating [Fe-S] clusters into specific proteins are also not known but might account for differences.
BL21(DE3) is considered a workhorse strain for recombinant protein production because of the efficient control provided by T7 RNA polymerase, whose gene is integrated into the genome, and the wide range of plasmid vectors containing T7 promoters used for protein synthesis or other biotechnological applications (34). The in-depth analysis of its genome sequence in 2009 provided a very important history of the progenitors of this strain as well as revealing several large deletions presumed to have been caused by UV irradiation that are specific to the BL21 lineage (see Table 4 of reference 22). Transcriptomics and metabolic modeling have provided additional insights into differences in the metabolic and transcriptional networks of this strain compared to other E. coli strains (34–36). Despite this wealth of knowledge, the genotype established in 2009 has not propagated to those listed by source companies, and as a consequence, the genotype listed in publications is typically incomplete [e.g., E. coli B F-fhuA2 dcm ompT hsdS(rB– mB–) gal (λ DE3)]. Thus, the function of genes of interest in BL21(DE3) should be verified depending on experimental needs.
In fact, a previous study (37) noted the limitation of BL21(DE3) in producing other metal-containing anaerobic respiratory enzymes. In this case, the deficiency in producing some of these enzymes could be tracked to a nonsense mutation in the gene encoding the anaerobic transcription factor FNR and as a consequence led to inefficient synthesis of proteins required for nickel transport. In addition, poor activity of some enzymes was also caused by a large (17,247-bp) deletion that removed the high-affinity molybdate transport system, compromising the ability of BL21(DE3) to make the molybdenum cofactor necessary for function of several anaerobic respiratory enzymes. Of note, defects in activity for formate dehydrogenases N and H were still detected even when these other systems were restored, suggesting that additional components in metalloenzyme synthesis are limiting in BL21(DE3). However, it seems unlikely that Suf-dependent [Fe-S] cluster assembly was the step that was impaired, since the Suf pathway is expressed at lower levels under anaerobic conditions (24). We also recently discovered that the disordered N terminus of BchL plays a protective role by binding and interacting with its [4Fe-4S] cluster (E. I. Corless and E. Antony, unpublished data). Mutations in this region that perturbed this protective role destabilized the BchL protein. Whether the Suf machinery evolved to handle such sensitive [Fe-S]-carrying proteins remains to be investigated.
In summary, BL21(DE3) is a highly utilized host strain for producing [Fe-S] proteins, and our correction of the deletion within the suf operon coupled with elevating its expression should provide researchers with another option for production of [Fe-S] proteins. As our knowledge for [Fe-S] cluster biogenesis pathway preferences for target proteins continues to expand, we will continue to optimize our SufFeScient strain of BL21(DE3) to further improve [Fe-S] protein yields.
MATERIALS AND METHODS
Reagents and buffers.
Chemicals were purchased from Sigma-Millipore Inc. (St. Louis, MO), Research Products International Inc. (Mount Prospect, IL), and Gold Biotechnology Inc. (St. Louis, MO). Oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). Enzymes were purchased from New England BioLabs (Ipswich, MA). All reagents and buffers used for protein purification were thoroughly degassed using alternating cycles of vacuum and nitrogen pressure on a home-built Schlenk line apparatus. Anaerobic conditions were maintained via airtight syringes, excess reductant, and a vinyl glove box (Coy Laboratories, MI) under an atmosphere of nitrogen (95%) and hydrogen (5%).
Strain construction.
