Abstract
Fungal pathogens represent a major human threat affecting more than a billion people worldwide. Invasive infections are on the rise, which is of considerable concern because they are accompanied by an escalation of antifungal resistance. Deciphering the mechanisms underlying virulence traits and drug resistance strongly relies on genetic manipulation techniques such as generating mutant strains carrying specific mutations, or gene deletions. However, these processes have often been time-consuming and cumbersome in fungi due to a number of complications, depending on the species (e.g., diploid genomes, lack of a sexual cycle, low efficiency of transformation and/or homologous recombination, lack of cloning vectors, nonconventional codon usage, and paucity of dominant selectable markers). These issues are increasingly being addressed by applying clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9 mediated genetic manipulation to medically relevant fungi. Here, we summarize the state of the art of CRISPR–Cas9 applications in four major human fungal pathogen lineages: Candida spp., Cryptococcus neoformans, Aspergillus fumigatus, and Mucorales. We highlight the different ways in which CRISPR has been customized to address the critical issues in different species, including different strategies to deliver the CRISPR–Cas9 elements, their transient or permanent expression, use of codon-optimized CAS9, and methods of marker recycling and scarless editing. Some approaches facilitate a more efficient use of homology-directed repair in fungi in which nonhomologous end joining is more commonly used to repair double-strand breaks (DSBs). Moreover, we highlight the most promising future perspectives, including gene drives, programmable base editors, and nonediting applications, some of which are currently available only in model fungi but may be adapted for future applications in pathogenic species. Finally, this review discusses how the further evolution of CRISPR technology will allow mycologists to tackle the multifaceted issue of fungal pathogenesis.
Introduction
The most common fungal diseases are superficial infections, affecting almost two billion people worldwide [1]. Despite being much less common, invasive fungal infections are nonetheless associated with high mortality rates. Fungal pathogens are therefore increasingly recognized as a major human threat. According to current (likely under-) estimates, 1.6 million people die every year from invasive diseases caused by different fungal species (mainly Cryptococcus, Candida, Aspergillus, and Pneumocystis), almost the same as from tuberculosis [2]. We are also currently facing a concerning escalation of antifungal resistance, with reports of azole-resistant Aspergillus fumigatus and the spread of multidrug-resistant Candida auris [3,4]. In this context, there is an urgent need for efficient genetic manipulation tools to further our understanding of the biology and pathophysiology of fungal pathogens and to decipher drug resistance mechanisms, which are critical for developing novel therapeutic strategies. However, genetic manipulation has often been time-consuming and cumbersome in fungi, particularly in species with diploid genomes that lack a sexual cycle. A low efficiency of transformation and/or homologous recombination, the absence of natural plasmids, and the lack of cloning vectors or the limited number of dominant markers available for selection are additional factors hampering this process [5,6].
Over the last decade, the groundbreaking discovery of clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 revolutionized genome editing [7]. Normally involved in bacterial adaptive immunity against bacteriophages, the discovery that Cas9 endonuclease could not only target infecting viral DNA but virtually any DNA paved the road for the application of this technique to eukaryotic cells, including mammalian cells, plants, and fungi (including the model yeast Saccharomyces cerevisiae) [8]. In this review we describe the state of the art of CRISPR–Cas9 applications in four major human fungal pathogens, Candida spp., Cryptococcus neoformans, A. fumigatus, and Mucorales, dissecting the customized strategies for the delivery of functional CRISPR–Cas9 elements. Moreover, we discuss the most promising future perspectives and their further development that will allow mycologists to face the challenge of fungal pathogenesis.
The CRISPR–Cas9 toolbox: How does it work?
The potential of CRISPR-associated protein Cas9 for genome editing was first reported in 2012 [7,9]. Cas9, a type II RNA-guided endonuclease, introduces a double-stranded break (DSB) 3 bp upstream of a protospacer adjacent motif (PAM, NGG for Streptococcus pyogenes Cas9). Cas9 is directed to the site by a single-guide RNA (sgRNA), encompassing both target specificity and Cas9 binding activity—mediated in the native bacterial system by the CRISPR-RNA (crRNA) and the trans-activating crRNA (tracrRNA), respectively. The DSB can be repaired by nonhomologous end joining (NHEJ), usually resulting in insertions or deletions leading to frame shift of the reading frame and premature stop codons. Microhomology-mediated end joining (MMEJ) is an alternative, also error-prone repair pathway requiring short homology regions (2–40 bp) [10]. If a donor template with appropriate homology regions is available (e.g. the second allele in diploid genomes, or exogenous DNA), repair may occur by homology-directed repair (HDR). Precise editing exploits HDR machinery by providing the cells with donor DNAs harboring the intended mutation flanked by homology arms. Although conventional methods can also be used to introduce mutations by HDR, DSBs significantly increase the rate of these events by up to 4,000-fold [11–15]. As a result, introducing Cas9-mediated DSBs opens up the possibility to exploit HDR even in species in which this pathway is normally not efficient.
Tips and tricks of CRISPR–Cas9 in fungi
Expression of CAS9, codon optimization, and nuclear localization of the protein are crucial for the success of CRISPR–Cas9 gene editing. In addition, the expression of a sgRNA suitable for interaction with Cas9 is pivotal. Not surprisingly, there is no one method for introducing these elements that is suitable for every fungal species. Firstly, whereas the versions of Cas9 with codons optimized for human cells work in C. neoformans and A. fumigatus [16,17], further modifications are required in many Candida species, in which CTG is translated as serine [18]. The choice of the regulatory elements to drive expression of CAS9 and sgRNA is another potential concern. Despite some flexibility among closely related species, species-specific promoters are usually more effective [19,20]. The sgRNA is usually expressed from an RNA pol III promoter because it is a nucleus-localized noncoding RNA molecule. Although RNA pol III promoters of some species have been characterized (e.g., SNR52 promoter in C. albicans), mining a genome to find de novo RNA pol III promoters is a cumbersome process of trial and error, which can only be partially streamlined if transcriptome data are available to guide the search. For this reason, suitable RNA pol II promoters can be used instead, provided that the sgRNA molecule is flanked by RNA elements (typically ribozymes or tRNAs) that will mediate the removal of the 5’ cap and the 3’ poly(A) tail [21].
An additional level of complexity is introduced by the delivery system chosen. Unlike S. cerevisiae, no natural plasmids are found in fungal pathogens, and circular vectors are not always stable (e.g., most vectors need to be linearized before being transformed into C. albicans and C. neoformans). In Candida glabrata, Candida parapsilosis, and A. fumigatus, the availability of autonomously replicating sequences allows the use of episomal plasmids [22,23]. In Mucorales, by contrast, plasmids usually do not integrate, and when they do, they may lead to unexpected genomic changes or recircularization [24]. One way to get around these problems is to use in vitro assembled ribonucleoprotein complexes (RNP) [25–28]. In this approach, cells are directly transformed with the Cas9–sgRNA complex, thus bypassing the need for any species-specific expression or delivery tailoring. However, the higher cost makes this approach unsuitable for large-scale efforts.
Regardless of the way that the DSB is induced, the outcome depends on the repair strategy. Fungal species differ intrinsically in DNA-repair mechanisms. C. albicans and C. parapsilosis, like S. cerevisiae, rely mostly on HDR, whereas Candida lusitaniae, A. fumigatus, and Mucorales rely mostly on NHEJ, and Candida glabrata and Candida tropicalis use both HDR and NHEJ [16,20,29–31]. Disrupting NHEJ-related genes increases HDR efficiency in some species (e.g., Ku70/Lig4 deletion increases efficiency from 25% to 81% in C. lusitaniae haploid cells) [20]. However, this strategy is not ideal, because NHEJ-defective strains may display reduced virulence [17]. Even in species with high levels of HDR, CRISPR–Cas9 increases the efficiency of this process so much that shorter sequences can be used to direct homologous recombination. As an example, homology arms as short as 50 bp arms resulted in 61% efficiency for deleting ADE2 in C. albicans [30], and homology arms of 30 bp resulted in almost a 100% gene-editing efficiency in Candida parapsilosis [29]. Very often ADE2, a gene involved in the adenine biosynthetic pathway, is selected as proof of principle target when implementing CRISPR–Cas9 in a new species. In fact, ade2 mutants are easy to screen because the interruption of the pathway results in the accumulation of a red to pink pigment that colors the colonies.
Finally, general recommendations for guide design to ensure efficiency and reduce off-target effects also apply to these fungi [32]. Numerous guide-design software programs are now available, some allowing uploading of custom genomes, such as EUPaGDT (http://grna.ctegd.uga.edu) or CHOPCHOP (http://chopchop.cbu.uib.no/).
Current methods available for genome editing in fungal pathogens
Figs 1 to 3 illustrate representative methods for CRISPR-genome editing developed for the fungal pathogens discussed below.
