Abstract
Background:
Mutations in the isocitrate dehydrogenase 1 (IDH1) and IDH2 genes are important for both the integrated diagnosis and the prognosis of diffuse gliomas. The p.R132H mutation of IDH1 is the most frequently observed IDH mutation, while IDH2 mutations were relatively rarely studied. The aim of the study was to determine the pathological and genetic characteristics of lower-grade gliomas that carry IDH2 mutations.
Methods:
Data from 238 adult patients with lower-grade gliomas were retrospectively analyzed. The status of IDH1/2 gene mutations, telomerase reverse transcriptase (TERT) promoter mutations, O6-methylguanine-DNA-methyltransferase (MGMT) promoter methylation, 1p/19q co-deletion and the expressions of IDH1 R132H, alpha-thalassemia X-linked mental retardation, and p53 were evaluated. Progression-free survival (PFS) and overall survival (OS) were calculated via Kaplan-Meier estimation using the log-rank test.
Results:
Totally, 71% (169/238) of patients were positive for IDH mutations, including 12 patients harboring mutations in IDH2. Among the 12 patients with IDH2 mutations, ten patients harbored the R172K mutation, one patient harbored the R172S mutation and one harbored the R172W mutation. Of these, 11 tumors occurred in the frontal lobe and showed morphology typical of oligodendroglioma. The proportion of grade II tumors was higher than that of grade III tumors in IDH2 mutant-gliomas. IDH2 mutations were frequently associated with TERT promoter mutations, 1p/19q co-deletion and MGMT promoter methylation. IDH2 mutations were associated with better outcomes compared with IDH wild-type gliomas (P < 0.05). However, the PFS and OS did not differ from that of IDH1 mutant patients (P = 0.95 and P = 0.60, respectively).
Conclusions:
IDH2 mutations are more frequent in oligodendrogliomas and associated with a better prognosis. IDH2 mutations may segregate in distinct clinico-pathological and genetic subtypes of gliomas, and therefore may merit routine investigation.
Keywords: Isocitrate dehydrogenase 1, Isocitrate dehydrogenase 2, Telomerase reverse transcriptase, Glioma, Oligodendroglioma
Introduction
Diffuse gliomas are the most common and variably aggressive type of primary brain tumor.[1,2] Mutations in the isocitrate dehydrogenase 1 (IDH1) or IDH2 gene have recently been identified in a large proportion of diffuse astrocytomas, oligodendrogliomas, and secondary glioblastomas, and patients with tumors harboring these mutations were found to have better outcomes than those with wild-type IDH genes.[3] The status of the IDH gene has consequently been accepted as an important factor for the integrated diagnosis and prognosis of diffuse gliomas in the 2016 World Health Organization classification of tumors of the central nervous system (2016 WHO).[1] The p.R132H (c.395G>A, exon 4 codon 132) mutation in IDH1 was most frequently observed among all IDH mutant gliomas.[4–7] By contrast, mutations in the IDH2 gene, as well as their associated pathological features and other genetic alterations in diffuse gliomas were detected at a lower frequency and were relatively less studied.
To determine the pathological and genetic characteristics, as well as their clinical courses of diffuse gliomas harboring IDH2 mutations, we examined 238 lower-grade gliomas in adult patients using Sanger sequencing for mutations in codon 132 of IDH1 and codon 172 of IDH2. We also determined other molecular markers (p53, alpha-thalassemia X-linked mental retardation [ATRX], telomerase reverse transcriptase [TERT], Lys-27-Met mutations in histone 3 genes [H2K27M], and O6-methylguanine-DNA-methyltransferase [MGMT]) to identify their relationship with IDH2 mutations in gliomas. In this study, we demonstrated that IDH2 mutations are more frequent in oligodendrogliomas, which are associated with a better prognosis. IDH2 mutations may segregate in distinct histological, genetic, and molecular subtypes of gliomas, and therefore may merit routine clinical investigation.
