This work provides insights into the phosphorylation of the DnaA initiator protein in Streptomyces coelicolor and suggests a novel bacterial regulatory mechanism for initiation of chromosome replication. Although phosphorylation of DnaA has been reported earlier, its biological role was unknown. This work shows that upon phosphorylation, the cooperative binding of the replication origin by DnaA may be disturbed. We found that AfsK kinase is responsible for phosphorylation of DnaA. Upon upregulation of AfsK, chromosome replication occurred further from the hyphal tip. Orthologs of AfsK are exclusively found in mycelial actinomycetes that are related to Streptomyces and exhibit a complex life cycle. We propose that the AfsK-mediated regulatory pathway serves as a nonessential, energy-saving mechanism in S. coelicolor.
KEYWORDS: AfsK, DnaA, Streptomyces, phosphorylation, replication
ABSTRACT
In all organisms, chromosome replication is regulated mainly at the initiation step. Most of the knowledge about the mechanisms that regulate replication initiation in bacteria has come from studies on rod-shaped bacteria, such as Escherichia coli and Bacillus subtilis. Streptomyces is a bacterial genus that is characterized by distinctive features and a complex life cycle that shares some properties with the developmental cycle of filamentous fungi. The unusual lifestyle of streptomycetes suggests that these bacteria use various mechanisms to control key cellular processes. Here, we provide the first insights into the phosphorylation of the bacterial replication initiator protein, DnaA, from Streptomyces coelicolor. We suggest that phosphorylation of DnaA triggers a conformational change that increases its ATPase activity and decreases its affinity for the replication origin, thereby blocking the formation of a functional orisome. We suggest that the phosphorylation of DnaA is catalyzed by Ser/Thr kinase AfsK, which was shown to regulate the polar growth of S. coelicolor. Together, our results reveal that phosphorylation of the DnaA initiator protein functions as a negative regulatory mechanism to control the initiation of chromosome replication in a manner that presumably depends on the cellular localization of the protein.
IMPORTANCE This work provides insights into the phosphorylation of the DnaA initiator protein in Streptomyces coelicolor and suggests a novel bacterial regulatory mechanism for initiation of chromosome replication. Although phosphorylation of DnaA has been reported earlier, its biological role was unknown. This work shows that upon phosphorylation, the cooperative binding of the replication origin by DnaA may be disturbed. We found that AfsK kinase is responsible for phosphorylation of DnaA. Upon upregulation of AfsK, chromosome replication occurred further from the hyphal tip. Orthologs of AfsK are exclusively found in mycelial actinomycetes that are related to Streptomyces and exhibit a complex life cycle. We propose that the AfsK-mediated regulatory pathway serves as a nonessential, energy-saving mechanism in S. coelicolor.
INTRODUCTION
Streptomyces spp. are Gram-positive, mycelium-forming, soil bacteria. In addition to their highly developed secondary metabolism and opportunities for industrial exploitation, these bacteria are relevant to basic research given their unique and complex life cycle, which shares some developmental features with the eukaryotic filamentous fungi (1). One of the most remarkable features of Streptomyces is the unidirectional cell extension at the hyphal tips. During the development of Streptomyces colonies, vegetative hyphae, which contain long multinucleoid compartments, undergo morphological differentiation. Upon nutrient depletion, the aerial hyphae grow away from vegetative mycelium and are subsequently converted into chains of spores. In addition to this unique life cycle, Streptomyces spp. also exhibit distinctive features in their chromosomes, which are large (ca. 8 to 10 Mbp), linear, and GC-rich (up to 80%) molecules with a centrally localized origin of chromosomal replication (oriC) and terminal repeats at both ends (2). In both vegetative and aerial hyphae, chromosome replication initiates asynchronously (not every chromosome localized within a single cellular compartment undergoes replication) and finishes before the segregation of chromosomes into spores (3, 4).
Chromosome replication is a key event in the bacterial cell cycle that is mainly controlled at the initiation step. It begins at a strictly defined chromosomal locus, oriC, which differs among bacteria in terms of its length and the number and arrangement of the unique motifs (DnaA boxes) that are bound by the replication initiator protein, DnaA. Streptomyces origins are considered to be relatively long (800 to 900 bp) compared to those of other bacteria (e.g., that of Escherichia coli is 250 bp, while that of Caulobacter crescentus is 405 bp) (5, 6).
DnaA initiator protein consists of four functional domains that are involved mainly in protein-DNA and protein-protein interactions. DnaA binds ATP and ADP, but only ATP-DnaA is able to initiate replication. DnaA domain I participates in inter-DnaA interactions and the binding of accessory proteins (7). Domain II is a flexible linker that joins domains I and III (8); it is the least conserved domain that varies in its sequence and length (e.g., it is approximately 220 amino acids [aa] in Streptomyces coelicolor and ∼60 aa in E. coli). Domain III mediates inter-DnaA interactions in addition to carrying various AAA+ ATPase motifs (including the Walker A and B motifs, which facilitate single-stranded DNA binding) and sequences that are involved in protein-membrane interactions. The C-terminal domain IV contains a helix-turn-helix fold that binds double-stranded DnaA box sequences (9).
During the initiation process, multiple DnaA molecules bind oriC to form a filamentous nucleoprotein complex (orisome), which forces local unwinding of the DNA helix (10). The double-stranded DNA opens at a DNA unwinding element (DUE), which is usually localized within an AT-rich segment of oriC. The initiation process proceeds with the recruitment of additional proteins (replicative helicase and primase), leading to further DNA unwinding and RNA priming. Recruitment of DNA polymerase III and formation of the replication complex (replisome) enable bidirectional DNA replication (11).
The initiation of chromosome replication must be tightly and precisely controlled. To date, most of the knowledge regarding the molecular mechanisms that control the bacterial initiation of chromosome replication comes from studies in E. coli, Bacillus subtilis, and C. crescentus (12). In bacteria, two major orisome components are subject to these regulatory mechanisms: oriC and DnaA. OriC may be modulated by altering its accessibility for the DnaA protein, e.g., in E. coli, methylation of adenine residues within oriC (by the Dam methylase), and subsequent SeqA protein binding inhibits replication initiation by preventing formation of functional orisome (13–15). The protein level of DnaA may be regulated by methylation-dependent transcriptional repression (14, 16). Additionally, the DnaA protein downregulates transcription of its own gene by binding to DnaA boxes localized in the dnaA promoter region (17, 18). Posttranscriptionally, the DnaA protein level may be controlled by proteolysis, as recently demonstrated in C. crescentus (19, 20). The activity of DnaA is regulated by various mechanisms, including interactions with other proteins, such as Soj and YabA in B. subtilis (21, 22).