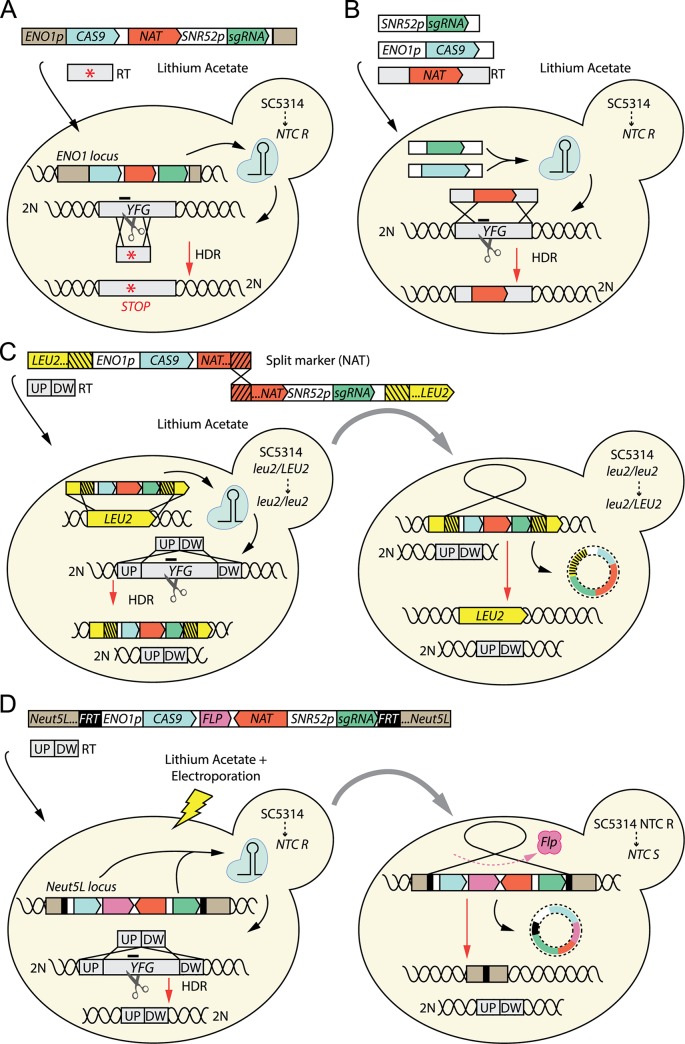
Fig 1. CRISPR–Cas9 methods in Candida albicans.
The vignettes illustrate four representative methods used for CRISPR–Cas9 genetic manipulation of C. albicans. The constructs expressing Cas9 and the sgRNA and the RT are depicted. Promoter sequences are in white, unless they constitute part of the upstream homology arm for the integration in the genomic locus, in which case they have the same color as the gene targeted for integration. The transformation method is also indicated. The targeted site on YFG is indicated by a dash. The ploidy of the alleles is indicated next to the DNA helix. (A) CAS9, the sgRNA, and the NAT cassette are stably integrated at the ENO1 locus, and HDR mediates the introduction of an RT carrying a stop codon following Cas9-induced cleavage [18]. (B) Transformation of linear DNA fragments into the cell results in transient expression of Cas9 and sgRNA, and HDR results in the integration of a NAT cassette into the targeted locus [33]. A further modification of this system allows for marker recycling [34]. (C) In the LEUpOUT approach, a split-marker strategy enables the in vivo reconstitution of the selectable cassette for the expression of the CRISPR elements. Integration of the reconstituted cassette disrupts the functional LEU2 locus of a LEU2/leu2 heterozygous strain. HDR between the targeted locus and the RT following Cas9 cut results in the deletion of the gene. HDR between directed repeats flanking the cassette (depicted as black lines on yellow background) mediates the removal of CRISPR elements and the restoration of the LEU2 ORF, which can be selected on leucine dropout medium [35]. Note that the leu2 deleted allele originally present in the LEU2/leu2 heterozygous strain remains untouched during the entire procedure, and so it is not depicted in the scheme. (D) An improved version of the method depicted in (A) allows for marker recycling. A cassette expressing the CRISPR elements and containing a FLP recombinase is flanked by FRT sequences (targets of Flp) and homology arms that mediate its integration in the Neut5L locus. HDR with an RT results in the target gene deletion. Activation of the inducible promoter driving the expression of Flp is followed by excision of the cassette, leaving only an FRT sequence in the Neut5L locus [30]. CRISPR, clustered regularly interspaced short palindromic repeats; DW, downstream; HDR, homology-directed repair; NTC R, Nourseothricin resistant; NTC S, Nourseothricin susceptible; ORF, open reading frame; RT, repair template; sgRNA, single-guide RNA; UP, upstream; YFG, your favorite gene.
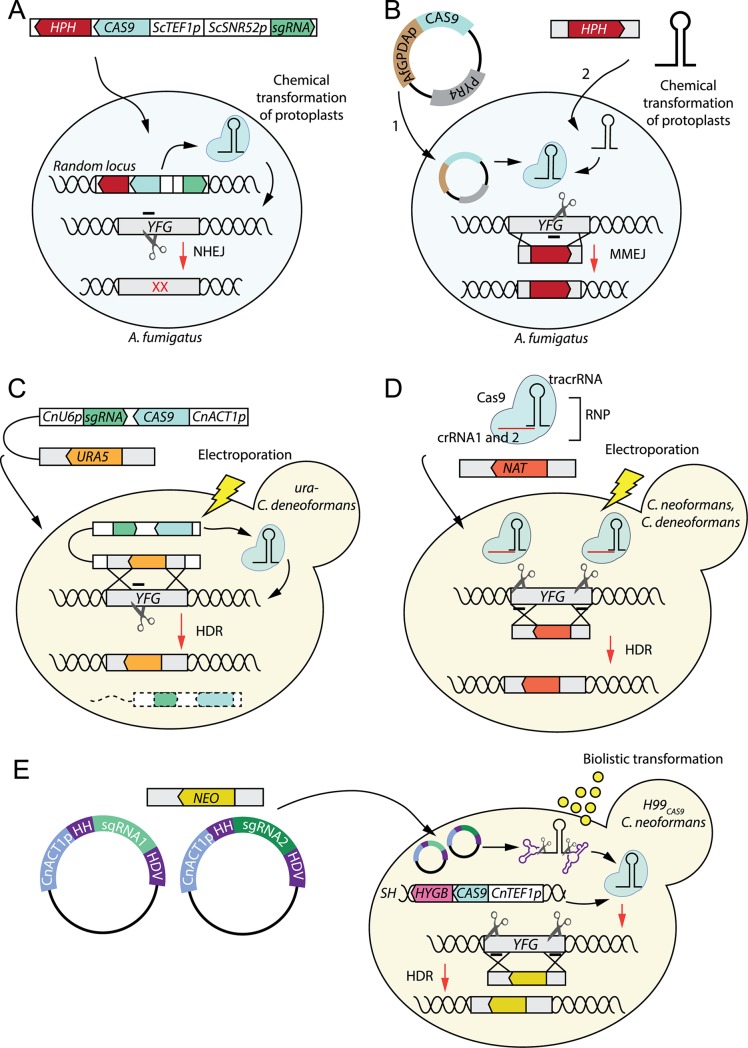
Fig 3. CRISPR–Cas9 methods in Aspergillus fumigatus, Cryptococcus neoformans and C. deneoformans.
The vignettes illustrate representative methods for CRISPR–Cas9 genetic manipulation of A. fumigatus (A–B), C. neoformans, and C. deneoformans (C–E). All the species depicted here are haploid. (A) In A. fumigatus, the CRISPR elements are integrated randomly in the genome of protoplasts and selected due to the presence of an HPH cassette. NHEJ results in indels at the cleavage site [16]. (B) MMEJ can be used to integrate an HPH cassette into a desired locus by using short homology arms. The strain is first transformed with a plasmid expressing Cas9 and containing a PYR4 marker and then with an in vitro transcribed sgRNA and the HPH cassette [45]. (C) In this “suicide” system, a cassette containing both the CRISPR elements and a URA5 marker flanked by homology arms to the target site can be transformed into an uracil auxotrophic strain of C. deneoformans. The double-strand break induced by the transiently expressed Cas9 is repaired by HDR with the URA5 cassette, and the crossover results in the release of the DNA of the CRISPR elements that induced the cleavage in the first place, which may be degraded (hence the name “suicide” system) [17]. (D) An RNP method was developed for C. neoformans and C. deneoformans, similar to the one adapted to Candida species. The cells are transformed with a mixture of two RNPs, each targeting a different end of the target gene, thus maximizing the chances of gene deletion [27]. (E) CAS9 can be integrated in the Safe Haven on C. neoformans H99 and selected due to the presence of a HYGB cassette. This strain is then be transformed with two transient plasmids expressing two sgRNAs flanked by ribozymes from the native ACT1 promoter, targeting the beginning and the end of the target gene. A NEO cassette flanked by homology arms to the target gene is used for HDR [53]. Af, Aspergillus fumigatus; Cn, Cryptococcus neoformans; CRISPR, clustered regularly interspaced short palindromic repeats; crRNA, CRISPR-RNA; HDR, homology-directed repair; HDV, human hepatitis delta virus; HH, hammerhead; MMEJ, microhomology-mediated end joining; NHEJ, nonhomologous end joining RNP, RNA–Cas9 protein complex; Sc, Saccharomyces cerevisiae; sgRNA, single-guide RNA; tracrRNA, trans-activating RNA; YFG, your favorite gene.
Candida species
In 2015, Vyas and colleagues reported the first CRISPR–Cas9 method in C. albicans [18], which relied on the integration of CAS9 and sgRNA either as a single cassette at the ENO1 locus (solo system) or as two separate cassettes at the ENO1 and RP10 loci, respectively (duet system). Transformed strains were selected using Nourseothricin, due to the presence of the NAT gene in the cassette(s). Expression of a CTG-codon–optimized CAS9 is driven from the promoter at the integration site, whereas the sgRNA is expressed from the RNA pol III promoter CaSNR52. HDR-mediated gene editing was achieved by using a donor DNA with 50 bp homology arms to introduce premature stop codons in the ADE2 gene, with variable efficiencies (20% to 80%), depending on the vector used (solo or duet) (Fig 1A). The system was also used to simultaneously target members of a gene family with a single sgRNA and disrupt essential genes by generating conditional alleles. However, in the original system, the selection marker could not be recycled, making sequential editing in the same background virtually impossible, and stable CAS9 integration in the genome raised concerns about potential off-target effects.