The genotypes of strains used in this study are shown in Table 1. A BL21(DE3) strain derivative (PK11465) was constructed to produce a functional sufABCDSE operon. This was accomplished using P1 vir transduction to move a cat-PsufA allele (24) from strain PK10028 to BL21(DE3) and selecting for growth on tryptone-yeast extract (TYE) agar plates containing 10 μg/ml of chloramphenicol and 10 mM citrate. After streak purification, colony PCR and DNA sequencing were carried out to confirm the genotype. Using the same method, BL21(DE3) strain derivative PK11466 was constructed to produce a functional sufABCDSE operon, but one that also lacks transcriptional repression by Fur; the cat-PsufA(−26ATA−24 changed to −26TAT−24) allele, which contains a mutation within the Fur binding site, from PK10882 was moved to BL21(DE3) via P1 vir transduction.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| BL21(DE3) | F− hsdS gal λDE3 | Laboratory stock |
| MG1655 | E. coli K-12 λ− F− rph-1 | Laboratory stock |
| PK10882 | zdh-3632::cat, −26ATA−24 bp relative to sufA TSS changed to −26TAT−24 | 24 |
| PK11466 [BL21(DE3)Suf++] | BL21(DE3) but zdh-3632::cat, −26ATA−24 bp relative to sufA TSS changed to −26TAT−24 | This study |
| PK10028 | zdh-3632::cat | 24 |
| PK11465 [BL21(DE3)Suf+] | BL21(DE3) but zdh-3632::cat | This study |
| PK13230 | MG1655 but with sufB-3×FLAG-TAA-FRT-cat-FRT | This study |
| PK13231 | MG1655 but with sufS-3×FLAG-TAA-FRT-cat-FRT | This study |
| PK13232 | BL21(DE3) but with sufA/B-3×FLAG-TAA-FRT-cat-FRT | This study |
| PK13233 | BL21(DE3) but with sufS-3×FLAG-TAA-FRT-cat-FRT | This study |
| PK13235 | BL21(DE3) but with sufB-3×FLAG-TAA-FRT-cat-FRT derived from PK13230 | This study |
| PK13237 | BL21(DE3) but with sufS-3×FLAG-TAA-FRT-cat-FRT derived from PK13231 | This study |
| ZY-5 | Rhodobacter sphaeroides strain with deleted bchL used for Pchlide preparation | 33 |
| BW25993 | lacIq hsdR514 ΔaraBADAH33 ΔrhaBADL478 | 38 |
| PK10046 | BW25993 ΔiscSUA-hscBA-fdx::kan | This study |
| CAG18469 | MG1655 gua-26::Tn10(Tetr) | 41 |
| PK13354 | CAG18469 ΔiscSUA-hscBA-fdx::kan | This study |
| PK13343 | PK11466 ΔiscSUA-hscBA-fdx | This study |
| PK8202 | MG1655 kan-PydfZ-lacZ | 25 |
| PK12368 | BL21(DE3) kan-PydfZ-lacZ pRZ7404 | This study |
| PK12372 | PK11466 kan-PydfZ-lacZ pRZ7404 | This study |
| PK13351 | PK13343 kan-PydfZ-lacZ pRZ7404 | This study |
| NiCo21(DE3) | Commercial strain | New England BioLabs |
| Lemo21(DE3) | Commercial strain | New England BioLabs |
| C41(DE3) | Commercial strain | Lucigen |
| Rosetta2(DE3)pLysS | Commercial strain | EMD-Millipore-Novagen |
| BLR(DE3)pLysS | Commercial strain | EMD-Millipore-Novagen |
| BL21(DE3)Ai | Commercial strain | Thermo Fisher |
| BL21(DE3)codon plus | Commercial strain | Agilent Technologies |
| Plasmids | ||
| pIND4-3×FLAG(n) | kan, 3×FLAG | T. Donohue |
| pPK8629 | pIND4-3×FLAG(n) but with TAA inserted after 3×FLAG | This study |
| pPK8630 | pPK8629 but with FRT-cat-FRT cloned into BamHI and NdeI restriction sites | This study |
| pKD32 | FRT-cat-FRT | B. Wanner |
| pKD13 | FRT-kan-FRT | B. Wanner |
| pKD46 | Red recombinase expression plasmid | B. Wanner |
| pRZ7404 | Apr; HindIII-BamHI of fnr (−521 to +1115 of fnr) cloned into same sites of pBR322 | This study |
| BchL RSF-Duet1 | Plasmid used to overproduce BchL protein with an N-terminal 6×histidine tag and a 3C protease cleavage site | This study |
| BchB RSF-Duet1 | Plasmid used to overproduce BchB protein with an N-terminal 6×histidine tag and a 3C protease cleavage site | This study |