Methods
Ethical approval
The study was approved by the Ethics Committee of the Xuanwu Hospital, Capital Medical University. The authors certify that they have obtained the appropriate patients’ consent form. In the form, the patients have given their consent for their images and other clinical information to be reported in the journal.
Patients
Data from a series of 238 patients with lower-grade gliomas (grade II and grade III) subtypes from the Xuanwu Hospital, Capital Medical University. Histological diagnoses were made based on formalin-fixed, paraffin-embedded hematoxylin and eosin-stained sections and were reviewed by two neuropathologists following the criteria of 2016 WHO classification.[1]
Immunohistochemical staining
Sections for immunohistochemical staining were made using a Leica Bond automated staining processor using antibodies against IDH1 R132H (clone H09, 1:500 dilution; DiaNova, Germany), ATRX (HPA001906, 1:100; Sigma Aldrich, St. Louis, MO, USA), p53 (clone DO-7, 1:100 dilution; Dako, Glostrup, Denmark), H3K27M (ABE419, 1:1000 dilution; Millipore, Billerica, MA, USA), Olig-2 (AB9610, 1:250 dilution; Millipore), Glial fibrillary acidic protein (polyclonal, 1:1000 dilution; Dako, USA), and Ki-67 (MIB-1, 1:50 dilution; Labvision, USA). The standard for judging IDH1 R132H, H3K27M, ATRX, and p53 staining was the same as in a previous study.[8]
DNA extraction
The presence of tumor tissue in the samples was histologically confirmed and an appropriate area comprising tumor tissue was selected for pyrosequencing (PSQ) and Sanger sequencing. Sections of each specimen with a thickness of 4 μm were cut from paraffin-embedded tissue, treated twice with xylene, and washed twice with ethanol. Genomic DNA was extracted using the DNeasy Tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Quantification was performed on a Nanodrop 2000 spectrophotometer (Thermo-Fisher Scientific, USA).
Analysis of IDH1/2 and TERT promoter mutations
The mutational status of IDH1/2 and TERT promoter was determined using Sanger sequencing. Exon4 of IDH1 and IDH2 was sequenced after amplification by polymerase chain reaction (PCR) using the IDH1 forward primer 5′-ACCAAATGGCACCATACGA-3′ and reverse primer 5′-TTC ATACCTTGCTTAATGGGTGT-3′, and the IDH2 forward primer 5′-GCTGCAGTGGGACCACTATT-3′ and reverse primer 5′- TGTGGCCTTGTACTGCAGAG-3′, respectively. The program for PCR amplification was as described previously.[9] Sequences covering the mutational hotspots in the TERT promoter, C228T and C250T were recovered using TERT forward primer 5′-CAGCGCTGCCTGAAACTC-3′ and reverse primer 5′-GTCCTGCCCCTTCACCTT-3′ in standard buffer, with 2 μL DNA and 1 μL DNA polymerase. The reaction mixture was subjected to 40 cycles of amplification (denaturation at 98°C for 10 s, annealing at 60°C for 15 s, and extension at 68°C for 1 min). The PCR products were analyzed by Sanger sequencing using 3730xl DNA Analyzer Technology (Applied Biosystems, USA).
Analysis of 1p/19q co-deletion
The analysis of the 1p and 19q co-deletion was conducted using fluorescence in situ hybridization. Sections with a thickness of 4 μm were cut from archival formalin-fixed, paraffin-embedded blocks, which were deparaffinized, dehydrated, digested in 0.3% pepsin solution and denatured at 85°C, followed by detection using fluorochrome-labeled 1p and 19q probes included dual-color probes localizing to 1p36/1q25 and 19q13/19p13 (Guangzhou LBP Pharmaceutical Technology Co., Ltd, China). All steps were conducted according to the kit's instructions. The stained sections were observed under a Lecia DM4000 M LED fluorescence microscope with appropriated filters. A total of 200 interphase non-overlapping nuclei were assessed in each case. The tumor was considered to have 1p/19q co-deletion if the signal ratio of 1p:1q to 19q:19p was ≤0.7.