Min and colleagues addressed the problem by transiently expressing CAS9 and the sgRNA in C. albicans [33]. The repair template contained an NAT cassette, which was used to replace the target gene (Fig 1B). Deletion of both alleles occurred either by independent recombination at both or by recombination at one allele followed by allelic gene conversion. Though this method avoided integrating CAS9, it was not until the following year that serial mutagenesis in the same background could be achieved [34,35]. Huang and colleagues [34] developed an ingenious marker-recycling model called CRISPR-mediated marker excision (CRIME), in which the selection marker used to delete the gene of interest is flanked by directly repeated sequences. Following the deletion of the target gene, the CRISPR–Cas9 system is used again and induces a DSB into the marker itself, leading to marker excision by recombination between the repeats [34].
Nguyen and colleagues [35] developed alternative strategies to allow marker recycling in different C. albicans genetic backgrounds, namely the LEUpOUT system—which requires an LEU2/leu2Δ parental strain—or the HIS–FLP system, applicable to any strain. In both cases, the transient integration of a cassette containing both a selection marker and CAS9 and sgRNA at a single locus (either LEU2 or HIS)—allowing the deletion of the gene of interest—is followed by the excision of the cassette by either recombination between direct repeats (LEUpOUT) or activation of an FLP recombinase [35] (Fig 1C). Markerless deletion in a single transformation and the absence of a cloning step are the main advantages of this method, as well as the possibility to generate reconstituted strains.
Most C. albicans editing strategies used an RNA pol III promoter to express the sgRNA. However, Ng and colleagues [21] showed that efficiency of CRISPR–Cas9 in C. albicans could be further improved by up to 10 times when the sgRNA is expressed from the strong RNA pol II (ADH) promoter and is flanked by an upstream tRNA sequence and a downstream human hepatitis delta virus (HDV) ribozyme to release any added 5’ cap and 3’ poly(A) tail.
Vyas and colleagues [30] also developed a method for marker recycling. The CAS9, sgRNA, and NAT sequences are integrated at the NEUT5L locus and subsequently removed due to the activation of a FLP recombinase (Fig 1D). Transformation with a donor DNA juxtaposing 50 bp upstream and downstream of a given ORF results in an unmarked deletion mutant. Because the marker selecting for the presence of the CRISPR–Cas9 elements is recyclable and no marker integration occurs at target gene, the system allows the construction of multiple deletion mutants in virtually any genetic background.
In 2017, Shapiro and colleagues [36] adapted the gene-drive principle described in S. cerevisiae to C. albicans, thanks to the discovery of mating-competent, C. albicans haploids [37–38]. Firstly, a cassette expressing CAS9 and two sgRNAs is integrated at the NEUT5L locus in a haploid cell. The sgRNAs are flanked by sequences homologous to the target gene. Following mating, an intact copy of the target gene is introduced. The sgRNAs direct Cas9 to the target gene, introducing DSBs that are repaired by HDR using the CAS9 and sgRNA cassette. This step will occur in any subsequent mating event, acting as a “drive” allele. When haploid cells containing the drive allele are mated to wild-type cells, the incoming wild-type allele is cut by Cas9 and deleted by replacement with the sgRNA module via HDR. Mating of haploid Candida cells deleted for specific ORFs therefore rapidly leads to homozygous diploid double-deletion mutants. This platform allows the analysis of genetic interaction by combining deletions of two genes in the same background. However, the stable integration of CAS9 within the genome requires monitoring of possible off-target effects.
CRISPR–Cas9 methods have also been implemented in non-albicans Candida species. Like C. albicans, the C. parapsilosis sensu lato complex species, C tropicalis, C. lusitaniae, and C. auris are members of the CTG clade. Norton and colleagues developed the first transient CRISPR–Cas9 expression system for gene deletion in the genetically intractable C. lusitaniae [20]. Editing efficiencies of 36% were achieved using 1 kb homology arms to direct integration of a NAT cassette at the target gene, using C. lusitaniae-specific promoters to express CAS9 and sgRNA. Efficiency increased to 81% after deleting the NHEJ-associated genes KU70 and LIG4, suggesting that this DNA-repair pathway plays a major role in C. lusitaniae.
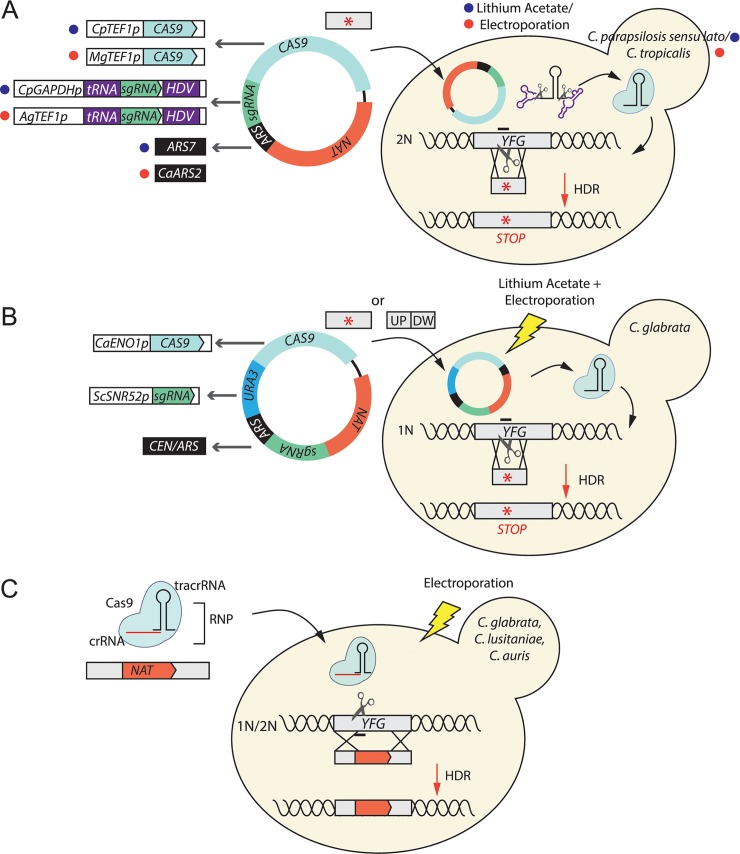
In 2017, Lombardi and colleagues [39] implemented a plasmid-based CRISPR–Cas9 technology in C. parapsilosis, in which the episomal pRIBO vector harbors the NAT marker, CAS9 expressed from the C. parapsilosis CpTEF1 promoter, and an sgRNA expression cassette. The sgRNA is expressed from the C. parapsilosis RNA pol II promoter GAPDH and flanked by a hammerhead (HH) and HDV ribozymes. Cotransformation of the replicating plasmid with a short donor DNA containing two in-frame stop codons flanked with 40-bp homology arms allowed for high-efficiency gene disruption. Despite variation in efficiency across strains, this method offers the possibility to edit any genetic background (e.g., clinical isolates). The plasmid is easily lost in the absence of selection, allowing sequential gene editing in the same genetic background. Zoppo and colleagues [40] showed that the system can also be used for gene editing in the sibling species Candida orthopsilosis. This system was recently improved by replacing the HH ribozyme with a tRNA molecule, which is recognized and cleaved by the yeast ribonuclease Z [29] (Fig 2A). This streamlined the cloning process, in that only the 20 bp guide region needs to be replaced to target a new gene. The editing system functions in C. parapsilosis, C. orthopsilosis, and Candida metapsilosis [29,41]. At the same time, a different plasmid was constructed that can be used in gene editing in C. tropicalis [29]. Notably, adaptation to C. tropicalis required not only changing the regulatory elements driving the expression of CAS9, NAT, and sgRNA—which were derived from Candida dubliniensis, Ashbya gossypii, and Meyerozyma guilliermondii, respectively—but also using a different autonomously replicating sequence, ARS2 from C. albicans [29,42] (Fig 2A).
Fig 2. CRISPR–Cas9 methods in non-albicans Candida species.
The vignettes illustrate representative methods for implementation of CRISPR–Cas9 in non-albicans species, including C. parapsilosis sensu lato, C. tropicalis, C. glabrata, C. lusitaniae, and C. auris. The ploidy of the species is indicated next to the DNA helices. (A) Two different selectable episomal plasmids allow the scarless introduction of premature stop codons by HDR in C. parapsilosis sensu lato and C. tropicalis. The relevant species-specific promoters and ARSs are depicted on the left side (blue dot: C. parapsilosis sensu lato; red dot: C. tropicalis). The sgRNA is expressed from an RNA pol II promoter, and the release of the mature molecule is mediated by the cleavage of the upstream tRNA molecule and the downstream HDV ribozyme. The plasmids are easily lost in the absence of selection [29]. (B) A similar approach can be used for gene editing and deletion in C. glabrata. In this case, the sgRNA is expressed from the Saccharomyces cerevisiae RNA pol III SNR52 promoter [30]. (C) The use of CRISPR RNPs, in which the crRNA and tracrRNA are assembled in vitro with Cas9 and then electroporated into the cells, results in the integration of a NAT cassette into the targeted gene in C. glabrata, C. lusitaniae, and C. auris [25]. Although this method does not require species-specific adaptation of the regulatory elements necessary for the expression of the CRISPR elements, marker recycling is not possible. Ag, Ashbya gossypii; ARS, autonomously replicating sequence; Ca, Candida albicans; Cp, C. parapsilosis; CRISPR, clustered regularly interspaced short palindromic repeats; crRNA, CRISPR-RNA; DW, downstream; HDR, homology-directed repair; HDV, human hepatitis delta virus; Mg, Meyerozyma guilliermondii; RNP, RNA–Cas9 protein complex; Sc, Saccharomyces cerevisiae; sgRNA, single-guide RNA; tracrRNA, trans-activating RNA; UP, upstream; YFG, your favorite gene.