| BchN pET-Duet1 | Plasmid used to overproduce BchN protein | This study |
Derivatives of MG1655 and BL21(DE3) were constructed to produce chromosomally derived, epitope-tagged variants of SufB [SufA/B in the case of BL21(DE3)] or SufS. First, a TAA stop codon was inserted directly after the 3×FLAG sequence on pIND4-3×FLAG(n) using QuikChange (Stratagene) to form pPK8629. Following PCR amplification of cat flanked by FLP recognition target (FRT) sites from pKD32 using primers containing BamHI and NdeI restriction sites, the PCR fragment was cloned into the same sites of pPK8629 to make pPK8630. To recombine 3×FLAG-TAA-FRT-cat-FRT directly before the native stop codons of the SufA/B fusion protein or SufS in BL21(DE3), this construct was PCR amplified with primers containing homology to either region of the chromosome, electroporated into a derivative of BL21(DE3) that harbored pKD46, and selected for chloramphenicol resistance (forming strains PK13232 and PK13233). Epitope-tagged variants of SufB and SufS were made in a similar manner in MG1655 (forming strains PK13230 and PK13231). All alleles were verified by DNA sequencing. Finally, P1 vir transduction was used to introduce sufB-3×FLAG-TAA-FRT-cat-FRT and sufS-3×FLAG-TAA-FRT-cat-FRT from the MG1655 derivatives PK13230 and PK13231 into BL21(DE3), forming strains PK13235 and PK13237, respectively.
Genetically linking ΔiscSUA-hscBA-fdx::kan and gua-26::Tn10(Tetr) involved multiple steps. First, the coding region encompassing iscSUA-hscBA-fdx in BW25993/pKD46 was replaced with kan flanked by FLP recognition target sites from pKD13 as previously described (24, 38), creating strain PK10046. P1 vir transduction was used to move ΔiscSUA-hscBA-fdx::kan from PK10046 to strain CAG18469 [MG1655 gua-26::Tn10(TetR)]; colonies were selected on TYE agar containing 40 μg/ml of kanamycin, 10 μg/ml of tetracycline, and 10 mM citrate, creating strain PK13354. After streak purification, P1 vir transduction was again used to move both the ΔiscSUA-hscBA-fdx::kan and gua-26::Tn10(Tetr) alleles into MG1655 and BL21(DE3). Colonies were initially selected for on medium containing 10 μg/ml of tetracycline and 10 mM citrate and were then screened for Kanr to establish genetic linkage.
Finally, derivatives of BL21(DE3) and PK11466 were constructed to evaluate FNR activity. First, ΔiscSUA-hscBA-fdx::kan was P1 vir transduced into PK11466 and colonies were selected on TYE agar plates containing 40 μg/ml of kanamycin and 10 mM citrate. After streak purification, kan was removed by transformation with pCP20, encoding FLP recombinase, and screening for kanamycin sensitivity (38), forming strain PK13343. Next, the kan-PydfZ-lacZ allele was P1 vir transduced from strain PK8202 into strains BL21(DE3), PK11466, and PK13343; the resulting transductants were transformed with pRZ7404, forming strains PK12368, PK12372, and PK13351, respectively.
Western blot analysis.
Cultures were grown aerobically at 37°C to an optical density at 600 nm (OD600) of 0.1 in M9 minimal medium containing 0.2% glucose, 0.2% Casamino Acids, 1 mM MgSO4, 10 μg/ml of ferric ammonium citrate, 4 μg/ml of thiamine, and 0.1 mM CaCl2. Aliquots (1 ml) of cells were pelleted and levels of SufB-FLAG, SufA/B-FLAG, or SufS-FLAG were measured by Western blotting as previously described (25, 39), except that purified anti-DYKDDDDK epitope tag antibody (Biolegend) was used.
β-Galactosidase assays.