MGMT promoter methylation
The PSQ methylation assay was performed using the PyroMark Q96 CpG MGMT Kit (Qiagen), according to the manufacturer's instructions and previous reports.[10]
Statistical analysis
Statistical analysis was carried out using SPSS 17.0 (IBM Corp., USA). Fisher exact test and χ2 test were used to test for associations or differences between IDH1/2 mutations, 1p/19q co-deletion, TERT promoter mutations and loss of ATRX. The Kaplan-Meier analyses were performed for survival data using the log-rank test. Differences were considered statistically significant at P < 0.05
Results
Clinical features of diffuse gliomas with IDH2 mutations
Among the 238 adult patients with lower-grade gliomas, 169 patients (71%) harbored IDH mutation, including 157 patients (66%, 157/238) with IDH1 and 12 patients (5%, 12/238) with IDH2 mutations. Their characteristics are shown in Table 1. Of these 12 IDH2-mutant patients, seven were males and five were females. The patients’ age at the time of diagnosis ranged from 26 to 75 years, with a median age of 44.5 years and average age of 42.7 years. Seizures and headaches were common initial symptoms. While 11 cases (11/12) located in the frontal lobe, only one case was in the parietal lobe [Table 1].
Table 1.
Data of 12 lower-grade gliomas with IDH2 mutation.
Pathological and molecular features of diffuse gliomas with IDH2 mutations
Among the 12 IDH2 mutant gliomas, initial histological diagnosis was oligodendroglioma (grade II) in eight patients, anaplastic oligodendroglioma (grade III) in three patients, and anaplastic astrocytoma in one patient. Histologically low-grade tumors (grade II) were more frequent than high-grade ones (grade III) among the IDH2-mutant gliomas.
Ten patients harbored the IDH2 R172K (c.515G>A) mutation, one patient harbored the IDH2 R172W (c.514A>T) mutation, and one patient harbored the IDH2 R172S (c.516G>T) mutation [Figure 1]. The IDH2 mutations were frequently associated with TERT promoter mutation (9/12, four patients with C228T mutation and five patients with C250T mutation) and 1p/19q co-deletion (11/12), and were negatively associated with loss of ATRX expression (2/12) and p53 overexpression (2/12). Only in one patient, the tumor showed the morphology of anaplastic astrocytoma, harbored IDH2 R172S mutation and also revealed a loss of ATRX expression and p53 overexpression. This patient was diagnosed with anaplastic astrocytoma, IDH-mutant, WHO grade III. None of the IDH2-mutant gliomas was positive for the H3K27M mutation. Among the ten patients that were available for detecting MGMT promoter methylation, nine patients were found to harbor MGMT promoter methylation [Figure 2].
Figure 1.
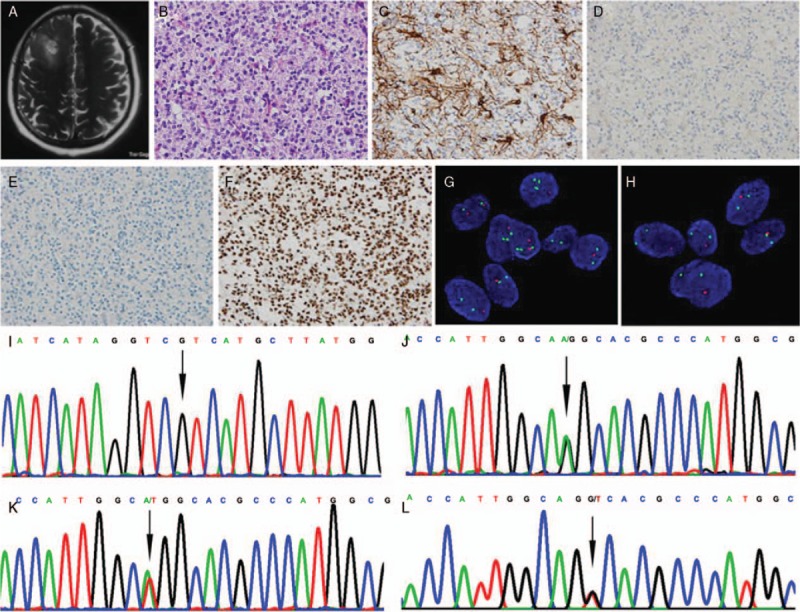