Streptomycetes are unique in generating multinucleoid cellular compartments in which asynchronous DNA replication takes place; therefore, the members of this genus may have unique regulatory mechanisms responsible for modulating chromosome replication initiation during their complex life cycle. Global proteomic analysis of S. coelicolor M600 as a model organism revealed that DnaA might be phosphorylated at threonine 486, located within domain III (23). This apparently unique posttranslational modification of the initiator protein may represent a novel mechanism for controlling replication initiation in bacteria. Here, we provide further insight into the biological function of DnaA phosphorylation.
RESULTS
S. coelicolor DnaA is phosphorylated during replicatively active stages of the life cycle.
Although one phosphoproteomic study found that S. coelicolor DnaA is phosphorylated at threonine 486 (23), a second study did not find evidence that DnaA is phosphorylated (24).
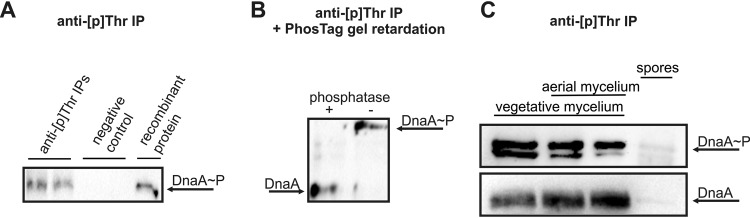
To examine whether DnaA is indeed phosphorylated, we used immunoprecipitation (IP) to purify phosphorylated DnaA. Protein extracts from S. coelicolor liquid cultures were prepared as previously described (24), and 1 mg of cellular protein was subjected to IP with a polyclonal anti-phospho-Thr (anti-[p]Thr) antibody, followed by SDS-PAGE separation and Western blot analysis with anti-DnaA rabbit antiserum (Fig. 1A). To exclude the possibility that DnaA was copurified with other immunoprecipitated proteins, we performed an additional experiment where IP-eluted proteins were separated into two equal portions, one of which was subjected to phosphatase treatment. Proteins from both samples were electrophoresed with the Phos-tag low-molecular-weight compound, which slows down the gel migration of phosphorylated proteins (25). Western blot analysis was performed, and we examined whether slower-migrating DnaA proteins were present in the IP fraction that had not been treated with phosphatase. Our results confirmed that DnaA is phosphorylated in submerged and exponentially growing S. coelicolor cells (Fig. 1B).
FIG 1.
Phosphorylation of DnaA during Streptomyces coelicolor life cycle. (A) Western blot analysis of DnaA proteins in lysates obtained from liquid-grown cultures subjected to immunoprecipitation (IP) with the anti-phospho-Thr (anti-[p]Thr) antibody. Negative controls were conducted without the antibody. Recombinant DnaAHis6 was used as a positive control for the identification of DnaA. (B) Experiments were performed with phosphatase treatment and addition of the Phos-tag compound to the polyacrylamide gel. The presence of a slowly migrating phospho-DnaA band (DnaA~P) excludes the possibility that DnaA was simply copurified during IP. (C) Detection of DnaA phosphorylation in extracts from solid medium-grown cultures of S. coelicolor at the vegetative hyphal stage, vegetative/aerial hyphal stage, and after sporulation. (Upper) Arrow indicates the DnaA phosphoprotein. (Lower) The presence of DnaA protein at each examined growth stage was analyzed by anti-DnaA Western blotting performed on the crude cellular extracts.
As S. coelicolor undergoes its full life cycle on solid media but not in submerged cultures and our results suggested that phosphorylation can be seen in exponentially growing cells, we examined the life cycle dependence of this phosphorylation. We used IP to test for the presence of phosphorylated DnaA in extracts obtained from cells grown on solid medium at the vegetative hyphal, aerial hyphal, and spore stages. To prevent spore germination, we incubated freshly collected S. coelicolor spores with anthranilic acid prior to lysis (26). Interestingly, the phosphorylation of DnaA protein was observed during the replicatively active stages of growth but not in spores (in which DNA replication does not occur) (Fig. 1C, upper). However, further analysis revealed that the DnaA protein was not detected in spores (Fig. 1C, bottom).
Together, our results confirm that DnaA can be phosphorylated in S. coelicolor and further show that this occurs during growth stages in which DNA replication is active.
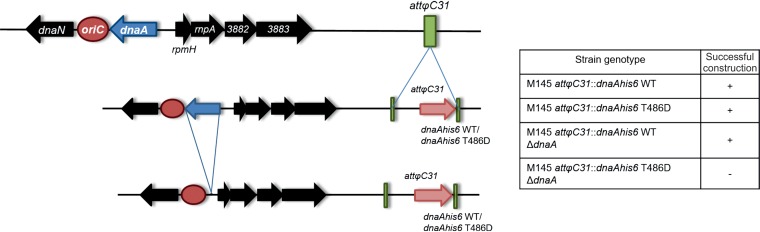
S. coelicolor cannot tolerate a single dnaA allele encoding a pseudophosphorylated protein.
To test the effect of DnaA phosphorylation in vivo, we attempted to construct S. coelicolor mutant strains harboring different versions of DnaA. We first used the integrative vector to introduce a dnaAhis6 gene encoding the wild-type protein, a pseudophosphorylated T486D mutant (in which substitution of aspartic acid for the threonine mimics the phosphorylated state), or a phosphoablative T486A mutational variant into the chromosomal attφC31 site. We then sought to delete the native dnaA gene from the generated strains.
We successfully generated a strain that harbored only the wild-type dnaAhis6 sequence but were unable to obtain cells that produced only the pseudophosphorylated protein (Fig. 2). This suggests that phosphorylation of the initiator protein leads to the formation of a replication-inactive DnaA pool. We abandoned our efforts to generate a strain expressing only the phosphoablative dnaAhis6 T486A gene, as discussed further in the next section.
FIG 2.
Construction of S. coelicolor dnaA mutant strains. (Left) Schematic representation of the stepwise strategy used to generate the S. coelicolor mutant strains. The DnaA protein-coding genes were introduced, along with the native promoter region, into the chromosomal attϕC31 attachment site, followed by attempted deletion of the native dnaA gene. (Right) Strain genotypes are listed in the table. S. coelicolor mutants were prepared using a PCR-targeting protocol (see the supplemental material).
Phosphorylation of DnaA protein alters the mutual arrangement of domains III and IV and influences protein activity.