Cultures were grown as described for Western blot analysis except anaerobically in screw-cap tubes, and final concentrations of 12.5 μg/ml of nicotinic acid and 50 μg/ml of ampicillin were present in the growth medium. Spectinomycin was added to culture samples at a final concentration of 100 μg/ml to terminate further protein synthesis, and cells were kept on ice until assayed for β-galactosidase activity as previously described (40).
Generation of protein synthesis plasmids.
Plasmids used to produce BchL, BchN, and BchB were generated from PCR-amplified Rhodobacter sphaeroides genomic DNA. BchL and BchB open reading frames were engineered to carry an N-terminal polyhistidine (6×His) tag and a 3C protease recognition site and were cloned into pRSF-Duet1 using BamHI/NotI and SacI/SalI restriction sites, respectively. BchN contained no modifications and was cloned into a pET-Duet1 vector using NdeI/KpnI restriction sites.
Generation of Pchlide.
Pchlide was isolated from supernatants of cultures of Rhodobacter sphaeroides strain ZY-5 harboring a deletion of bchL (a kind gift from Carl Bauer, Indiana University) (33). Ten milliliters of an overnight culture grown with shaking (200 rpm) in RCV 2/3 medium with 50 μg/ml of kanamycin at 34°C was used to inoculate a 125-ml culture with RCV 2/3 medium containing 5.0 μg/ml of kanamycin in a foil-wrapped 250-ml flask and grown for 24 h at 34°C and 130 rpm. Cells were removed by centrifugation at 4,392 × g, and the green supernatant was stored at 4°C. The cell pellet was then resuspended in 500 ml of fresh RCV 2/3 medium with 5.0 μg/ml of kanamycin and grown with shaking (130 rpm) for 24 h at 34°C in the dark, and again the supernatant was separated from the cells by centrifugation. This process was repeated twice, and the combined supernatant containing Pchlide was centrifuged at 4,392 × g and 4°C for 1 h to remove any remaining cells, filtered through a white nylon 0.44-μm filter (Millipore; catalog number HNWP04700), and extracted in 500-ml aliquots with 1/3 volume of diethyl ether in a 1-liter separatory funnel, taking care to vent built-up pressure from volatile ether after vigorous shaking. The organic phase containing the green Pchlide was removed, centrifuged at 4,122 × g and 4°C for 5 min to further separate any lipid or aqueous contamination, and then decanted and evaporated to dryness under a stream of nitrogen. Water droplets that formed on the inside and outside of the vessel were allowed to evaporate inside the fume hood. The resulting dark green flaky material was then resuspended in 250 μl of dimethyl sulfoxide (DMSO)–500 ml of extract. The suspended Pchlide was aliquoted and stored in amber-colored 1.5-ml conical tubes (Fisher Scientific; catalog no. 05-408-134) at 4°C. Concentration of Pchlide was determined using 4 dilutions, 3 replicates each in 80% acetone, using the molar extinction coefficient 30,400 M−1 cm−1.
Protein synthesis and purification.
Bacterial cells freshly transformed with the appropriate plasmid for recombinant protein production were used to inoculate 1 liter of Luria broth medium supplemented with an appropriate antibiotic (100 μg/ml of ampicillin and/or 50 μg/ml of kanamycin), 1 mM ferric citrate, and 1 mM l-cysteine in 2.8-liter baffled flasks and were grown aerobically with shaking (200 rpm) at 37°C. Protein synthesis was induced at a culture OD600 of 0.6 with 35 μM IPTG, and then cultures were shifted to 25°C with shaking at 150 rpm overnight. Cells were harvested after a 3-h incubation with sodium dithionite (0.3 g/liter of growth) in 1-liter airtight centrifuge tubes at 17°C with no shaking. All subsequent steps were performed in the glove box or in airtight septum-sealed bottles under positive nitrogen pressure unless otherwise noted. Cell pellets were resuspended in degassed STD buffer (100 mM HEPES [pH 7.5], 150 mM NaCl, 1.7 mM sodium dithionite), transferred to a septum-sealed glass bottle, and stored at −20°C until lysis.