Representative clinico-neuropathological images of patients with lower-grade gliomas. (A) A patient (No. 9) with IDH2 mutant glioma showed a high-signal space-occupying lesion was on right frontal lobe on axial T2-weighted image of MRI. (B) The oligodendroglioma features of “fried eggs” nucleus and “chicken claw” vessels were observed, and mitotic figures were common (hematoxylin and eosin staining, original magnification ×400). (C–F) Immunohistochemical staining, original magnification ×400; tumor cells were immunopositive for GFAP (C), negative for IDH1 R132H (D) and p53 (E), and immunopositive for ATRX (F). (G and H) The tumor cells were harboring 1p/19q co-deletion detected by FISH (G: 1p loss of heterozygosity, H: 19q loss of heterozygosity; original magnification ×1000). (I and J) Sanger sequencing showed tumors were IDH1 wild-type (I: arrow, IDH1 wild-type) and IDH2 mutation (J: arrow, IDH2 R172K, AGG>AAG). (K) A patient (No. 11) showed IDH2 R172W mutation (Sanger sequencing, arrow, IDH2 R172W, AGG>TGG). (L) A patient (No. 12) showed IDH2 R172S mutation (Sanger sequencing, arrow, IDH2 R172S, AGG>AGT). ATRX: Alpha-thalassemia X-linked mental retardation; FISH: Fluorescence in situ hybridization; GFAP: Glial fibrillary acidic protein; IDH: Isocitrate dehydrogenase.
Figure 2.

Clinical and molecular characteristics of 12 lower-grade gliomas with IDH2 mutation. ATRX: Alpha-thalassemia X-linked mental retardation; H3K27M: Histone H3K27M; IDH: Isocitrate dehydrogenase; MGMT: O6-methylguanine-DNA-methyltransferase; TERT: Telomerase reverse transcriptase.
IDH2 mutations were associated with better prognosis
IDH mutational status and survival data were available for 214 patients. Kaplan-Meier survival analysis revealed significantly longer progress free survival (PFS) and overall survival (OS) of patients carrying the IDH mutation than those with IDH wild-type tumors (log-rank test, PFS: P = 0.049; OS: P < 0.001). The median PFS and OS of patients with IDH wild-type gliomas were 45 and 83 months, respectively, whereas the PFS of patients with IDH2-mutant gliomas was 96 months and only one patient died (18 months after diagnosis). However, there was no significant difference between IDH1 and IDH2 mutant patients (log-rank test, PFS: P = 0.575; OS: P = 0.773) [Figure 3].
Figure 3.
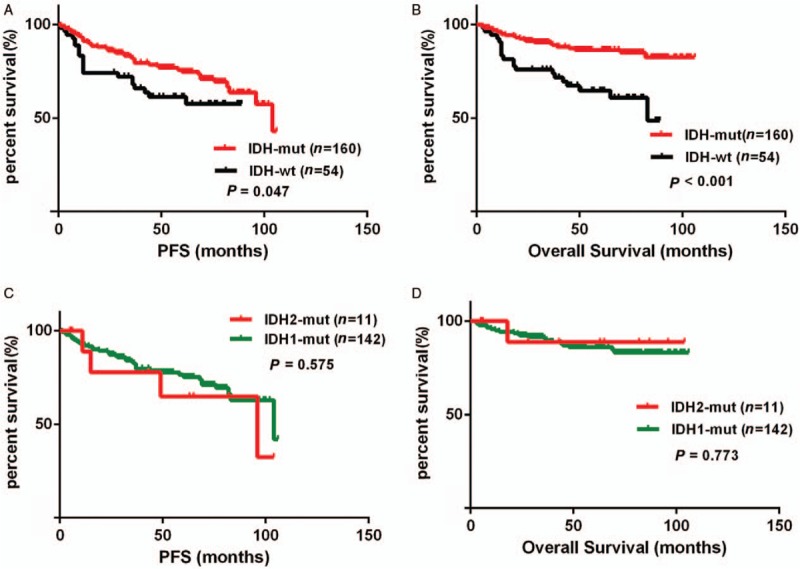
Kaplan-Meier survival curves and Log-rank tests for IDH-mutant lower-grade gliomas. The IDH mutation was associated with longer PFS (A) and OS (B). However, there was no significant difference between IDH1 and IDH2 mutant cases on PFS (C) or OS (D). IDH: Isocitrate dehydrogenase; MUT: Mutation; OS: Overall survival; PFS: Progress free survival; WT: Wild-type.