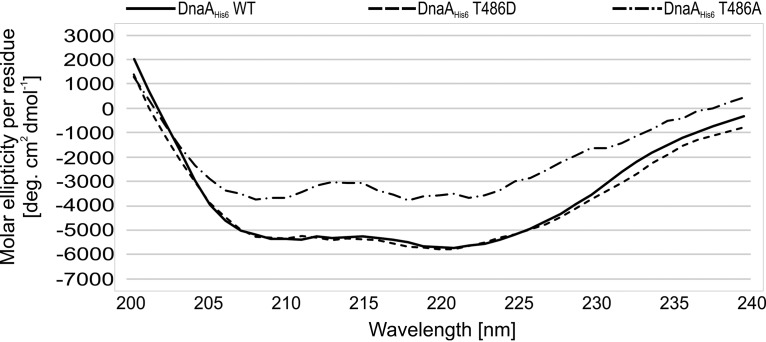
To test how phosphorylation influences DnaA activity, we purified wild-type DnaAHis6, pseudophosphorylated DnaAHis6 T486D, and phosphoablative DnaAHis6 T486A mutant proteins as previously described (27). Surprisingly, we had difficulty purifying the T486A mutant. To test whether alanine substitution impacted the protein structure, we measured the circular dichroism (CD) spectra of the purified proteins. The measured CD spectra were very similar between the wild-type and phosphomimetic mutant proteins, whereas striking differences were observed for the phosphoablative mutant (Fig. 3). This suggests that alanine substitution for threonine 486 causes structural disorder, whereas aspartic acid substitution does not. Therefore, we did not include the DnaAHis6 T486A mutant in our subsequent analyses.
FIG 3.
CD spectra of DnaA protein and its mutants. CD spectra of purified DnaAHis6 WT, DnaAHis6 T486D, and DnaAHis6 T486A proteins. Spectra were recorded in the wavelength range from 200 to 240 nm in 2-nm intervals. The data are presented as molar ellipticity per amino acid residue.
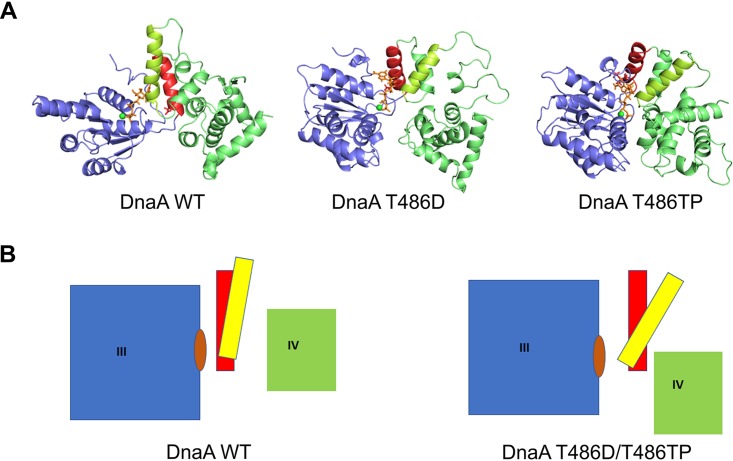
To determine the possible influence of phosphorylation on the protein structure of DnaA, we performed molecular modeling of S. coelicolor DnaA protein based on the structure of DnaA protein from Aquifex aeolicus (PDB entry 3R8F) as a template and molecular dynamics (MD) simulation for wild-type DnaA, T486D mutant DnaA, and T486-phosphorylated (T486TP) variant of DnaA from S. coelicolor. Our molecular dynamics simulations revealed that the N-terminal part of domain III (residues 316 to 484) (Fig. 4, shown in blue) is very rigid in all of the analyzed proteins, exhibiting structural fluctuation of less than 2 Å for the whole domain, regardless of substitution at position 486. Similar rigidity was observed for domain IV (Fig. 4, green). However, the region that links domains III and IV (localized within domain III, residues 485 to 512), which is composed of a few short helices, displayed a high degree of conformational dynamics. For the wild-type sequence, domains III and IV were separated from each other by a broad cleft. According to the model, a deep and narrow nucleotide-binding pocket is located between the rigid N-terminal portion of domain III and the short helices of the sequence linking domains III and IV. These helices exhibit high conformational dynamics but retain a specific, almost parallel orientation (Fig. 4, red and yellow colors). In the T486D mutant and T486TP form of DnaA, the residues are bulkier than threonine and push the red helix (residues 485 to 489) of the linker region away from the N terminus of domain III. This small displacement of the red helix leads to larger displacement of the yellow helix (residues 520 to 534), which pushes domain IV closer to domain III and widens the nucleotide-binding pocket for both studied mutants. In both T486D and T486TP models, we observe the same type of structural differences compared to the wild-type protein model. However, in the case of the T486TP model, these differences seem to be even more prominent than in the DnaA T486D model (Fig. 4). These differences may result from a different positioning of additional negative charges in both protein models (in T486D charge is localized on one oxygen atom of aspartic acid, whereas in the case of T486TP negative charge is delocalized between oxygen atoms in the phosphate group).
FIG 4.
Molecular modeling and molecular dynamics simulations for DnaA phosphorylation. (A) Structural models based on structure of DnaA protein from Aquifex aeolicus (domains III and IV) for variants of S. coelicolor DnaA. Unphosphorylated wild type, DnaA WT; pseudophosphorylated mutant, DnaA T486D; and phosphorylated variant, DnaA T486TP. Structures from the last frame of each molecular dynamics simulation are shown. (B) Schematic representation of the structural rearrangements observed among the studied variants (left, scheme for wild-type DnaA; right, scheme for both pseudophosphorylated T486D and phosphorylated T486P variants). Color code: domain III, blue; helices formed at the C-terminal part of domain III, red and yellow; domain IV, green; ATP molecule, orange. Residue 486 is located at the bottom of the red helix.
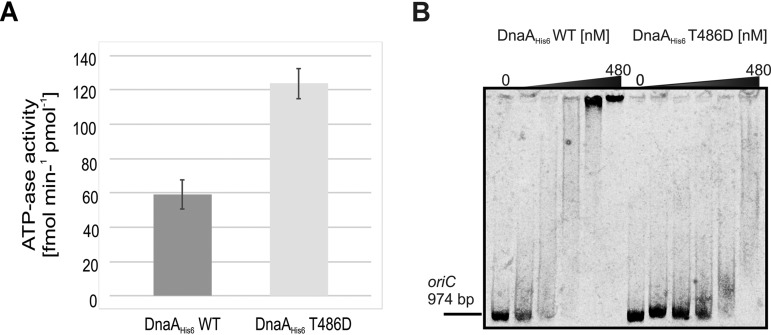
The localization of conserved T486 within DnaA domain III and the results of our molecular modeling suggest that the ATPase activity mediated by this domain is affected by phosphorylation (see Discussion). To test this hypothesis, we used the malachite green assay to analyze the intrinsic ATPase activity of DnaA. Under our experimental conditions, the wild-type protein generated free phosphate (Pi) from ATP at an average rate of 59.2 fmol min−1 per pmol of DnaAHis6, whereas the pseudophosphorylated mutant exhibited approximately 2-fold more ATPase activity, generating an average of 123.7 fmol of Pi min−1 per pmol of DnaAHis6 T486D (Fig. 5A).