Cell lysis was performed using lysozyme (0.5 mg/ml) for 30 min at room temperature in septum-sealed bottles. Cells were then sonicated inside a glove box for 3 min on ice (Branson sonifier, 50% duty cycle, 60 s on and 60 s off for 3 cycles). Cell lysates were clarified via centrifugation (37,157 × g for 60 min) and loaded onto a nickel-nitrilotriacetic acid (Ni2+-NTA) column, equilibrated with STD buffer. After washing with STD buffer containing 20 mM imidazole, bound proteins were eluted into a septum-sealed bottle using 30 ml of STD buffer containing 250 mM imidazole. For BchN-BchB, the eluted proteins were concentrated using an Amicon 15-ml 30-kDa-molecular-weight-cutoff spin concentrator (Sigma-Millipore Inc., St. Louis, MO), by centrifugation for 8 min at 4,122 × g and 4°C. For BchL, protein eluted from the Ni2+-NTA affinity column was subsequently purified over a 10-ml Q-Sepharose column (GE Healthcare). The Q column was first sequentially equilibrated with 150 mM HEPES (pH 7.5) containing 1 M NaCl, followed by 150 mM HEPES (pH 7.5; no NaCl). BchL pooled from the affinity step was then loaded and the column was subsequently washed with STD buffer. Bound BchL was eluted using a 0.15 to 1 M NaCl gradient in 100 ml of STD buffer, and fractions were collected inside the glove box. Fractions containing BchL protein were concentrated using a spin concentrator as described above. After isolation, the concentration of BchL is typically 100 μM and that of BchNB is 40 μM. Proteins were aliquoted into 1.2-ml cryotubes (catalog number 430487; Corning) and capped in the glove box. Sealed tubes were removed from the glove box, flash frozen using liquid nitrogen, and stored under liquid nitrogen.
Comparison of protein induction and purification using SDS-PAGE analysis.
Uninduced and induced samples (1.0 ml and 0.5 ml, respectively) were centrifuged in a tabletop centrifuge (13,222 × g, 60 s, and 4°C), and the supernatant was decanted. Sample cell pellets were resuspended in 100 μl of water. Tenfold dilutions of resuspended cells were used to record the OD600, and this value was used to normalize all sample optical densities. Equivalent units of OD600 of undiluted samples were then diluted to 100 μl, mixed with 100 μl of 2 × SDS Laemmli sample buffer, and boiled for 10 min, and 10 μl was analyzed by SDS-PAGE using 10% SDS-polyacrylamide gels. A PageRuler Plus prestained ladder (Thermo Scientific) was used as a protein size ladder for reference.
Assay for substrate reduction by DPOR.
Reduction of Pchlide to Chlide was measured spectroscopically by mixing BchN-BchB protein (3 μM tetramer), BchL protein (9 μM dimer), and 35 μM Pchlide, in the absence or presence of ATP (3 mM), in STD buffer plus 10 mM MgCl2. Reactions (40 μl) were quenched with 160 μl of 100% acetone (80% [vol/vol] final concentration). Precipitated proteins were removed by centrifugation in a tabletop centrifuge (13,226 × g for 4 min). The supernatant (160 μl) was transferred to a cyclic olefin half-area well plate (catalog number 4680; Corning), and absorbance scans from 600 nm to 725 nm were recorded on a SpectraMax i3x plate reader (Molecular Devices). Chlide appearance was quantified using its molar extinction coefficient, 74,900 M−1 cm−1, at 666 nm.
Protein and iron content determination.
Protein concentrations were determined using the Bradford assay reagent (Bio-Rad) and bovine serum albumin (Gold Biotechnology Inc.) as a standard. Iron content was determined by colorimetric assay using 2′2′-dipyridyl absorbance at 520 nm and Fe(NH4)2(SO4)2 (Fisher) as a standard after sample denaturation and iron reduction (60 min of exposure to 5% HCl and boiling, followed by exposure to excess [10%] hydroxylamine).
Growth curve generation.