Discussion
Mutations in the IDH1 and IDH2 genes have been found frequently in diffuse gliomas. Integrated diagnosis of diffuse glioma requires the assessment of mutations in IDH1/IDH2 and co-deletion of 1p/19q.[1]IDH1 is localized in the cytoplasm, while IDH2 is found in the mitochondrial matrix.[11,12] The IDH1 R132H mutation is by far the most frequent (noted in >90% of patients) mutation observed in diffuse gliomas among all IDH mutations, and the genetic and epigenetic landscape of IDH1 mutant gliomas has been studied extensively.[13] In contrast to IDH1, IDH2 mutations are relatively rare (3%–5%) and the functional role of IDH2 is less clear. As reported previously, IDH1 is localized in the cytoplasm and peroxisomes, while IDH2 is localized in the mitochondria and participates in the tricarboxylic acid cycle to produce energy. The energy production in IDH2-mutated gliomas may favor oxidative phosphorylation over aerobic glycolysis. However, this hypothesis needs to be verified by focusing on metabolic pathways and general characteristics with more IDH2-mutant gliomas.[13] Biochemical investigations showed that IDH1 and IDH2 mutations differ in D-2-hydroxyglutarate production in gliomas, and thus may impact different cellular pathways and exert different tumorigenic effects.[14] Therefore, in this study, we investigated the clinical and pathological characteristics of diffuse gliomas with tumors harboring IDH2 mutations.
In our study, 12 patients (5%) were harboring IDH2 mutations. The IDH2 R172K mutation, accounting for 83.3%, was the most frequent mutation type in IDH2, which was consistent with previous studies.[15] Notably, mutations in IDH1 and IDH2 were mutually exclusive in gliomas. IDH2 mutations were mainly associated with tumors having the morphological features of oligodendroglioma. Our data also indicated that IDH2 mutations were mainly found in WHO grade II gliomas, which was different from previous reports.[16] The presence of IDH2 mutations did not correlate with the presence of TP53 mutations and ATRX loss, but a highly significant positive correlation was observed with the presence of 1p/19q co-deletion and TERT promoter mutations. According to 2016 WHO classification,[1] most cases (11/12) confirmed the integrated diagnosis of oligodendroglioma or anaplastic oligodendroglioma, IDH mutant, and 1p/19q co-deletion. Only one patient, with the onset age of 75 years, showed the features of anaplastic astrocytoma, which was accompanied by ATRX and TP53 mutations. Furthermore, we also found a patient with the IDH2 R172K mutation in combination 1p/19q co-deletion, as well as a wild-type TERT promoter and loss of ATRX expression, which was similar to a previously described study.[17] Loss of ATRX expression is a characteristic alteration of astrocytoma, and it is virtually mutually exclusive with 1p/19q co-deletion. These patients may have shown false-positive results for 1p/19q co-deletion and have a complex c-kit and platelet derived growth factor receptor alpha (PDGFRA) amplification, homozygous cyclin dependent kinase inhibitor 2A (CDKN2A) and CDKN2B deletions, as well as a loss of heterozygosity on chromosome 17p (including TP53).[17] Therefore, the diagnosis of oligodendroglioma needs further verification.