FIG 5.
In vitro analysis of how phosphorylation influences DnaA initiator protein activity. (A) The malachite green assay was used to compare the ATPase activity of wild-type (DnaAHis6 WT) and pseudophosphorylated (DnaAHis6 T486D) recombinant DnaA proteins (both at 200 nM). The chart presents the data obtained from three technical repeats of each experiment. The average Pi generation rates were 59.2 ± 9.2 fmol min−1 per pmol of DnaAHis6 for the wild type and 123.7 ± 9.6 fmol min−1 per pmol of DnaAHis6 for T486D. (B) Electrophoretic mobility shift assay. OriC region from S. coelicolor was PCR amplified using near-infrared labeled primers. The linear DNA fragment (50 fmol) was incubated with increasing concentrations of wild-type and pseudophosphorylated DnaAHis6 recombinant protein (concentrations in subsequent lanes were 0, 30, 60, 120, 240, and 480 nM). The DNA fragments and nucleoprotein complexes were analyzed by 3.5% polyacrylamide gel electrophoresis.
We also performed gel retardation experiments to test the DNA-binding activity of DnaA proteins to a PCR-generated fragment containing the entire oriC region. Our pseudophosphorylated DnaA mutant exhibited lower affinity toward the oriC region than the wild-type protein, as shown by electrophoretic mobility shift experiments (Fig. 5B).
Taken together, our in vitro results show that phosphorylation of DnaA elevates its ATPase activity and weakens its affinity toward the full-length oriC region. These findings further support our hypothesis that phosphorylation of DnaA leads to formation of an initiation-inactive pool of the initiator protein.
The kinase AfsK phosphorylates DnaA.
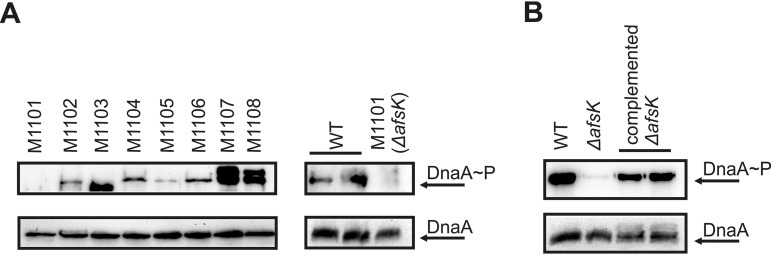
We next tried to identify the kinase responsible for DnaA phosphorylation. S. coelicolor harbors 34 genes identified as encoding putative eukaryotic-like serine/threonine protein kinases (STPKs), only a few of which have been studied to date (28). We used a number of S. coelicolor kinase deletion mutants (kindly provided by Mark Buttner) to test the phosphorylation status of DnaA protein (kinase deletion mutants, designations, and genotypes are listed in Table S1 in the supplemental material). Interestingly, among the analyzed mutants, we did not observe a DnaA signal in the anti-[p]Thr immunoprecipitates obtained from the strain lacking AfsK kinase, while the DnaA protein was detected in all analyzed mutants, including the ΔafsK mutant (Fig. 6A). In the next step, we complemented the afsK mutant, and this restored DnaA phosphorylation (Fig. 6B).
FIG 6.
Identification of the kinase involved in DnaA phosphorylation. (A, upper) Western blot analysis of the DnaA protein in anti-[p]Thr-immunoprecipitated samples. S. coelicolor wild-type (WT) and various kinase deletion strains (as indicated; genotypes are listed in Table S1) were grown in liquid medium. Cell extracts were prepared, as previously described (23), from two biological replicates of each strain and subjected to IP. The arrow indicates the DnaA phosphoprotein. (B, upper) Complementation of the ΔafsK mutant restores DnaA phosphorylation. The afsK gene present on the pKF256 plasmid (29) was introduced into the ΔafsK mutant strain. Anti-[p]Thr IP was conducted in two biological replicates as described above, and DnaA was assessed by Western blotting. The arrow indicates the DnaA phosphoprotein. (A and B, lower) The presence of DnaA protein in each examined strain was analyzed by anti-DnaA Western blotting performed on the crude cellular extracts.
Together, these results suggest that DnaA is phosphorylated by AfsK kinase. This protein is known to regulate the polar growth and branching of S. coelicolor (29).
Elevated cellular levels of AfsK influence tip-proximal replisome localization.
Previous studies showed that AfsK exerts strong effects on cell polarity, tip extension, and the initiation of new hyphal branches (29). This protein exhibits a specific subcellular localization pattern, with accumulation at the tips of vegetative hyphae. AfsK was shown to phosphorylate the DivIVA protein, which is responsible for the apical growth of S. coelicolor. Ectopic overexpression of afsK leads to phosphorylation of DivIVA, disassembly of DivIVA-containing apical growth complexes (polarisomes), arrest of tip extension, and eventually formation of new hyphal branches. The AfsK-mediated dismantling of polarisomes was proposed to function as a cellular response mechanism to inhibition of cell wall synthesis (29). Our discovery that this kinase is also responsible for DnaA phosphorylation, which alters DnaA activity, led us to examine the possible biological consequence(s) of DnaA phosphorylation.