BL21(DE3) and BL21(DE3)Suf++ were transformed with BchL plasmid and plated on LB agar containing 50 μg/ml of kanamycin. Six individual colonies from both strains were used to inoculate 5-ml cultures (LB and 50 μg/ml of kanamycin). Overnight cultures were normalized to an OD600 of 0.01 with LB containing 50 μg/ml of kanamycin. Two microliters of OD600-corrected starter cultures was used to inoculate 200 μl of medium in a polystyrene 96-well cell culture plate (Cole-Palmer). Six wells containing only 200 μl of LB and 50 μg/ml of kanamycin served as a blank and negative control. Cells were grown in a SpectraMax i3x plate reader (Molecular Devices Inc.) at 37°C, shaken for 10 s before each read. OD600 was measured every 2.5 min. Blank measurements were averaged, and individual growth curves were corrected by subtracting averaged blank values at each time point and then averaged after correction for Fig. 3d. Doubling rates (r) were calculated from values at 2 and 4 h as follows: r = (ln[OD2/OD1])/(t2 − t1). Doubling time was calculated using ln(2)/r.
ACKNOWLEDGMENTS
E.I.C. and E.L.M. performed experiments. E.A. and P.J.K. conceived the experiments. All authors contributed to the writing of the manuscript.
This work was supported by a grant from the Department of Energy, Office of Basic Energy Sciences (DE-SC0017866), to E.A. and NIH grant R01-GM115894 to P.J.K.
REFERENCES
- 1.Beinert H. 2000. Iron-sulfur proteins: ancient structures, still full of surprises. J Biol Inorg Chem 5:2–15. doi: 10.1007/s007750050002. [DOI] [PubMed] [Google Scholar]
- 2.Fontecave M. 2006. Iron-sulfur clusters: ever-expanding roles. Nat Chem Biol 2:171–174. doi: 10.1038/nchembio0406-171. [DOI] [PubMed] [Google Scholar]
- 3.Kiley PJ, Beinert H. 2003. The role of Fe-S proteins in sensing and regulation in bacteria. Curr Opin Microbiol 6:181–185. doi: 10.1016/s1369-5274(03)00039-0. [DOI] [PubMed] [Google Scholar]
- 4.Boyd ES, Thomas KM, Dai Y, Boyd JM, Outten FW. 2014. Interplay between oxygen and Fe-S cluster biogenesis: insights from the Suf pathway. Biochemistry 53:5834–5847. doi: 10.1021/bi500488r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanc B, Gerez C, Ollagnier de Choudens S. 2015. Assembly of Fe/S proteins in bacterial systems: biochemistry of the bacterial ISC system. Biochim Biophys Acta 1853:1436–1447. doi: 10.1016/j.bbamcr.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Outten FW. 2015. Recent advances in the Suf Fe-S cluster biogenesis pathway: beyond the Proteobacteria. Biochim Biophys Acta 1853:1464–1469. doi: 10.1016/j.bbamcr.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Py B, Barras F. 2010. Building Fe-S proteins: bacterial strategies. Nat Rev Microbiol 8:436–446. doi: 10.1038/nrmicro2356. [DOI] [PubMed] [Google Scholar]
- 8.Perard J, Ollagnier de Choudens S. 2018. Iron-sulfur clusters biogenesis by the SUF machinery: close to the molecular mechanism understanding. J Biol Inorg Chem 23:581–596. doi: 10.1007/s00775-017-1527-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rocha AG, Dancis A. 2016. Life without Fe-S clusters. Mol Microbiol 99:821–826. doi: 10.1111/mmi.13273. [DOI] [PubMed] [Google Scholar]
- 10.Vinella D, Brochier-Armanet C, Loiseau L, Talla E, Barras F. 2009. Iron-sulfur (Fe/S) protein biogenesis: phylogenomic and genetic studies of A-type carriers. PLoS Genet 5:e1000497. doi: 10.1371/journal.pgen.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka N, Kanazawa M, Tonosaki K, Yokoyama N, Kuzuyama T, Takahashi Y. 2016. Novel features of the ISC machinery revealed by characterization of Escherichia coli mutants that survive without iron-sulfur clusters. Mol Microbiol 99:835–848. doi: 10.1111/mmi.13271. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi Y, Tokumoto U. 2002. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem 277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 13.Tokumoto U, Kitamura S, Fukuyama K, Takahashi Y. 2004. Interchangeability and distinct properties of bacterial Fe-S cluster assembly systems: functional replacement of the isc and suf operons in Escherichia coli with the nifSU-like operon from Helicobacter pylori. J Biochem 136:199–209. doi: 10.1093/jb/mvh104. [DOI] [PubMed] [Google Scholar]