We also found that IDH2 mutations are associated with TERT promoter mutations and MGMT promoter methylation. Earlier studies suggested that IDH-mutant gliomas with TERT promoter mutations have a better outcome than corresponding TERT wild-type tumors.[18–20] Patients with gliomas containing a methylated MGMT promoter benefited from adjuvant therapy.[18,21–23] According to the Chinese Glioma Cooperative Group recommendations, the survival patterns can also be refined by sub-grouping by oligodendroglial, astrocytic, or glioblastoma molecular signatures, which could be served as a valuable source of information for comprehensive and precise treatment.[24] Furthermore, studies found that immunological tumor microenvironment differs in association with IDH mutation status in diffuse gliomas, which may be relevant for immune-modulatory treatment strategies.[25] Patients with oligodendroglial tumors were found to have a better prognosis (median OS of 8 years) compared with patients with astrocytic tumors (median OS of 5 years).[13] Therefore, our data suggest that patients with IDH2 mutations should be sensitive to adjuvant therapy and have a better prognosis. As expected, patients with IDH2-mutant tumors in our cohort had longer PFS and OS than IDH wild-type patients. However, there was no significantly statistical difference. Moreover, there was no significant difference in prognosis between patients with IDH1- and IDH2-mutant tumors in our cohort. This may be due to small number of IDH2-mutant cases. In addition, it could be the fact that many oligodendroglioma patients with IDH1 mutations also have a good prognosis. Therefore, additional data and further research are needed to accurately describe the prognostic implications of IDH2 mutations.
In conclusion, our results describe the clinical and pathological characteristics of IDH2 mutant gliomas. IDH2 mutations are more frequent in oligodendrogliomas and associate with a better prognosis. IDH2 mutations may segregate in distinct clinico-pathological and genetic subtypes of gliomas, and therefore may merit routine investigation.
Funding
This work was supported by grants from the Capital Health Research and Development of Special Program (No. 2014-2-2013), the Beijing Excellent Talent Training Project Grant (No. 201600026833ZK07), the National Science-Technology Support Plan (No. 2014BAI04B02), and the Project of Beijing Municipal Health Commission (No. PXM2019_026283_000002).
Conflicts of interest
None.
Footnotes
How to cite this article: Wang LM, Li Z, Piao YS, Cai YN, Zhang LY, Ge HJ, Xu WW, Lu DH. Clinico-neuropathological features of isocitrate dehydrogenase 2 gene mutations in lower-grade gliomas. Chin Med J 2019;132:2920–2926. doi: 10.1097/CM9.0000000000000565
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of tumors of the central nervous system: a summary. Acta Neuropathological 2016; 131:803–820.. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Olar A, Wani KM, Alfaro-Munoz KD, Heathcock LE, van Thuijl HF, Gilbert MR, et al. IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta Neuropathological 2015; 129:585–596.. doi: 10.1007/s00401-015-1398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appin CL, Brat DJ. Biomarker-driven diagnosis of diffuse gliomas. Mol Aspects Med 2015; 45:87–96.. doi: 10.1016/j.mam.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009; 360:765–773.. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visani M, Acquaviva G, Marucci G, Paccapelo A, Mura A, Franceschi E, et al. Non-canonical IDH1 and IDH2 mutations: a clonal and relevant event in an Italian cohort of gliomas classified according to the 2016 World Health Organization (WHO) criteria. J Neurooncol 2017; 135:245–254.. doi: 10.1007/s11060-017-2571-0. [DOI] [PubMed] [Google Scholar]
- 6.Gravendeel LA, Kloosterhof NK, Bralten LB, van Marion R, Dubbink HJ, Dinjens W, et al. Segregation of non-p.R132H mutations in IDH1 in distinct molecular subtypes of glioma. Hum Mutat 2010; 31:E1186–E1199.. doi: 10.1002/humu.21201. [DOI] [PubMed] [Google Scholar]