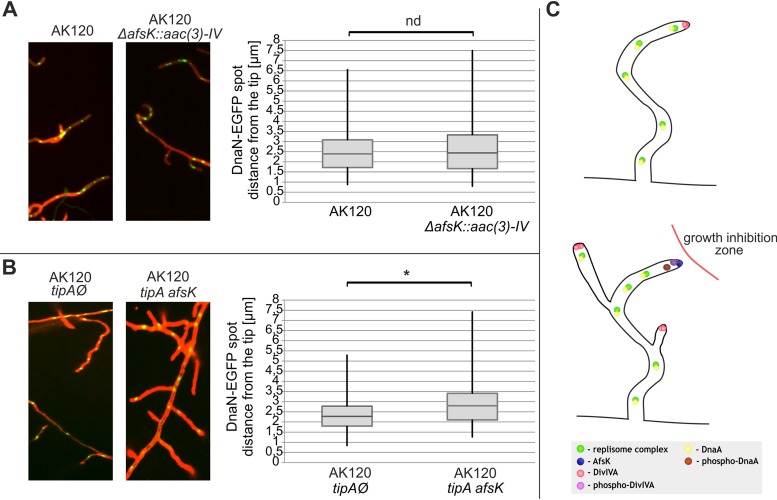
To test whether AfsK influences replication in vivo, we created S. coelicolor mutant strains in which afsK had been deleted or a thiostrepton-inducible afsK gene had been introduced, both using strain AK120 (Table S1) producing DnaN-EGFP (strain kindly provided by Agnieszka Kois-Ostrowska and Dagmara Jakimowicz). DnaN, also known as a DNA polymerase III β-clamp, is often fused with enhanced green fluorescence protein (EGFP) and used to visualize replisomes in bacteria. The appearance and disappearance of DnaN-EGFP fluorescent foci is considered to correspond to the initiation and termination, respectively, of replication (30). These strains allowed us to monitor replisome dynamics in cells that either lacked the afsK gene or overproduced AfsK proteins. The phenotypes of the obtained strains (data not shown) resembled those observed in previous studies. Deletion of afsK was found to alter the tip-to-branch distribution, increasing the average distance compared to that in the control strain, while overproduction of AfsK was associated with the emergence of multiple irregularly shaped branches in the hyphal structure (29).
Since AfsK is primarily localized at the tips of hyphae, we first analyzed the positioning of the tip-proximal replisome by performing fluorescence microscopy and calculating the tip-to-replisome distances. Deletion of afsK did not alter the localization of the closest-to-tip replisome, as the median measurements were 2.40 μm and 2.45 μm for the control strain and afsK mutant, respectively (Fig. 7A). In contrast, overproduction of AfsK significantly shifted the median measured distance from 2.28 μm to 2.74 μm for the control strain and AfsK overproduction mutant, respectively (Fig. 7B). No other striking difference was observed, leading us to conclude that AfsK, which is primarily associated with the hyphal tip, influences only the nearest available orisome and, consequently, the formation of replisome at the tip.
FIG 7.
Localization of tip-proximal replisomes in afsK mutant strains. (A and B) Representative images of DnaN-EGFP fluorescence in the wild-type strain (AK120) and ΔafsK mutant [AK120 ΔafsK::aac(3*)-IV] (A) and in the control strain (empty vector, AK120 tipAØ) and induced afsK expression strain (AK120 tipA afsK) (B). Images show an overlay of DnaN-EGFP fluorescence (green) and WGA-Texas Red staining (red, used for cell wall visualization). The distances between the hyphal tip and tip-proximal DnaN-EGFP foci were measured with the AxioVision Rel. 4.8 software and plotted on the diagrams presented in panels A and B. Box plots show the range of obtained results (vertical line) with the median of distribution (horizontal line) and the first and third quartiles (horizontal borders of boxes) indicated. Significant differences (P < 0.05) are marked by an asterisk; nd, not significantly different. The numbers of measurements were 90 each in panel A (AK120 and ΔafsK mutant) and 130 each in panel B (AK120 tipAØ and AK120 tipA afsK mutants). (C) Schematic representation of events occurring upon hyphal growth inhibition (mimicked by AfsK overproduction). AfsK negatively regulates polar growth by phosphorylation of DivIVA polarisome protein, dismantling of apical growth complexes, and emergence of new hyphal branches as a consequence (29). Additionally, AfsK inhibits chromosome replication by phosphorylation of DnaA initiator protein in closest-to-tip localization.
DISCUSSION
The lifestyle of streptomycetes is unusual among bacteria and is, in many ways, similar to that of filamentous fungi. Streptomyces spp. are predominantly soil-dwelling bacteria that grow as branching hyphae and disperse through spores. Unlike many rod-shaped bacteria, such as E. coli, cell wall extension occurs by the incorporation of peptidoglycan precursors at the tips of the growing hyphae (31). Another distinctive feature of streptomycetes is the occurrence of multinucleoid cellular compartments that are divided into unigenomic spores at the end of the life cycle. This feature suggests that apical hyphal growth must be strictly coordinated with cell cycle events, such as DNA replication and chromosome segregation. Although both processes have been studied extensively in Streptomyces and their dynamics have been well established (3, 4, 32), the cellular mechanisms responsible for choosing the chromosome that will undergo replication within a given compartment remain unknown.
In this study, we provide insights into the phosphorylation of the DnaA initiator protein in S. coelicolor and suggest a novel bacterial regulatory mechanism for initiation of chromosome replication. Although a previous phosphoproteomic study suggested phosphorylation of DnaA protein at threonine residue 486 (23), the biological role of this modification remained unknown. Here, we show that the phosphorylation of DnaA in S. coelicolor functions as a site-specific mechanism (acting at the hyphal tips) that negatively regulates the initiation of chromosome replication. Interestingly, threonine 486 is a highly conserved residue in bacterial initiator proteins (see Fig. S1 in the supplemental material), and its localization within domain III suggests that this phosphorylation influences key activities of DnaA. Our molecular modeling further showed that this phosphorylation is predicted to yield conformational changes within the region that links domains III and IV, and that the studied phosphorylation mutant presumably exhibits widening of the nucleotide-binding pocket (Fig. 4). The presence of ATP at the protein exterior could increase the nucleotide exchange rate and the efficiency of ATP hydrolysis. Indeed, the presence of the additional negative charge derived from aspartic acid in the phosphomimetic T486D mutation doubled the in vitro ATPase activity of DnaA (Fig. 5A). It is well known that DnaA binds both ADP and ATP, but the latter is crucial for functional orisome formation and DNA unwinding (33). Thus, DnaA phosphorylation may shift the ATP-DnaA/ADP-DnaA ratio toward the ADP-bound form (due to higher ATPase activity) and possibly create a pool of initiation-inactive DnaA. This could explain our observation that the pseudophosphorylated T486D mutant exhibited a lower affinity toward the oriC region (Fig. 5B). We suggest that, upon phosphorylation, the cooperative binding of the replication origin (33, 34) is disturbed by overrepresentation of DnaA-ADP molecules. Thus, when DnaA is phosphorylated, the functional orisome presumably is not formed. The negative regulatory function of DnaA phosphorylation is further supported by our failure to construct an S. coelicolor mutant strain expressing only the pseudophosphorylated dnaA allele (Fig. 2).