CRISPR–Cas9 can also be used in C. glabrata, a species that is not part of the CTG clade and is more closely related to S. cerevisiae than to C. albicans. Enkler and colleagues [19] transformed a triple auxotrophic strain (his3, trp1 and leu2) of C. glabrata with a LEU2 plasmid expressing the sgRNA and a TRP1 plasmid expressing Cas9. By exploiting NHEJ—the preferential DNA-repair mechanism in this species—this strategy allowed up to 100% efficient ADE2 gene disruption when using C. glabrata CYC1 and RNAH1 promoters to express CAS9 and the sgRNA, respectively. Although 500-bp homology arms are usually recommended for HDR in C. glabrata, this method was used successfully to insert either short (57 bp) or large (1,202 bp) cassettes by HDR using only 20-bp or 200-bp homology arms, though 200-bp homology arms performed better for the small insert [43]. Vyas and colleagues [30] also generated an editing system in which CAS9, sgRNA, and NAT sequences are harbored on an episomal plasmid that can be used also in prototrophic clinical isolates of C. glabrata (Fig 2B). Transformation of the cells resulted in ade2 mutants both with and without a repair template designed to introduce premature stop codons in the gene. Recently, Maroc and colleagues described a CRISPR vector containing the sgRNA cassette and expressing CAS9 under the control of the MET3 promoter, which can be used to induce mutations by either NHEJ or HDR in C. glabrata [44]. Since Cas9 expression is inducible, the authors foresee that this system may also be used for identifying essential genes in this asexual yeast.
As an alternative to expressing CAS9 and the sgRNA in the cell, Grahl and colleagues [25] assembled Cas9 and the specific crRNA:tracrRNA duplex RNA in vitro to form an RNP (Fig 2C). The RNP was then transformed into C. lusitaniae, C. auris, and C. glabrata to induce gene deletion by homologous recombination of a selection marker flanked by 0.5–1 kb homology arms. Efficiencies ranged from 60% to 70%. However, this strategy does not allow marker recycling.
The implementation of the CRISPR–Cas9 technology in Candida species boosted our ability to decipher cellular signaling associated with virulence and the underlying genetic interactions. CRISPR–Cas9 used in combination with C. albicans mating-competent haploids allowed the investigation of epistatic relationships between members of gene families involved in fungal virulence (e.g., adhesins and efflux pumps) by generating libraries of homozygous double gene-deletion mutants [36]. This technology also streamlined the process of linking specific genes or SNPs (either heterozygous or homozygous) to relevant phenotypes in Candida species (e.g., resistance or sensitivity to drugs or cell-wall stressors [41,45–47].
Aspergillus fumigatus
Gene manipulation in A. fumigatus is hampered by low efficiency of HDR (3%–5%), and NHEJ-impaired strains and 1-kb homology arms are traditionally used for site-specific integration [48]. Moreover, a reliable strategy of sexual reproduction in A. fumigatus has not been established, making mutagenesis of multiple genes by sexual crossing a challenging task, especially in clinical isolates. The first CRISPR–Cas9 method developed for A. fumigatus exploited NHEJ for gene disruption of the pks gene—involved in melanin production—by randomly integrating a cassette containing the hygromycin marker (HPH), the sgRNA expressed from the S. cerevisiae SNR52 promoter, and the human optimized CAS9 gene expressed from a TEF1 promoter in the genome [16] (Fig 3A). Albino mutants were obtained with a 25%–53% frequency. Interestingly, unintended NHEJ-mediated integration of the transforming DNA to the Cas9 cut site was often observed. Notably, CAS9 expression in the absence of a sgRNA did not alter growth rate or virulence in a murine model.
The following year, Zhang and colleagues [49] exploited MMEJ to efficiently (95%–100%) integrate desired sequences in-frame into a target gene via very short (approximately 35-bp) homology arms. They used strains constitutively expressing a human optimized CAS9 gene from the fungal GPDA promoter on an autonomously replicating plasmid with a PYR4 selectable marker. The sgRNA targeting the gene of interest, whose expression is driven by the native A. fumigatus RNA pol III U6 promoter, can either be transformed as a linear cassette or directly provided to the cells as an RNA molecule following in vitro transcription. An HPH cassette flanked by microhomology regions can then be targeted to the cleavage site (Fig 3B). The same approach can be used to insert a green fluorescent protein (GFP) tag (although the selection for positive transformants is based on the random integration of the HPH cassette) and to create multiple mutants. Notably, this method can be used in clinical isolates [49].
In line with other Aspergillus species, Weber and colleagues [50] used the RNA pol II GPDA promoter to drive the expression of a HH–HDV ribozyme-flanked sgRNA. The authors adopted an ingenious split-marker approach to induce a single-nucleotide deletion in a NHEJ-defective strain containing an integrated tetracycline-inducible CAS9. An integrating plasmid harbors the sgRNA expression cassette along with the two halves of the split marker (the pyrithiamine resistance cassette (PTRA) of A. oryzae) interrupted by the same guide and PAM sequence that is being targeted in the genome. CRISPR–Cas9 mediated cleavage of the native target occurs in parallel with the cleavage of the “twin site” on the split marker, resulting in the reconstitution of the functional selection marker. The desired mutation can then be introduced by HDR with a donor DNA. Although inducible expression of CAS9 lowers the risks of off-target mutations, it still remains in the genome.
Al Abdallah and colleagues [26] first used the RNP complex in conjunction with MMEJ for gene deletion in A. fumigatus. By using two sgRNAs to target Cas9-induced DSBs upstream and downstream of the PKSP gene and an HPH marker flanked with 35–50 bp homology arms as donor DNA, gene deletion occurred with an efficiency of >90% in both NHEJ-competent and NHEJ-defective strains. Although applicable to virtually any A. fumigatus isolates, this approach does not allow marker recycling, because the target gene is replaced by the HPH marker. Umeyema and colleagues [51] used a similar strategy to introduce two single point mutations previously identified in clinical isolates in the CYP51A gene to assess their role in azole resistance. In this case, the donor DNA was the full gene sequence containing the desired mutation(s), fused to the HPH marker. Therefore, despite several improvements, CRISPR–Cas9 in A. fumigatus still suffers from the absence of methods for marker recycling.
Single-base CRISPR–Cas9 editing has been used to gain further insights into the emerging phenomenon of azole resistance in A. fumigatus. The association of amino acid substitutions with azole resistance (e.g., Gly138Ser in Cyp51A [51]) also led to the discovery of a non-cyp51A mediated resistance mechanism relying on a mutated version of the SVF1 gene [52].
Cryptococcus species
Site-directed mutagenesis has been challenging in Basidiomycetes such as pathogenic Cryptococcus species for a long time. The main target species are C. neoformans (formerly known as C. neoformans var. grubii, serotype A), C. deneoformans (formerly known as Cr. neoformans var. neoformans, serotype D), and C. gattii [53]. Chemical transformation is not effective in these yeasts, and exogenous DNA is usually introduced into the cells by either biolistic transformation or electroporation. However, electroporated cells are unstable, and the efficiency of HDR ranges from 0.00001% to 0.001% [54,55]. By contrast, HDR following biolistic transformation rises to 1%–10%, and transformed cells are more stable [54], making this technique the gold standard for gene targeting in Cryptococcus. Unfortunately, biolistic transformation requires expensive equipment. In addition, although NHEJ-defective mutants have higher frequencies of HDR, their reduced virulence prevents their use in pathogenesis studies [56,57].
The implementation of CRISPR–Cas9 in Cryptococcus had a dramatic effect on increasing the efficiency of HDR, so much so that, in the last two years, electroporation is again being used for genetic manipulation. In the system developed by Wang and colleagues [17] for C. deneoformans, the sgRNA is expressed from a native cryptococcal RNA pol III U6 promoter, and a C. neoformans ACT1 promoter drives the expression of a human codon-optimized CAS9 gene. Notably, electroporation of linearized URA5 vectors harboring the cassettes in a ura-strain resulted in 82%–88% pink auxotrophic colonies when ADE2 was targeted by NHEJ (or NHEJ-equivalent) repair. Cotransformation with a 1-kb donor DNA also allowed a single codon change mediated by HDR. The ADE2 gene was also disrupted by replacing with the hygromycin marker HYGB flanked by 1-kb homology arms on each side, although HDR occurred only in 8 out of 20 pink transformants, suggesting that the NHEJ pathway may be prevalent. Although the vectors were not integrated in the genome, the cassettes were stable in C. deneoformans even in the absence of selective pressure, increasing the risk of off-target effects and making gene complementation virtually impossible (the newly introduced wild-type (WT) copy of the disrupted gene will be recognized and cleaved).
In order to fix this problem, Wang and colleagues [17] further developed a “suicide” system that allows for the elimination of CRISPR components (Fig 3C). In this design, homology arms flank the URA5 marker on the vector, driving its integration by HDR into the cleaved target gene. As the CRISPR and sgRNA elements are located on one side of the homology arm, they may be resolved by double crossover in HDR and eventually degraded. When put to the test, the “suicide” system efficiently disrupted the gene of interest and roughly 50% of the mutants lost the CRISPR elements, thus allowing subsequent complementation of the disruption.
Arras and colleagues [58] used biolistic transformation to integrate a HYGB cassette containing S. pyogenes CAS9 expressed from a native TEF1 promoter into the C. neoformans H99 Safe Haven region on chromosome 1 (Fig 3E). This strain can be subsequently transformed with a vector containing the G418 resistance marker NEO and expressing the sgRNA from the C. neoformans RNA pol II ACT1 promoter, flanked with HH and HDV ribozymes [59].
Cotransformation of the vector transiently expressing the sgRNA and a cassette carrying approximately 1-kb homology arms on both sides led to the HDR-mediated gene disruption with an efficiency of 70%. Despite the concerns about the stable expression of Cas9, this system simplifies genetic manipulation in that it requires only the generation of the sgRNA vector and donor DNA constructs for targeting a gene of interest.