- 14.Outten FW, Djaman O, Storz G. 2004. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol 52:861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 15.Roche B, Aussel L, Ezraty B, Mandin P, Py B, Barras F. 2013. Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim Biophys Acta 1827:455–469. doi: 10.1016/j.bbabio.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 16.Mettert EL, Kiley PJ. 2015. How is Fe-S cluster formation regulated? Annu Rev Microbiol 69:505–526. doi: 10.1146/annurev-micro-091014-104457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai CL, Tainer JA. 2018. Robust production, crystallization, structure determination, and analysis of [Fe-S] proteins: uncovering control of electron shuttling and gating in the respiratory metabolism of molybdopterin guanine dinucleotide enzymes. Methods Enzymol 599:157–196. doi: 10.1016/bs.mie.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura M, Saeki K, Takahashi Y. 1999. Hyperproduction of recombinant ferredoxins in Escherichia coli by coexpression of the ORF1-ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster. J Biochem 126:10–18. doi: 10.1093/oxfordjournals.jbchem.a022409. [DOI] [PubMed] [Google Scholar]
- 19.Kriek M, Peters L, Takahashi Y, Roach PL. 2003. Effect of iron-sulfur cluster assembly proteins on the expression of Escherichia coli lipoic acid synthase. Protein Expr Purif 28:241–245. doi: 10.1016/s1046-5928(02)00680-0. [DOI] [PubMed] [Google Scholar]
- 20.Grawert T, Kaiser J, Zepeck F, Laupitz R, Hecht S, Amslinger S, Schramek N, Schleicher E, Weber S, Haslbeck M, Buchner J, Rieder C, Arigoni D, Bacher A, Eisenreich W, Rohdich F. 2004. IspH protein of Escherichia coli: studies on iron-sulfur cluster implementation and catalysis. J Am Chem Soc 126:12847–12855. doi: 10.1021/ja0471727. [DOI] [PubMed] [Google Scholar]
- 21.Daegelen P, Studier FW, Lenski RE, Cure S, Kim JF. 2009. Tracing ancestors and relatives of Escherichia coli B, and the derivation of B strains REL606 and BL21(DE3). J Mol Biol 394:634–643. doi: 10.1016/j.jmb.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 22.Studier FW, Daegelen P, Lenski RE, Maslov S, Kim JF. 2009. Understanding the differences between genome sequences of Escherichia coli B strains REL606 and BL21(DE3) and comparison of the E. coli B and K-12 genomes. J Mol Biol 394:653–680. doi: 10.1016/j.jmb.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 23.Jeong H, Barbe V, Lee CH, Vallenet D, Yu DS, Choi SH, Couloux A, Lee SW, Yoon SH, Cattolico L, Hur CG, Park HS, Segurens B, Kim SC, Oh TK, Lenski RE, Studier FW, Daegelen P, Kim JF. 2009. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J Mol Biol 394:644–652. doi: 10.1016/j.jmb.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 24.Mettert EL, Kiley PJ. 2014. Coordinate regulation of the Suf and Isc Fe-S cluster biogenesis pathways by IscR is essential for viability of Escherichia coli. J Bacteriol 196:4315–4323. doi: 10.1128/JB.01975-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mettert EL, Outten FW, Wanta B, Kiley PJ. 2008. The impact of O2 on the Fe-S cluster biogenesis requirements of Escherichia coli FNR. J Mol Biol 384:798–811. doi: 10.1016/j.jmb.2008.