We also show that DnaA phosphorylation in S. coelicolor occurs during the growth phases, in which DNA replication takes place. More interestingly, the DnaA protein was not identified in spores. (Fig. 1C). Since most cellular processes (including chromosome replication) are inactive in Streptomyces spores, this finding suggests that DnaA phosphorylation is primarily responsible for modulating the intensity of origin-firing events during the vegetative phase. We speculated that DnaA phosphorylation somehow corresponds to the complex growth mode of Streptomyces (e.g., tip extension and branching). Our data suggest that phosphorylation of DnaA is site specific. We found that the AfsK kinase associated with the polar growth of hyphae is responsible for phosphorylation of DnaA protein (Fig. 6). As previously reported, the specific localization of AfsK at the tip of the hyphae leads to phosphorylation of the DivIVA polarisome protein. Following this phosphorylation, the tip-associated DivIVA-containing protein complexes are disassembled and further growth of the hyphal tip is inhibited. Elevated AfsK activity and increased DivIVA phosphorylation have been observed following the antibiotic-induced inhibition of cell wall synthesis (29). Thus, AfsK-mediated DivIVA phosphorylation has been suggested to act as a cellular response to growth obstacles or the presence of cell wall synthesis-inhibiting molecules (29). We found that chromosome replication, which was visualized by the appearance of polymerase III β-clamp (a replisome marker) foci, occurred further from the hyphal tip upon upregulation of AfsK (Fig. 7B). Replisomes appear upon the DnaA-triggered initiation of chromosome replication, and the current data do not suggest that AfsK interacts directly with replisome proteins. We propose that the observed differences in the distance between the tip of the cell and the first replisome result from an inability to form a functional, tip-proximal orisome in the strain overproducing AfsK. This may be explained by a higher concentration of phosphorylated DnaA at the cell tip, where the DnaA molecules presumably localize with the overproduced AfsK. However, we cannot exclude the possibility that the observed differences in the tip-proximal replisome localization could result from phosphorylation of AfsK substrates other than DnaA. Surprisingly, deletion of the afsK gene had no effect on the positioning of the tip-proximal replisome. Although a previous study showed that the ΔafsK strain was uniquely characterized by a prolonged distance between the tip and first lateral branch, we did not observe any difference in the localization of the replisome in this strain versus the wild-type strain (Fig. 7A).
Since DnaA phosphorylation appears to occur predominantly at the tips, the phosphorylated fraction of DnaA protein should be relatively small with respect to the cellular pool of the initiator protein, which is presumably a reason why we were unable to accurately estimate the percentage of phosphorylated molecules. Moreover, it explains why phosphorylated DnaA was identified in only one of the two major phosphoproteomic studies of S. coelicolor (23, 24). We do not yet know how S. coelicolor cells deal with phosphorylated DnaA molecules. The absence of initiator protein in spores suggests that both nonphosphorylated and phosphorylated DnaA molecules undergo extensive proteolysis upon termination of the reproductive phase. However, here we did not identify any protease that could be involved in this process.
Notably, orthologs of AfsK are exclusively found in mycelial actinomycetes that are closely related to Streptomyces and exhibit a complex life cycle. Based on our present results we propose that, in these bacteria, the AfsK-mediated regulatory pathway serves as an additional, nonessential energy-saving mechanism during the inhibition of hyphal growth. Moving beyond the previous reports showing that AfsK negatively regulates polar growth, we suggest that AfsK prevents the initiation of chromosome replication within growth-inhibited apical cellular compartments (Fig. 7C).
MATERIALS AND METHODS
Bacterial strains and general methods.
The utilized bacterial strains, plasmids, and primers are listed in Tables S1 and S2 in the supplemental material. Buffers, media, culture conditions, and general genetic manipulations were as described previously for E. coli (35) and Streptomyces (36).
IP.
S. coelicolor strains were cultured either in liquid organic medium 79 without dextrose (37) for 24 to 26 h or on MS agar plates covered with cellophane discs and grown to the desired stage of the life cycle (36). Cells were harvested by centrifugation and resuspended in lysis buffer, which was prepared as described previously (23). Crude protein extracts were subjected to IP with polyclonal anti-phospho-Thr (anti-[p]Thr) antibodies (for the ΔafsK complementation strain, 1:16 dilution; number 61-8300; Thermo Scientific; for all other experiments, 1:50 dilution; number 9381; Cell Signaling Technology). For each sample, 200 μl of diluted antibodies was coupled to 50 μl Pierce protein A magnetic beads (Thermo Scientific) during a 20-min incubation at 20°C, and the sample was washed three times with phosphate-buffered saline with Tween 20 (PBST) buffer. The extracted total proteins (1 mg) were applied to antibody-coupled beads, and IP was conducted overnight at 4°C. Washing and elution steps were carried out according to the instructions provided with the magnetic beads.
Phos-tag gel retardation and Western blotting.
The phospho-DnaA gel retardation experiment was carried out using 50 μM Phos-tag and 0.1 mM Mn2+ gel additives, according to manufacturer’s instructions (FUJIFILM Wako Chemicals, USA). Briefly, immunoprecipitated protein samples were aliquoted into equal-volume portions (10 μl). One portion was subjected to phosphatase treatment with 400 U of lambda protein phosphatase (New England BioLabs). The reaction was carried out in the provided buffer (final volume, 20 μl) for 30 min at 37°C and stopped by a 5-min incubation with Laemmli sample buffer at 95°C. Proteins were separated by 10% Phos-tag gel SDS-PAGE. After electrophoresis, the gel was incubated for 10 min in TBS buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl) containing 10 mM EDTA and then washed three times with TBS buffer. Proteins were transferred to a nitrocellulose membrane (Millipore), which was blocked for 1 h with 5% (wt/vol) nonfat dry milk in TBST buffer (TBS with 0.05% Tween 20). The membrane was washed with TBST buffer, incubated for 1.5 h at 20°C with anti-DnaA antiserum from rabbit (diluted 1:3,000), washed three times with TBST, and incubated for 1 h at 20°C with goat anti-rabbit IgG conjugated to horseradish peroxidase (1:10,000; Santa Cruz Biotechnology). The membrane was washed six times in TBST solution, the proteins were visualized with the SuperSignal West Pico Plus chemiluminescence substrate (Thermo Scientific), and the results were captured using a ChemiDoc XRS system (Bio-Rad). For Western blot analysis of experiments that did not involve the Phos-tag additive, we omitted the TBS/EDTA incubation step. For Western blot analysis of crude cellular extracts, we used 10 μg of total cellular protein for each strain analyzed.
Protein purification.
Purification of recombinant DnaAHis6 wild-type and mutant proteins was conducted as previously described (27).
Circular dichroism.
Spectra were measured with Jasco JD715 equipment. Samples of purified proteins DnaAHis6 WT, DnaAHis6 T486D, and DnaAHis6 T486A (0.16 mg/ml, 0.25 mg/ml, and 0.25 mg/ml, respectively) were subjected to buffer exchange into CDS buffer (20 mM phosphate buffer, pH 7.5), and CD spectrum of each protein was recorded immediately. Spectra were recorded in a wavelength range from 200 to 240 nm using a path length of 1 mm.