In 2018, Fan and Lin [60] developed TRACE (transient CRISPR–Cas9 coupled with electroporation) for both C. neoformans and C. deneoformans. Cryptococcal cells are electroporated with two linear cassettes with no selectable markers, in which CAS9 and the sgRNA are under the control of native C. neoformans regulatory elements (GPD1 promoter for the human optimized CAS9 gene and U6 promoter for the sgRNA targeting ADE2), together with a NAT cassette carrying 1-kb homology arms. Pink transformants were obtained with a 50%–90% efficiency depending on the strain, although further analyses showed that not all of the colonies were consistent with HDR-mediated integration of the cassette into ADE2. Moreover, both CAS9 and the sgRNA were found randomly integrated into the genome in some of the transformants. Lowering the amount of Cas9 and sgRNA provided reduced the frequency of the unwanted integration events. TRACE can also be used to simultaneously target members of a gene family and to ectopically complement a mutated gene by introducing a WT copy into the Cas9-cleaved Safe Haven region (SH2).
Two additional approaches for C. neoformans and C. deneoformans were published by Wang in 2018 [27] (Fig 3D). Both methods rely on the simultaneous use of two guides, targeting the 5’ and the 3’ of the gene of interest, in an attempt to facilitate deletion by HDR with a NAT cassette carrying 800 to 900 bp homology arms. In the first approach, two plasmid vectors are transformed, each containing a S. pyogenes CAS9 and one of the two sgRNAs expressed from C. neoformans TEF1 and U6 promoters. In the second approach, CRISPR elements are provided to the cells via electroporation as an RNP complex. Although more transformants were obtained using the plasmids, the RNP method remains a valuable approach, as it potentially allows the implementation of CRISPR–Cas9 in species for which expression cassettes are not available yet (e.g., C. gattii and C. deuterogattii).
Mucorales species
The generation of stable gene replacements by HDR is a challenging process in Mucorales, because plasmids tend to not integrate regardless of the size of the homology arms. The first CRISPR–Cas9 method available uses the RNP approach to generate stable mutants in Mucor circinelloides [28]. Different to other RNP methods, assembly of Cas9 with the sgRNA occurs in vivo, inside the fungal protoplasts. The Cas9-induced DSB resulted in 100% efficiency for both gene disruption by NHEJ and gene deletion by HDR-mediated integration of a PYRG cassette flanked by homology arms. Notably, NHEJ led to an unexpected >2.3kb-long deletion upstream of the PAM in the resulting mutants, possibly causing unwanted effects (i.e., the gene adjacent to the targeted one was also affected by the deletion). The HDR pathway is therefore preferable, despite resulting in the stable integration of the section marker. Despite very low transformation efficiency, this method provides stable mutants, streamlining the genetic manipulation of these genetically intractable fungi. Earlier this year, Wang and colleagues demonstrated that a CRISPR–Cas9 plasmid formerly developed for Cryptococcus species could be used for NHEJ-mediated gene disruption in Rhizopus delemar [61]. The implementation of CRISPR–Cas9 technology in Mucorales provides a much needed window into the biology of these emerging opportunistic pathogens for which robust genetic tools are lacking.
Perspectives and future directions
Genetic manipulation of fungal pathogens has been a milestone in deepening our understanding of virulence and host–pathogen interactions. The generation of single-gene mutant libraries (e.g., in C. neoformans [62], C. albicans [63], and C. parapsilosis [64], as well as other approaches like promoter replacement [65], transposon mutagenesis to study gene essentiality [66], and heterozygous libraries generated for haploinsufficiency-based genetic interaction studies in C. albicans [67] allowed the dissection of the molecular players of fungal biology and pathogenicity.
CRISPR–Cas9 methods are now available for most of the major fungal species causing infections, facilitating genetic engineering by increasing the rate of homologous recombination as never seen before. Unique guide sequences are available to target >90% of the annotated ORFs in both C. albicans and C. glabrata genomes [18,30]. Different genetic changes can be introduced (e.g., deletion, single-nucleotide mutation, stop codon insertion, barcoding, fluorescent tagging, etc.) in coding as well as noncoding regions. Ongoing research in model eukaryotic organisms like S. cerevisiae keeps expanding the boundaries of potential CRISPR applications [11,68]. An interesting refinement is the possibility of engineering heterozygous mutations in diploid species, which allows the study of heterozygous variants or essential genes. This was first achieved in mammalian cell lines using mixed repair templates carrying a different set of mutations or, alternatively, by exploiting the cut-to-mutation distance (the efficiency of mutation incorporation rapidly falls with increasing distance from the Cas9 cleavage site) [69]. Recently, heterozygous mutations were introduced in C. parapsilosis and C. orthopsilosis by applying the same principle [29,41].
Larger chromosomal deletions (>30 kb) and chromosome fusion have been achieved in S. cerevisiae through NHEJ repair between endpoints of the DNA breaks, which can facilitate the study of gene clusters or minimal genome research, as well as the study of replication, recombination, and segregation [70,71,72]. In principle, this may open up the possibility to generate CRISPR–Cas9–induced genome rearrangements in C. albicans, in which specific whole-chromosome aneuploidies have been associated with drug resistance and/or cross-adaptation to different drugs [73]. In S. cerevisiae, the introduction of a DSB by Cas9 can result in extensive loss of heterozygosity (LOH) events consistent with HDR [74]. Whereas this needs to be taken into account when manipulating polyploid and highly heterozygous genomes, it could also be exploited to study the impact of LOH on C. albicans virulence. In fact, exposure to the mammalian host increases the frequency of LOH in C. albicans, likely selecting for specific mutations. In addition, LOH is associated with reduced drug susceptibility in strains sequentially isolated from the same patient [75].
Although most of our knowledge on microbial pathogenesis was built on extensive analyses of one or few laboratory strains, it is becoming increasingly clear that genotype–phenotype correlation should be validated using multiple isolates. An example is the regulatory network controlling biofilm production and hyphal formation in C. albicans; circuit diversification appears to be widespread in different isolates, as attested by the fact that loss of function of the same transcription factor can have a different impact on the network depending on the strain [76]. Many of the CRISPR–Cas9 systems developed for pathogenic fungi are applicable to any genetic background, thus allowing the generation of mutations in different clinical isolates and a more robust validation of gene function.
Targeting gene families is also possible by exploiting the homology within coding regions of paralogs. For example, Wijsman used 11 unique guide sequences to delete 21 hexose transporters in S. cerevisiae, in only 3 transformation steps [77]. Multiplexing is time-saving and triggers less chromosomal rearrangements than the Loxp/Cre system, ensuring high fidelity when addressing extensive genome editing strategies [77]. CRISPR–Cas9 based gene-drive strategies had been developed in S. cerevisiae and C. albicans [36,37,78], in which the selfish element itself is used as donor DNA and replaces the WT allele after Cas9 cleavage. In diploid organisms with sexual reproduction, all progeny cells will inherit the gene-drive, bypassing mendelian inheritance [37].
CRISPR–Cas9 made the construction of genome-scale mutant libraries easier than ever before, thus streamlining downstream applications such as genome-wide variant engineering [79] or gene interaction screens [80], which are useful in identifying fungal-specific genes involved in essential biological processes that could represent potential new therapeutic targets. Finally, directed evolution studies are another perspective that can be achieved for example by a CRISPR-guided nickase fused to an error-prone DNA polymerase, dramatically increasing the mutation rate [81]. Such strategies favoring directed evolution could facilitate the identification of new variant genotypes with successful virulence or antifungal resistance traits.
Besides Cas9, at least 10 different Cas enzymes have been described, differing in structure, PAM recognition sites, or type of cleavage mechanisms, allowing new targeting options [82]. Examples are eSpCas9, SpCas9-HF1 and HypaCas9 (engineered variants of Cas9 with a higher specificity [83–85]), Cas12a (less off-target effect; PAM = TTN; appropriate for low GC-rich regions) [86], and xCas9 (broadest PAM compatibility; PAM = NG, GAA and GAT)[87]. On the other hand, developing CRISPR base editors paved the way for genome editing without introducing DSB DNA-breaks by inducing gene conversions such as C to T, G to A [88], or A-T to G-C [89]. Anzalone and colleagues further improved this strategy by combining a catalytically impaired Cas9 (only able to nick one DNA strand) with a reverse transcriptase; the complex is driven to the target site by a prime editing guide RNA (pegRNA) that also encodes the edit [90]. This method enables precise targeted insertions and deletions and all 12 possible classes of point mutations (including transversion mutations) without inducing DSBs, therefore minimizing side effects, and at the same time not relying on HDR with a donor DNA to edit the genome. While this is particularly relevant in mammalian cells, it could also be applied to fungi in which the efficiency of HDR remains low, even following a Cas9-induced DSB.
One more step has also been reached by editing RNAs using Cas13 enzymes, making nonpermanent changes of the genome possible [91].
Nonediting applications using Cas9 have also emerged over the years [92]. Fusion of Cas proteins with reporters allows live-cell imaging of DNA and RNA [93]. A catalytically dead variant with no nuclease activity (dCas9) but retaining its binding activity has been used to regulate gene expression in S. cerevisiae, allowing either activation (CRISPRa)—when fused to a transcriptional activator—or interference (CRISPRi), when fused to a repressor [94]. Similar systems have recently been developed for C. albicans, in which comprehensive genetic interaction maps could not previously be built using RNAi-based genetic knockdown, because RNAi is not effective in this species. The faster and cheaper genetic manipulation of fungal genome and expression facilitated by CRISPR–Cas9 is likely to have a profound impact on medical mycology. Loss of function, gene activation, and gene inhibition strategies can be scaled up as CRISPR–Cas9 libraries [95–97], and large-scale library screens can facilitate the identification of new targets for antifungal therapy, of which there is a desperate need [98].