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nomata J, Kitashima M, Inoue K, Fujita Y. 2006. Nitrogenase Fe protein-like Fe-S cluster is conserved in L-protein (BchL) of dark-operative protochlorophyllide reductase from Rhodobacter capsulatus. FEBS Lett 580:6151–6154. doi: 10.1016/j.febslet.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 27.Nomata J, Swem LR, Bauer CE, Fujita Y. 2005. Overexpression and characterization of dark-operative protochlorophyllide reductase from Rhodobacter capsulatus. Biochim Biophys Acta 1708:229–237. doi: 10.1016/j.bbabio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Nomata J, Ogawa T, Kitashima M, Inoue K, Fujita Y. 2008. NB-protein (BchN-BchB) of dark-operative protochlorophyllide reductase is the catalytic component containing oxygen-tolerant Fe-S clusters. FEBS Lett 582:1346–1350. doi: 10.1016/j.febslet.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Brocker MJ, Schomburg S, Heinz DW, Jahn D, Schubert WD, Moser J. 2010. Crystal structure of the nitrogenase-like dark operative protochlorophyllide oxidoreductase catalytic complex (ChlN/ChlB)2. J Biol Chem 285:27336–27345. doi: 10.1074/jbc.M110.126698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muraki N, Nomata J, Ebata K, Mizoguchi T, Shiba T, Tamiaki H, Kurisu G, Fujita Y. 2010. X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature 465:110–114. doi: 10.1038/nature08950. [DOI] [PubMed] [Google Scholar]
- 31.Moser J, Lange C, Krausze J, Rebelein J, Schubert WD, Ribbe MW, Heinz DW, Jahn D. 2013. Structure of ADP-aluminium fluoride-stabilized protochlorophyllide oxidoreductase complex. Proc Natl Acad Sci U S A 110:2094–2098. doi: 10.1073/pnas.1218303110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nomata J, Kondo T, Mizoguchi T, Tamiaki H, Itoh S, Fujita Y. 2014. Dark-operative protochlorophyllide oxidoreductase generates substrate radicals by an iron-sulphur cluster in bacteriochlorophyll biosynthesis. Sci Rep 4:5455. doi: 10.1038/srep05455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujita Y, Bauer CE. 2000. Reconstitution of light-independent protochlorophyllide reductase from purified Bchl and BchN-BchB subunits. In vitro confirmation of nitrogenase-like features of a bacteriochlorophyll biosynthesis enzyme. J Biol Chem 275:23583–23588. doi: 10.1074/jbc.M002904200. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Jeong H, Kim EY, Kim JF, Lee SY, Yoon SH. 2017. Genomic and transcriptomic landscape of Escherichia coli BL21(DE3). Nucleic Acids Res 45:5285–5293. doi: 10.1093/nar/gkx228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H, Kim S, Yoon SH. 2018. Metabolic network reconstruction and phenome analysis of the industrial microbe, Escherichia coli BL21(DE3). PLoS One 13:e0204375. doi: 10.1371/journal.pone.0204375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monk JM, Koza A, Campodonico MA, Machado D, Seoane JM, Palsson BO, Herrgard MJ, Feist AM. 2016. Multi-omics quantification of species variation of Escherichia coli links molecular features with strain phenotypes. Cell Syst 3:238–251.e12. doi: 10.1016/j.cels.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinske C, Bonn M, Kruger S, Lindenstrauss U, Sawers RG. 2011. Metabolic deficiences revealed in the biotechnologically important model bacterium Escherichia coli BL21(DE3). PLoS One 6:e22830. doi: 10.1371/journal.pone.0022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nesbit AD, Giel JL, Rose JC, Kiley PJ. 2009. Sequence-specific binding to a subset of IscR-regulated promoters does not require IscR Fe-S cluster ligation. J Mol Biol 387:28–41. doi: 10.1016/j.jmb.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 41.Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev 53:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]