In vitro analysis of DnaA activity.
EMSA (electrophoretic mobility shift assay) experiments were carried out as described previously (27), with some modifications. The 974-bp oriC fragments were amplified using a pair of near-infrared-labeled primers (Table S2). The indicated amounts of DnaAHis6 proteins and DNA fragment were incubated for 15 min at 20°C in DNA binding buffer (20 mM HEPES-KOH [pH 7.6], 5 mM magnesium acetate, 1 mM dithiothreitol, 5% [vol/vol] glycerol, 0.5 mg/ml bovine serum albumin [BSA], 5 mM ATP) in a final volume of 20 μl. The reaction products were subjected to 3.5% native PAGE at 4°C for 6 h. Near-infrared signals were detected using an Azure c600 imaging system (Azure Biosystems).
DnaAHis6 ATPase activity was measured using the malachite green assay, which was carried out using a PiColorLock gold phosphate detection system (Innova Biosciences) according to the manufacturer’s instructions. Briefly, 200 nM recombinant DnaAHis6 proteins was reacted at 37°C in DNA binding buffer (described above) supplemented with 10 mM ATP in a final volume of 1 ml. At the indicated time points, 50 μl of each sample was collected and mixed in a 96-well plate with an appropriate volume of malachite green reagent. After 30 min, absorbance at 620 nm was measured three times per sample/time point. The amount of inorganic phosphate generated was calculated by plotting the results against a standard curve.
Molecular modeling and MD simulations.
A structural model of Streptomyces coelicolor DnaA protein was built using the SWISS-MODEL (38) service by using the structure of DnaA protein from Aquifex aeolicus deposited in the PDB database (entry 3R8F) as a template and sequence of DnaA protein from Streptomyces coelicolor deposited in the UNIPROT database (P27902). The three-dimensional model covers residues 316 to 653. The position of the ATP molecule as well as magnesium ion was retained from the template to the modeled structure. For molecular dynamics (MD) simulations, the AmberTools v. 16 package was used (39). The system was prepared for MD simulations using standard protocols: charge neutralization (adding counter ions) and solvation in a TIP3P water rectangular box. All MD simulations were run at a constant temperature (300 K) using periodic boundary conditions as well as Ewald summation for electrostatic interactions. System density was equilibrated for about 10- to 20-ns running simulations under constant pressure, and all production simulations were run using a constant volume regime. Mutants of DnaA T486D and T486TP were built using standard AmberTools v. 16 protocols (39) by in silico mutation of the residue at position 486 to aspartic acid (D) or phosphorylated threonine (TP). In the case of all three studied variants, MD simulations lasted at least 200 ns. MD trajectories were analyzed using software from the AmberTools v.16 package. Three-dimensional structures were depicted using the PyMol package (PyMOL Molecular Graphics System, version 2.0; Schrödinger, LLC.).
Microscopy.
The indicated strains were inoculated from pregerminated spores at the acute-angle junction of coverslips, inserted at a 45° angle in MM agar containing 1% mannitol (36), and cultured at 30°C for 24 h. Staining was performed as previously described (40). Briefly, samples were fixed for 10 min with paraformaldehyde-glutaraldehyde, washed with PBS, and blocked with 2% BSA. WGA-Texas Red (10 μg/ml; Molecular Probes) was used for cell wall visualization. After five washes with PBS, coverslips were mounted on microscope slides covered with a BSA-glycerol mixture. The presence of fluorescence foci was analyzed using a Zeiss Axio Imager Z1 microscope equipped with a 100× objective. All images were analyzed using the AxioVision Rel. 4.8 software. The measured distances were subjected to Mann-Whitney statistical analysis due to their nonnormal distribution. Significance was accepted at a P value of <0.05.
Supplementary Material
ACKNOWLEDGMENTS
We declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
T.Ł., M.W., and J.Z.-C. designed the experiments. T.Ł. performed research. S.O. performed molecular modeling and molecular dynamics simulations. T.Ł., S.O., and J.Z.-C. wrote the manuscript. K.F. and M.W. performed critical revision.
This study was supported by the National Science Center (Poland), Preludium grant 2017/25/N/NZ1/01030 to T.Ł., and by Maestro grant 2012/04/A/NZ1/00057 to J.Z.-C. Computational resources used in this project were provided by the Informatics Center of the Metropolitan Academic Network (IC MAN-TASK) in Gdańsk, Poland.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, Klenk H-P, Clément C, Ouhdouch Y, van Wezel GP. 2016. Taxonomy, physiology, and natural products of actinobacteria. Microbiol Mol Biol Rev 80:1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley SD, Chater KF, Cerdeño-Tárraga A-M, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang C-H, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream M-A, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 3.Ruban-Ośmiałowska B, Jakimowicz D, Smulczyk-Krawczyszyn A, Chater KF, Zakrzewska-Czerwińska J. 2006. Replisome localization in vegetative and aerial hyphae of Streptomyces coelicolor. J Bacteriol 188:7311–7316. doi: 10.1128/JB.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kois-Ostrowska A, Strzałka A, Lipietta N, Tilley E, Zakrzewska-Czerwińska J, Herron P, Jakimowicz D. 2016. Unique function of the bacterial chromosome segregation machinery in apically growing streptomyces–targeting the chromosome to new hyphal tubes and its anchorage at the tips. PLoS Genet 12:e1006488. doi: 10.1371/journal.pgen.1006488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuller RS, Funnell BE, Kornberg A. 1984. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell 38:889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- 6.Wolański M, Donczew R, Zawilak-Pawlik A, Zakrzewska-Czerwińska J. 2014. oriC-encoded instructions for the initiation of bacterial chromosome replication. Front Microbiol 5:735. doi: 10.3389/fmicb.2014.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zawilak-Pawlik A, Nowaczyk M, Zakrzewska-Czerwińska J. 2017. The role of the N-terminal domains of bacterial initiator DnaA in the assembly and regulation of the bacterial replication initiation complex. Genes 8:136. doi: 10.3390/genes8050136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nozaki S, Ogawa T. 2008. Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology 154:3379–3384. doi: 10.1099/mic.0.2008/019745-0. [DOI] [PubMed] [Google Scholar]