The use of RNA–Cas9 complexes is particularly important in species refractory to genetic manipulation, making easier and more timely the study of emergent and multiresistant fungi. Although not yet applied to medical mycology, the diagnostics field already started harnessing the potential of CRISPR–Cas9. Myrhvold and colleagues, developed a Cas13-based nucleic acid detection platform, Specific Highly sensitive Enzymatic Reporter unLOCKing (SHERLOCK), coupled with detection by a lateral flow device as a point-of-care test to diagnose Zika and Dengue virus in clinical samples [99]. The extension of this technique to the high sensitivity and specificity detection of fungal DNA could radically improve the diagnosis of fungal infections, especially in low resource settings [100]. CRISPR–Cas9 and Next Generation Sequencing were combined in Finding Low Abundance Sequences by Hybridization (FLASH), a new method for multiplex detection of antimicrobial resistance in clinical samples of gram-positive bacteria and Plasmodium falciparum [101].
The potential applications of CRISPR-based methods developed in either model fungi (like S. cerevisiae) or bacteria, viruses, and mammalian cells to the study of fungal pathogenesis are summarized in Table 1.
Table 1. Potential translation of CRISPR-based methods developed for model organisms to the study of fungal pathogenesis.
| Method | Developed in | Potential application to fungal pathogens | Refs. |
|---|---|---|---|
| CRISPR-mediated large chromosomal deletions and fusions | S. cerevisiae | Study of the correlation existing between large chromosomal rearrangements and virulence and drug resistance (e.g., aneuploidy and LOH in C. albicans, aneuploidy in C. neoformans) | [70–72] |
| Directed evolution through CRISPR-guided DNA polymerase | Escherichia coli | Identification of new variant genotypes with virulence or antifungal resistance traits | [81] |
| CRISPR prime editing | Human cells | Precise DSB-free editing in species refractory to HDR | [90] |
| Cas13-based nucleic acid detection platform (e.g., SHERLOCK) | Zika and Dengue viruses | Rapid diagnosis of fungal infections in low resource settings based on the development of highly sensitive (attomolar range) kits for detection of fungal DNA | [99] |
| CRISPR–Cas9 based enrichment of sequences for NGS (e.g., FLASH) | Gram-positive bacteria, P. falciparum | Enrichment of fungal sequences associated with a trait of interest (e.g., antifungal resistance) prior to NGS, potentially resulting in antimicrobial resistance profiling directly from the patient sample | [101] |
CRISPR, clustered regularly interspaced short palindromic repeats; DSB, double-strand break; FLASH, Finding Low Abundance Sequences by Hybridization; HDR, homology-directed repair; LOH, loss of heterozygosity; NGS, next generation sequencing; SHERLOCK, Specific Highly Sensitive Enzymatic Reporter UnLOCKing
Conclusions
This review provides an updated overview of the state of the art of CRISPR–Cas9 genome editing in pathogenic fungi to both researchers and medical mycologists interested in using this technology. Though we focused on major fungal pathogens encountered in the clinical setting [102], CRISPR–Cas9 methods have also been developed in other human fungal pathogens not discussed here, such as Malassezia [103], Fusarium [104], and the dimorphic fungus Blastomyces dermatitidis [105]. The effectiveness and flexibility of CRISPR–Cas9 have the potential to have a dramatic impact on the field of medical mycology, both in the lab—by boosting genetic manipulation for studying fungal biology and pathogenesis, dissecting the role of virulence factors, and investigating new potential drug targets and host–pathogen interaction—and at the bedside, by improving the diagnosis of fungal infections and the monitoring of antifungal resistance [4]. Moreover, further applications currently developed in model fungi may be applied to fungal pathogens in the near future for studying host–fungi interactions or tackling antifungal resistance [106].
Funding Statement
The authors received no specific funding for this study.
References
- 1.Cole DC, Govender NP, Chakrabarti A, Sacarlal J, Denning DW. Improvement of fungal disease identification and management: combined health systems and public health approaches. Lancet Infect Dis. 2017. December;17(12):e412–e419. 10.1016/S1473-3099(17)30308-0 [DOI] [PubMed] [Google Scholar]
- 2.Bongomin F, Gago S, Oladele R, Denning D. Global and Multi-National Prevalence of Fungal Diseases—Estimate Precision. J Fungi. 2017. October 18;3(4):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhary A, Sharma C, Meis JF. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017. May 18;13(5):e1006290 10.1371/journal.ppat.1006290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher MC, Hawkins NJ, Sanglard D, Gurr SJ. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science. 2018. May 18;360(6390):739–742. 10.1126/science.aap7999 [DOI] [PubMed] [Google Scholar]
- 5.Samaranayake DP, Hanes SD. Milestones in Candida albicans gene manipulation. Fungal Genet Biol. 2011;48(9):858–865. 10.1016/j.fgb.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Defosse TA, Courdavault V, Coste AT, Clastre M, de Bernonville TD, Godon C, et al. A standardized toolkit for genetic engineering of CTG clade yeasts. J Microbiol Methods. 2018. January;144:152–156. 10.1016/j.mimet.2017.11.015 [DOI] [PubMed] [Google Scholar]
- 7.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012. August 17;337(6096):816–821 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dicarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013. April;41(7):4336–4343. 10.1093/nar/gkt135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012. September 25;109(39):E2579–2586. 10.1073/pnas.1208507109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceccaldi R, Rondinelli B, D’Andrea AD. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016. January;26(1):52–64. 10.1016/j.tcb.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raschmanová H, Weninger A, Glieder A, Kovar K, Vogl T. Implementing CRISPR-Cas technologies in conventional and non-conventional yeasts: Current state and future prospects. Biotechnol Adv. 2018;36(3):641–665. 10.1016/j.biotechadv.2018.01.006 [DOI] [PubMed] [Google Scholar]
- 12.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008. August;9(8):619–631. 10.1038/nrg2380 [DOI] [PubMed] [Google Scholar]
- 13.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994. December;14(12):8096–8106. 10.1128/mcb.14.12.8096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smih F, Rouet P, Romanienko PJ, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995. December 25;23(24):5012–5019. 10.1093/nar/23.24.5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Storici F, Durham CL, Gordenin DA, Resnick MA. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc Natl Acad Sci U S A. 2003. December 9;100(25):14994–14999. 10.1073/pnas.2036296100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuller KK, Chen S, Loros JJ, Dunlap JC. Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryot Cell. 2015. November;14(11):1073–1080. 10.1128/EC.00107-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Wei D, Zhu X, Pan J, Zhang P, Huo L, et al. A “suicide” CRISPR-Cas9 system to promote gene deletion and restoration by electroporation in Cryptococcus neoformans. Sci Rep. 2016. August 9;6:31145 10.1038/srep31145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vyas VK, Barrasa MI, Fink GR. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci Adv. 2015;1(3):e1500248 10.1126/sciadv.1500248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enkler L, Richer D, Marchand AL, Ferrandon D, Jossinet F. Genome engineering in the yeast pathogen Candida glabrata using the CRISPR-Cas9 system. Sci Rep. 2016. December 21;6(1):35766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norton EL, Sherwood RK, Bennett RJ. Development of a CRISPR-Cas9 System for Efficient Genome Editing of Candida lusitaniae. mSphere. 2017. June 21;2(3). pii: e00217–17. 10.1128/mSphere.00217-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng H, Dean N. Dramatic Improvement of CRISPR/Cas9 Editing in Candida albicans by Increased Single Guide RNA Expression. mSphere. 2017. April 19;2(2). pii: e00385–16. 10.1128/mSphere.00385-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nosek J, Adamíkovâ Ľ, Zemanová J, Tomáška Ľ, Zufferey R, Mamoun C Ben. Genetic manipulation of the pathogenic yeast Candida parapsilosis. Curr Genet. 2002. October;42(1):27–35. 10.1007/s00294-002-0326-7 [DOI] [PubMed] [Google Scholar]
- 23.Krappmann S. CRISPR-Cas9, the new kid on the block of fungal molecular biology. Med Mycol. 2017. January 1;55(1):16–23. 10.1093/mmy/myw097 [DOI] [PubMed] [Google Scholar]
- 24.Papp T, Csernetics Á, Nyilasi I, ÁBrÓk M, VÁgvÓlgyi C. Genetic Transformation of Zygomycetes Fungi In: Progress in Mycology. Dordrecht: Springer Netherlands; 2010. p. 75–94. [Google Scholar]