- 7.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015; 43:D805–D811.. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Li Z, Zhang M, Piao Y, Chen L, Liang H, et al. H3 K27M-mutant diffuse midline gliomas in different anatomical locations. Hum Pathol 2018; 78:89–96.. doi: 10.1016/j.humpath.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Sun J, Li Z, Chen L, Fu Y, Zhao L, et al. Gliosarcomas with the BRAF V600E mutation: a report of two cases and review of the literature. J Clin Pathol 2017; 70:1079–1083.. doi: 10.1136/jclinpath-2017-204620. [DOI] [PubMed] [Google Scholar]
- 10.Wang L, Li Z, Liu C, Chen L, Liu L, Hu Z, et al. Comparative assessment of three methods to analyze MGMT methylation status in a series of 350 gliomas and gangliogliomas. Pathol Res Pract 2017; 213:1489–1493.. doi: 10.1016/j.prp.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Philip B, Yu DX, Silvis MR, Shin CH, Robinson JP, Robinson GL, et al. Mutant IDH1 promotes glioma formation in vivo. Cell Rep 2018; 23:1553–1564.. doi: 10.1016/j.celrep.2018.03.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol 2016; 27:599–608.. doi: 10.1093/annonc/mdw013. [DOI] [PubMed] [Google Scholar]
- 13.Wang HY, Tang K, Liang TY, Zhang WZ, Li JY, Wang W, et al. The comparison of clinical and biological characteristics between IDH1 and IDH2 mutations in gliomas. J Exp Clin Cancer Res 2016; 35:86.doi: 10.1186/s13046-016-0362-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ward PS, Lu C, Cross JR, Abdel-Wahab O, Levine RL, Schwartz GK, et al. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem 2013; 288:3804–3815.. doi: 10.1074/jbc.M112.435495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol 2009; 118:469–474.. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 16.Appay R, Tabouret E, Macagno N, Touat M, Carpentier C, Colin C, et al. IDH2 mutations are commonly associated with 1p/19q codeletion in diffuse adult gliomas. Neuro Oncol 2018; 20:716–718.. doi: 10.1093/neuonc/noy014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballester LY, Huse JT, Tang G, Fuller GN. Molecular classification of adult diffuse gliomas: conflicting IDH1/IDH2, ATRX, and 1p/19q results. Hum Pathol 2017; 69:15–22.. doi: 10.1016/j.humpath.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 2015; 372:2499–2508.. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Killela PJ, Pirozzi CJ, Healy P, Reitman ZJ, Lipp E, Rasheed BA, et al. Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget 2014; 5:1515–1525.. doi: 10.18632/oncotarget.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan AK, Yao Y, Zhang Z, Chung NY, Liu JS, Li KK, et al. TERT promoter mutations contribute to subset prognostication of lower-grade gliomas. Mod Pathol 2015; 28:177–186.. doi: 10.1038/modpathol.2014.94. [DOI] [PubMed] [Google Scholar]
- 21.Arita H, Yamasaki K, Matsushita Y, Nakamura T, Shimokawa A, Takami H, et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol Commun 2016; 4:79.doi: 10.1186/s40478-016-0351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leu S, von Felten S, Frank S, Vassella E, Vajtai I, Taylor E, et al. IDH/MGMT-driven molecular classification of low-grade glioma is a strong predictor for long-term survival. Neuro Oncol 2013; 15:469–479.. doi: 10.1093/neuonc/nos317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamashita AS, Da CRM, Borodovsky A, Festuccia WT, Chan T, Riggins GJ. Demethylation and epigenetic modification with 5-azacytidine reduces IDH1 mutant glioma growth in combination with temozolomide. Neuro Oncol 2019; 21:189–200.. doi: 10.1093/neuonc/noy146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang T, Mao Y, Ma W, Mao Q, You Y, Yang X, et al. CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett 2016; 375:263–273.. doi: 10.1016/j.canlet.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 25.Berghoff AS, Kiesel B, Widhalm G, Wilhelm D, Rajky O, Kurscheid S, et al. Correlation of immune phenotype with IDH mutation in diffuse glioma. Neuro Oncol 2017; 19:1460–1468.. doi: 10.1093/neuonc/nox054. [DOI] [PMC free article] [PubMed] [Google Scholar]