- 9.Ozaki S, Katayama T. 2009. DnaA structure, function, and dynamics in the initiation at the chromosomal origin. Plasmid 62:71–82. doi: 10.1016/j.plasmid.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Duderstadt KE, Chuang K, Berger JM. 2011. DNA stretching by bacterial initiators promotes replication origin opening. Nature 478:209–213. doi: 10.1038/nature10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Y-B, Ratnakar PVAL, Mohanty BK, Bastia D. 1996. Direct physical interaction between DnaG primase and DnaB helicase of Escherichia coli is necessary for optimal synthesis of primer RNA. Proc Natl Acad Sci U S A 93:12902–12907. doi: 10.1073/pnas.93.23.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katayama T, Ozaki S, Keyamura K, Fujimitsu K. 2010. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol 8:163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 13.Campbell JL, Kleckner N. 1990. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62:967–979. doi: 10.1016/0092-8674(90)90271-F. [DOI] [PubMed] [Google Scholar]
- 14.Lu M, Campbell JL, Boye E, Kleckner N. 1994. SeqA: a negative modulator of replication initiation in E. coli. Cell 77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 15.Nievera C, Torgue JJ-C, Grimwade JE, Leonard AC. 2006. SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-RC. Mol Cell 24:581–592. doi: 10.1016/j.molcel.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bogan JA, Helmstetter CE. 1997. DNA sequestration and transcription in the oriC region of Escherichia coli. Mol Microbiol 26:889–896. doi: 10.1046/j.1365-2958.1997.6221989.x. [DOI] [PubMed] [Google Scholar]
- 17.Braun RE, O'Day K, Wright A. 1985. Autoregulation of the DNA replication gene dnaA in E. coli K-12. Cell 40:159–169. doi: 10.1016/0092-8674(85)90319-8. [DOI] [PubMed] [Google Scholar]
- 18.Messer W, Weigel C. 1997. DnaA initiator–also a transcription factor. Mol Microbiol 24:1–6. doi: 10.1046/j.1365-2958.1997.3171678.x. [DOI] [PubMed] [Google Scholar]
- 19.Jonas K, Liu J, Chien P, Laub MT. 2013. Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell 154:623–636. doi: 10.1016/j.cell.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorbatyuk B, Marczynski GT. 2005. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol 55:1233–1245. doi: 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- 21.Scholefield G, Errington J, Murray H. 2012. Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J 31:1542–1555. doi: 10.1038/emboj.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scholefield G, Murray H. 2013. YabA and DnaD inhibit helix assembly of the DNA replication initiation protein DnaA: regulation of DnaA by YabA and DnaD. Mol Microbiol 90:147–159. doi: 10.1111/mmi.12353. [DOI] [PubMed] [Google Scholar]
- 23.Parker JL, Jones AME, Serazetdinova L, Saalbach G, Bibb MJ, Naldrett MJ. 2010. Analysis of the phosphoproteome of the multicellular bacterium Streptomyces coelicolor A3(2) by protein/peptide fractionation, phosphopeptide enrichment and high-accuracy mass spectrometry. Proteomics 10:2486–2497. doi: 10.1002/pmic.201000090. [DOI] [PubMed] [Google Scholar]
- 24.Manteca A, Ye J, Sánchez J, Jensen ON. 2011. Phosphoproteome analysis of Streptomyces development reveals extensive protein phosphorylation accompanying bacterial differentiation. J Proteome Res 10:5481–5492. doi: 10.1021/pr200762y. [DOI] [PubMed] [Google Scholar]
- 25.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. 2006. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics 5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Aoki Y, Yoshida Y, Yoshida M, Kawaide H, Abe H, Natsume M. 2005. Anthranilic acid, a spore germination inhibitor of phytopathogenic Streptomyces sp. B-9-1 causing root tumor of melon. Actinomycetologica 19:48–54. doi: 10.3209/saj.19.48. [DOI] [Google Scholar]
- 27.Zawilak-Pawlik AM, Kois A, Zakrzewska-Czerwinska J. 2006. A simplified method for purification of recombinant soluble DnaA proteins. Protein Expr Purif 48:126–133. doi: 10.1016/j.pep.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 28.Petrickova K, Petricek M. 2003. Eukaryotic-type protein kinases in Streptomyces coelicolor: variations on a common theme. Microbiology 149:1609–1621. doi: 10.1099/mic.0.26275-0. [DOI] [PubMed] [Google Scholar]
- 29.Hempel AM, Cantlay S, Molle V, Wang S-B, Naldrett MJ, Parker JL, Richards DM, Jung Y-G, Buttner MJ, Flärdh K. 2012. The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proc Natl Acad Sci U S A 109:2371–2379. doi: 10.1073/pnas.1207409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trojanowski D, Hołówka J, Zakrzewska-Czerwińska J. 2018. Where and when bacterial chromosome replication starts: a single cell perspective. Front Microbiol 9:2819. doi: 10.3389/fmicb.2018.02819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flärdh K, Richards DM, Hempel AM, Howard M, Buttner MJ. 2012. Regulation of apical growth and hyphal branching in Streptomyces. Curr Opin Microbiol 15:737–743. doi: 10.1016/j.mib.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 32.Wolański M, Jakimowicz D, Zakrzewska-Czerwińska J. 2012. AdpA, key regulator for morphological differentiation regulates bacterial chromosome replication. Open Biol 2:120097. doi: 10.1098/rsob.120097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Speck C, Messer W. 2001. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J 20:1469–1476. doi: 10.1093/emboj/20.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majka J, Zakrzewska-Czerwiñska J, Messer W. 2001. Sequence recognition, cooperative interaction, and dimerization of the initiator protein DnaA of Streptomyces. J Biol Chem 276:6243–6252. doi: 10.1074/jbc.M007876200. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 36.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, UK. [Google Scholar]
- 37.Atlas RM. 2004. Handbook of microbiological media. CRC Press, Inc, Boca Raton, FL. [Google Scholar]
- 38.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:296–303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Case DA, Betz RM, Cerutti DS, Cheatham TE III, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, Homeyer N, Izadi S, Janowski P, Kaus J, Kovalenko A, Lee TS, LeGrand S, Li P, Lin C, Luchko T, Luo R, Madej B, Mermelstein D, Merz KM, Monard G, Nguyen H, Nguyen HT, Omelyan I, Onufriev A, Roe DR, Roitberg A, Sagui C, Simmerling CL, Botello-Smith WM, Swails J, Walker RC, Wang J, Wolf RM, Wu X, Xiao L, Kollman PA. 2016. Amber 16. University of California, San Francisco, CA. doi: 10.13140/rg.2.2.27958.70729. [DOI] [Google Scholar]
- 40.Jakimowicz D, Zydek P, Kois A, Zakrzewska-Czerwińska J, Chater KF. 2007. Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol Microbiol 65:625–641. doi: 10.1111/j.1365-2958.2007.05815.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.