- 25.Grahl N, Demers EG, Crocker AW, Hogan DA. Use of RNA-Protein Complexes for Genome Editing in Non- albicans Candida Species. mSphere. 2017. June 21;2(3). pii: e00218–17. 10.1128/mSphere.00218-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al Abdallah Q, Ge W, Fortwendel JR. A Simple and Universal System for Gene Manipulation in Aspergillus fumigatus: In Vitro-Assembled Cas9-Guide RNA Ribonucleoproteins Coupled with Microhomology Repair Templates. mSphere. 2018. June 13;3(3). pii: e00208–18. 10.1128/mSphereDirect.00208-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P. Two Distinct Approaches for CRISPR-Cas9-Mediated Gene Editing in Cryptococcus neoformans and Related Species. mSphere. 2018;3(3):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagy G, Szebenyi C, Csernetics Á, Vaz AG, Tóth EJ, Vágvölgyi C, et al. Development of a plasmid free CRISPR-Cas9 system for the genetic modification of Mucor circinelloides. Sci Rep. 2017. December 1;7(1):16800 10.1038/s41598-017-17118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lombardi L, Oliveira-Pacheco J, Butler G. Plasmid-Based CRISPR-Cas9 Gene Editing in Multiple Candida Species. mSphere. 2019. February 13;4(2):557926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vyas VK, Bushkin GG, Bernstein DA, Getz MA, Sewastianik M, Barrasa MI, et al. New CRISPR Mutagenesis Strategies Reveal Variation in Repair Mechanisms among Fungi. mSphere. 2018. April 25;3(2). pii: e00154–18. 10.1128/mSphere.00154-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meussen BJ, de Graaff LH, Sanders JPM, Weusthuis RA. Metabolic engineering of Rhizopus oryzae for the production of platform chemicals. Appl Microbiol Biotechnol. 2012. May 13;94(4):875–886. 10.1007/s00253-012-4033-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell AP. Location, location, location: Use of CRISPR-Cas9 for genome editing in human pathogenic fungi. PLoS Pathog. 2017. March 30;13(3):e1006209 10.1371/journal.ppat.1006209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Min K, Ichikawa Y, Woolford CA, Mitchell AP. Candida albicans Gene Deletion with a Transient CRISPR-Cas9 System. mSphere. 2017. March 15;2(2). pii: e00050–17. 10.1128/mSphere.00050-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang MY, Mitchell AP. Marker Recycling in Candida albicans through CRISPR-Cas9-Induced Marker Excision. mSphere. 2017;2(2):e00050–17. 10.1128/mSphere.00050-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen N, Quail MMF, Hernday AD. An Efficient, Rapid, and Recyclable System for CRISPR-Mediated Genome Editing in Candida albicans. mSphere. 2017. April 26;2(2). pii: e00149–17. 10.1128/mSphereDirect.00149-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shapiro RS, Chavez A, Porter CBM, Hamblin M, Kaas CS, DiCarlo JE, et al. A CRISPR-Cas9-based gene drive platform for genetic interaction analysis in Candida albicans. Nat Microbiol. 2018. January;3(1):73–82. 10.1038/s41564-017-0043-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DiCarlo JE, Chavez A, Dietz SL, Esvelt KM, Church GM. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat Biotechnol. 2015. December;33(12):1250–1255. 10.1038/nbt.3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hickman MA, Zeng G, Forche A, Hirakawa MP, Abbey D, Harrison BD, et al. The “obligate diploid” Candida albicans forms mating-competent haploids. Nature. 2013;494(7435):55–59. 10.1038/nature11865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lombardi L, Turner SA, Zhao F, Butler G. Gene editing in clinical isolates of Candida parapsilosis using CRISPR/Cas9. Sci Rep. 2017. August 14;7(1):8051 10.1038/s41598-017-08500-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zoppo M, Lombardi L, Rizzato C, Lupetti A, Bottai D, Papp C, et al. CORT0C04210 is required for Candida orthopsilosis adhesion to human buccal cells. Fungal Genet Biol. 2018. November;120:19–29. 10.1016/j.fgb.2018.09.001 [DOI] [PubMed] [Google Scholar]
- 41.Morio F, Lombardi L, Binder U, Loge C, Robert E, Graessle D, et al. Precise genome editing using a CRISPR-Cas9 method highlights the role of CoERG11 amino acid substitutions in azole resistance in Candida orthopsilosis. J Antimicrob Chemother. 2019. August 1;74(8):2230–2238. 10.1093/jac/dkz204 [DOI] [PubMed] [Google Scholar]
- 42.Cannon RD, Jenkinson HF, Shepherd MG. Isolation and nucleotide sequence of an autonomously replicating sequence (ARS) element functional in Candida albicans and Saccharomyces cerevisiae. Mol Gen Genet. 1990. April;221(2):210–218. 10.1007/bf00261723 [DOI] [PubMed] [Google Scholar]
- 43.Schwarzmüller T, Ma B, Hiller E, Istel F, Tscherner M, Brunke S, et al. Systematic Phenotyping of a Large-Scale Candida glabrata Deletion Collection Reveals Novel Antifungal Tolerance Genes. PLoS Pathog. 2014. June 19;10(6):e1004211 10.1371/journal.ppat.1004211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maroc L, Fairhead C. A new inducible CRISPR-Cas9 system useful for genome editing and study of double-strand break repair in Candida glabrata. Yeast. 2019. August 18 10.1002/yea.3440 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 45.de San Vicente KM, Schröder MS, Lombardi L, Iracane E, Butler G. Correlating Genotype and Phenotype in the Asexual Yeast Candida orthopsilosis Implicates ZCF29 in Sensitivity to Caffeine. G3 (Bethesda). 2019. September 4;9(9):3035–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kapoor M, Moloney M, Soltow QA, Pillar CM, Shaw KJ. Evaluation of Resistance Development to the Gwt1 inhibitor Manogepix (APX001A) in Candida Species. Antimicrob Agents Chemother. 2019. October 14. pii: AAC.01387-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rybak JM, Doorley LA, Nishimoto AT, Barker KS, Palmer GE, Rogers PD. Abrogation of Triazole Resistance upon Deletion of CDR1 in a Clinical Isolate of Candida auris. Antimicrob Agents Chemother. 2019. March 27;63(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krappmann S, Sasse C, Braus GH. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end- joining-deficient genetic background. Eukaryot Cell. 2006. January;5(1):212–215. 10.1128/EC.5.1.212-215.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang C, Meng X, Wei X, Lu L. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet Biol. 2016;86:47–57. 10.1016/j.fgb.2015.12.007 [DOI] [PubMed] [Google Scholar]
- 50.Weber J, Valiante V, Nødvig CS, Mattern DJ, Slotkowski RA, Mortensen UH, et al. Functional reconstitution of a fungal natural product gene cluster by advanced genome editing. ACS Synth Biol. 2017. January 20;6(1):62–68. 10.1021/acssynbio.6b00203 [DOI] [PubMed] [Google Scholar]
- 51.Umeyama T, Hayashi Y, Shimosaka H, Inukai T, Yamagoe S, Takatsuka S, et al. Cas9/CRISPR genome editing to demonstrate the contribution of Cyp51A Gly138Ser to azole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother. 2018. August 27;62(9). pii: e00894–18. 10.1128/AAC.00894-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ballard E, Weber J, Melchers WJG, Tammireddy S, Whitfield PD, Brakhage AA, Brown AJP, Verweij PE, Warris A. Recreation of in-host acquired single nucleotide polymorphisms by CRISPR-Cas9 reveals an uncharacterised gene playing a role in Aspergillus fumigatus azole resistance via a non-cyp51A mediated resistance mechanism. Fungal Genet Biol. 2019. September;130:98–106. 10.1016/j.fgb.2019.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hagen F, Khayhan K, Theelen B, Kolecka A, Polacheck I, Sionov E, et al. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol. 2015. May;78:16–48. 10.1016/j.fgb.2015.02.009 [DOI] [PubMed] [Google Scholar]
- 54.Lin X, Chacko N, Wang L, Pavuluri Y. Generation of stable mutants and targeted gene deletion strains in Cryptococcus neoformans through electroporation. Med Mycol. 2015. April 1;53(3):225–234. 10.1093/mmy/myu083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davidson RC, Cruz MC, Sia RAL, Allen B, Alspaugh JA, Heitman J. Gene Disruption by Biolistic Transformation in Serotype D Strains of Cryptococcus neoformans. Fungal Genet Biol. 2000. February;29(1):38–48. 10.1006/fgbi.1999.1180 [DOI] [PubMed] [Google Scholar]
- 56.Chen Y, Toffaletti DL, Tenor JL, Litvintseva AP, Fang C, Mitchell TG, et al. The Cryptococcus neoformans transcriptome at the site of human meningitis. MBio. 2014. February 4;5(1):e01087–13. 10.1128/mBio.01087-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goins CL, Gerik KJ, Lodge JK. Improvements to gene deletion in the fungal pathogen Cryptococcus neoformans: absence of Ku proteins increases homologous recombination, and co-transformation of independent DNA molecules allows rapid complementation of deletion phenotypes. Fungal Genet Biol. 2006. August;43(8):531–544. 10.1016/j.fgb.2006.02.007 [DOI] [PubMed] [Google Scholar]
- 58.Arras SDM, Chua SMH, Wizrah MSI, Faint JA, Yap AS, Fraser JA. Targeted genome editing via CRISPR in the pathogen Cryptococcus neoformans. PLoS ONE. 2016. October 6;11(10):e0164322 10.1371/journal.pone.0164322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao Y, Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol. 2014. April;56(4):343–349. 10.1111/jipb.12152 [DOI] [PubMed] [Google Scholar]
- 60.Fan Y, Lin X. Multiple applications of a transient CRISPR-Cas9 coupled with electroporation (TRACE) system in the cryptococcus neoformans species complex. Genetics. 2018. April;208(4):1357–1372. 10.1534/genetics.117.300656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bruni GO, Zhong K, Lee SC, Wang P. CRISPR-Cas9 induces point mutation in the mucormycosis fungus Rhizopus delemar.Fungal Genet Biol. 2019. March;124:1–7. 10.1016/j.fgb.2018.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee KT, So YS, Yang DH, Jung KW, Choi J, Lee DG, Kwon H, Jang J, Wang LL, Cha S, Meyers GL, Jeong E, Jin JH, Lee Y, Hong J, Bang S, Ji JH, Park G, Byun HJ, Park SW, Park YM, Adedoyin G, Kim T, Averette AF, Choi JS, Heitman J, Cheong E, Lee YH, Bahn YS. Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans. Nat Commun. 2016. September 28;7:12766 10.1038/ncomms12766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Noble S. M., French S., Kohn L. A., Chen V. & Johnson A. D. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42, 590–598 (2010). 10.1038/ng.605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holland LM, Schröder MS, Turner SA, Taff H, Andes D, Grózer Z, Gácser A, Ames L, Haynes K, Higgins DG, Butler G. Comparative phenotypic analysis of the major fungal pathogens Candida parapsilosis and Candida albicans. PLoS Pathog. 2014. September 18;10(9):e1004365 10.1371/journal.ppat.1004365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C, Martel N, Veronneau S, Lemieux S, Kauffman S, Becker J, Storms R, Boone C, Bussey H. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 50, 167–181 (2003). 10.1046/j.1365-2958.2003.03697.x [DOI] [PubMed] [Google Scholar]
- 66.Segal ES, Gritsenko V, Levitan A, Yadav B, Dror N, Steenwyk JL, Silberberg Y, Mielich K, Rokas A, Gow NAR, Kunze R, Sharan R, Berman J. Gene Essentiality Analyzed by In Vivo Transposon Mutagenesis and Machine Learning in a Stable Haploid Isolate of Candida albicans. MBio. 2018. October 30;9(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Glazier VE, Murante T, Koselny K, Murante D, Esqueda M, Wall GA, Wellington M, Hung CY, Kumar A, Krysan DJ. Systematic Complex Haploinsufficiency-Based Genetic Analysis of Candida albicans Transcription Factors: Tools and Applications to Virulence-Associated Phenotypes. G3 (Bethesda). 2018. March 28;8(4):1299–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018. August 31;361(6405):866–869. 10.1126/science.aat5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016. May 27;533(7601):125–129. 10.1038/nature17664 [DOI] [PubMed] [Google Scholar]
- 70.Hao H, Wang X, Jia H, Yu M, Zhang X, Tang H, et al. Large fragment deletion using a CRISPR/Cas9 system in Saccharomyces cerevisiae. Anal Biochem. 2016. September 15;509:118–123. 10.1016/j.ab.2016.07.008 [DOI] [PubMed] [Google Scholar]
- 71.Li Z-H, Liu M, Lyu X-M, Wang F-Q, Wei D-Z. CRISPR/Cpf1 facilitated large fragment deletion in Saccharomyces cerevisiae. J Basic Microbiol. 2018. December;58(12):1100–1104. 10.1002/jobm.201800195 [DOI] [PubMed] [Google Scholar]
- 72.Shao Y, Lu N, Xue X, Qin Z. Creating functional chromosome fusions in yeast with CRISPR-Cas9. Nat Protoc. 2019. August;14(8):2521–2545. 10.1038/s41596-019-0192-0 [DOI] [PubMed] [Google Scholar]
- 73.Yang F, Teoh F, Tan ASM, Cao Y, Pavelka N, Berman J. Aneuploidy Enables Cross-Adaptation to Unrelated Drugs. Mol Biol Evol. 2019. August 1;36(8):1768–1782. 10.1093/molbev/msz104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gorter de Vries AR, Couwenberg LGF, van den Broek M, de la Torre Cortés P, Ter Horst J, Pronk JT, Daran JG. Allele-specific genome editing using CRISPR-Cas9 is associated with loss of heterozygosity in diploid yeast. Nucleic Acids Res. 2019. February 20;47(3):1362–1372. 10.1093/nar/gky1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ford CB, Funt JM, Abbey D, Issi L, Guiducci C, Martinez DA, Delorey T, Li BY, White TC, Cuomo C, Rao RP, Berman J, Thompson DA, Regev A. The evolution of drug resistance in clinical isolates of Candida albicans. Elife. 2015. February 3;4:e00662 10.7554/eLife.00662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang MY, Woolford CA, May G, McManus CJ, Mitchell AP. Circuit diversification in a biofilm regulatory network. PLoS Pathog. 2019. May 22;15(5):e1007787 10.1371/journal.ppat.1007787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wijsman M, Swiat MA, Marques WL, Hettinga JK, van den Broek M, de la Torre Cortés P, et al. A toolkit for rapid CRISPR-SpCas9 assisted construction of hexose-transport-deficient Saccharomyces cerevisiae strains. FEMS Yeast Res. 2019. January 1;19(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burt A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc Biol Sci. 2003. May 7;270(1518):921–928. 10.1098/rspb.2002.2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sadhu MJ, Bloom JS, Day L, Siegel JJ, Kosuri S, Kruglyak L. Highly parallel genome variant engineering with CRISPR-Cas9. Nat Genet. 2018. April;50(4):510–514. 10.1038/s41588-018-0087-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Adames NR, Gallegos JE, Peccoud J. Yeast genetic interaction screens in the age of CRISPR/Cas. Curr Genet. 2019. April;65(2):307–327. 10.1007/s00294-018-0887-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Halperin SO, Tou CJ, Wong EB, Modavi C, Schaffer D V., Dueber JE. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature. 2018. August;560(7717):248–252. 10.1038/s41586-018-0384-8 [DOI] [PubMed] [Google Scholar]
- 82.Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018. May 15;9(1):1911 10.1038/s41467-018-04252-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016. January 1;351(6268):84–88. 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016. January 28;529(7587):490–495. 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen JS, Dagdas YS, Kleinstiver benjamin P, Welch MM, Sousa AA, Harrington LB, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017. October 19;550(7676):407–410. 10.1038/nature24268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015. October 22;163(3):759–771. 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018. April 5;556(7699):57–63. 10.1038/nature26155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016. May 20;533(7603):420–424. 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. Programmable base editing of T to G C in genomic DNA without DNA cleavage. Nature. 2017. October 25;551(7681):464–471. 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019. October 21 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, et al. RNA targeting with CRISPR-Cas13. Nature. 2017. October 12;550(7675):280–284. 10.1038/nature24049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Giersch RM, Finnigan GC. Yeast still a beast: Diverse applications of CRISPR/CAS editing technology in S. cerevisiae. Yale J Biol Med. 2017. December 19;90(4):643–651. [PMC free article] [PubMed] [Google Scholar]
- 93.Knight SC, Tjian R, Doudna JA. Genomes in Focus: Development and Applications of CRISPR-Cas9 Imaging Technologies. Angew Chem Int Ed Engl. 2018. April 9;57(16):4329–4337. 10.1002/anie.201709201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schultz C, Lian J, Zhao H. Metabolic Engineering of Saccharomyces cerevisiae Using a Trifunctional CRISPR/Cas System for Simultaneous Gene Activation, Interference, and Deletion. Methods Enzymol. 2018;608:265–276. 10.1016/bs.mie.2018.04.010 [DOI] [PubMed] [Google Scholar]
- 95.Wensing L, Sharma J, Uthayakumar D, Proteau Y, Chavez A, Shapiro RS. A CRISPR Interference Platform for Efficient Genetic Repression in Candida albicans. mSphere. 2019. February 13;4(1). pii: e00002–19. 10.1128/mSphere.00002-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Román E, Coman I, Prieto D, Alonso-Monge R, Pla J. Implementation of a CRISPR-Based System for Gene Regulation in Candida albicans. mSphere. 2019. February 13;4(1). pii: e00001–19. 10.1128/mSphere.00001-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Halder V, Porter CBM, Chavez A, Shapiro RS. Design, execution, and analysis of CRISPR-Cas9-based deletions and genetic interaction networks in the fungal pathogen Candida albicans. Nat Protoc. 2019. March;14(3):955–975. 10.1038/s41596-018-0122-6 [DOI] [PubMed] [Google Scholar]
- 98.Scorzoni L, de Paula E Silva AC, Marcos CM, Assato PA, de Melo WC, de Oliveira HC, Costa-Orlandi CB, Mendes-Giannini MJ, Fusco-Almeida AM. Antifungal Therapy: New Advances in the Understanding and Treatment of Mycosis. Front Microbiol. 2017. January 23;8:36 10.3389/fmicb.2017.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, Kellner MJ, Tan AL, Paul LM, Parham LA, Garcia KF, Barnes KG, Chak B, Mondini A, Nogueira ML, Isern S, Michael SF, Lorenzana I, Yozwiak NL, MacInnis BL, Bosch I, Gehrke L, Zhang F, Sabeti PC.Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018. April 27;360(6387):444–448. 10.1126/science.aas8836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Arastehfar A, Wickes BL, Ilkit M, Pincus DH, Daneshnia F, Pan W, Fang W, Boekhout T.Identification of Mycoses in Developing Countries.J Fungi (Basel). 2019. September 29;5(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Quan J, Langelier C, Kuchta A, Batson J, Teyssier N, Lyden A, Caldera S, McGeever A, Dimitrov B, King R, Wilheim J, Murphy M, Ares LP, Travisano KA, Sit R, Amato R, Mumbengegwi DR, Smith JL, Bennett A, Gosling R, Mourani PM, Calfee CS, Neff NF, Chow ED, Kim PS, Greenhouse B, DeRisi JL, Crawford ED.FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences.Nucleic Acids Res. 2019. August 22;47(14):e83 10.1093/nar/gkz418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bongomin F, Gago S, Oladele RO, Denning DW. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J Fungi (Basel). 2017. October 18;3(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ianiri G, Dagotto G, Sun S, Heitman J.Advancing Functional Genetics Through Agrobacterium-Mediated Insertional Mutagenesis and CRISPR/Cas9 in the Commensal and Pathogenic Yeast Malassezia. Genetics. 2019. August;212(4):1163–1179. 10.1534/genetics.119.302329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang Q, Coleman JJ.CRISPR/Cas9-mediated endogenous gene tagging in Fusarium oxysporum. CRISPR/Cas9-mediated endogenous gene tagging in Fusarium oxysporum. Fungal Genet Biol. 2019. May;126:17–24. 10.1016/j.fgb.2019.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kujoth GC, Sullivan TD, Merkhofer R, Lee TJ, Wang H, Brandhorst T, Wüthrich M, Klein BS. CRISPR/Cas9-Mediated Gene Disruption Reveals the Importance of Zinc Metabolism for Fitness of the Dimorphic Fungal Pathogen Blastomyces dermatitidis. MBio. 2018. April 3;9(2). pii: e00412–18. 10.1128/mBio.00412-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sanglard D. Finding the needle in a haystack: Mapping antifungal drug resistance in fungal pathogen by genomic approaches. Hogan DA, editor. PLoS Pathog. 2019. January 31;15(1):e1007478 10.1371/journal.ppat.1007478 [DOI] [PMC free article] [PubMed] [Google Scholar]