Abstract
Tumor-associated macrophages (TAMs) are the most abundant immune cells in the tumor microenvironment (TME) and are critical for cancer initiation and progression. MicroRNAs (miRNAs) could notably influence the phenotype of TAMs through various targets and signal pathways during cancer progression due to their post-transcriptional regulation. In this review, we discuss mainly the regulatory function of miRNAs on macrophage differentiation, functional polarization, and cellular crosstalk. Firstly, during the generation process, miRNAs take part in the differentiation from myeloid cells to mature macrophages, and this maturation process directly influences their recruitment into the TME, attracted by tumor cells. Secondly, macrophages in the TME can be either tumor-promoting or tumor-suppressing, depending on their functional polarization. Large numbers of miRNAs can influence the polarization of macrophages, which is crucial for tumor progression, including tumor cell invasion, intravasation, extravasation, and premetastatic site formation. Thirdly, crosstalk between tumor cells and macrophages is essential for TME formation and tumor progression, and miRNAs can be the mediator of communication in different forms, especially when encapsulated in microvesicles or exosomes. We also assess the potential value of certain macrophage-related miRNAs (MRMs) as diagnostic and prognostic markers, and discuss the possible development of MRM-based therapies.
Keywords: MicroRNA, Tumor microenvironment, Tumor-associated macrophage, Functional polarization
1. Introduction
Macrophages are one of the major components of the innate immune system and are responsible for pathogen clearance and antigen presentation. In the tumor microenvironment (TME), macrophages are among the most abundant immune cells recruited to the tumor site and play critical roles in various respects, including angiogenesis, tumor cell invasion, motility, intravasation, extravasation, survival, and premetastatic site formation (Noy and Pollard, 2014). These macrophages are termed tumor-associated macrophages (TAMs). Recent studies have revealed that macrophages in tissue are heterogenic and have different resources (Zhao et al., 2018). The major population of tissue-resident macrophages is established before birth, and their maintenance during adulthood depends on proliferation and longevity (Yona et al., 2013). This population can be replenished by monocytes derived from bone marrow, which differentiate into macrophages after injury, infection, or inflammation (Zhao et al., 2018). Plasticity is another significant characteristic of macrophages. In general, TAMs can be activated in classical (referred to as M1) or alternative (referred to as M2) pathway, which have different regulatory functions in the TME, according to distinct intracellular signaling pathways induced by different extracellular molecules (Martinez and Gordon, 2014).
MicroRNAs (miRNAs) are endogenous RNAs about 23 nucleotides long. They function as negative regulators of gene expression by increasing mRNA decay or by blocking the process of translation, establishing a new paradigm of gene regulation at the post-transcriptional level. Altered expression of the miRNA species is well established in TAMs (Baer et al., 2016; Zhao et al., 2016; Xi et al., 2018; Shidal et al., 2019). miRNAs are involved in tumor development, progression, and metastasis by targeting mRNAs of oncogenes or tumor suppressor genes. Moreover, some important miRNAs have the potential to be markers for diagnosis and prognosis or to serve as effective targets for therapeutic interventions in cancer.
In this review, we focus mainly on the role of miRNAs as molecular determinants in macrophage differentiation, TAM-mediated tumor immunity, and clinical application. Note that the function of some miRNAs in this review is based on experimental evidence from macrophages, not TAMs, and still needs verification in the TME.
2. miRNAs involved in the differentiation of myeloid cells into macrophages
Despite the complex origins of macrophages in tissue, most studies of miRNAs regulating the development of macrophages focus on those originating from hematopoietic stem cells (HSCs). The process of development from HSCs to macrophages involves successive differentiation steps, including from common myeloid progenitors (CMPs) to granulocyte-macrophage precursors (GMPs) and then to macrophage/dendritic cell (DC) progenitors (MDPs) (Geissmann et al., 2010). TAMs are unable to proliferate or survive for long in the TME (Cortez-Retamozo et al., 2012), so it is necessary to recruit precursor cells continuously to the tumor site to replenish the macrophage population.
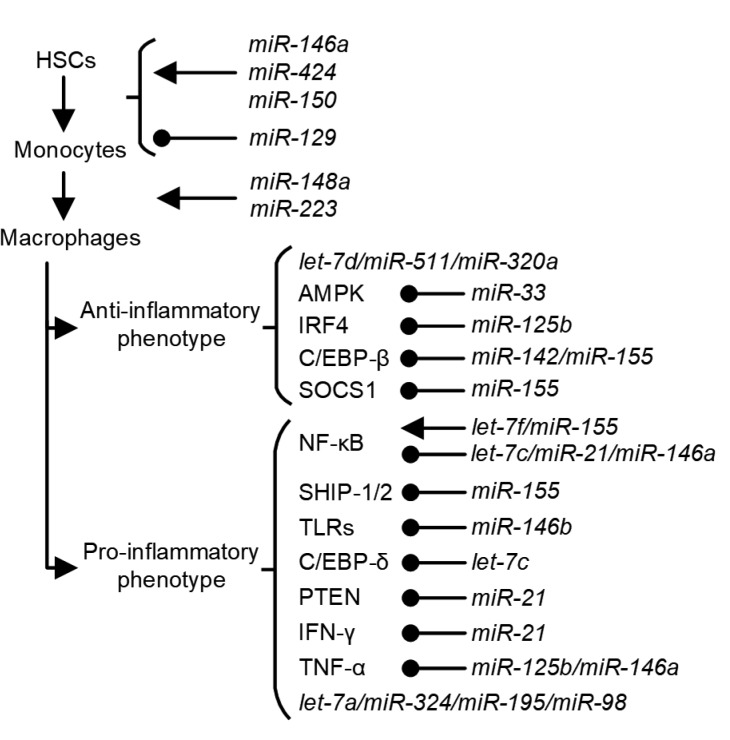
Transcription factor PU.1 is a key molecule in the differentiation of the myeloid lineage and participates in the transcription of several genes specifically expressed in myeloid cells. PU.1-controlled miRNAs are also necessary components of this differentiation process. One of the direct targets of PU.1 is miRNA-146a (miR-146a), which induces the differentiation of HSCs to peritoneal macrophages in mice models (Ghani et al., 2011). miR-424 is another target miRNA of PU.1. Its upregulation in myeloid cells by PU.1 impairs the translation of nuclear factor 1 A (NFIA). The downregulation of NFIA gives rise to the expression of macrophage colony-stimulating factor receptor (M-CSFR), which promotes differentiation towards the monocyte/macrophage lineage (Rosa et al., 2007). Runt-related transcription factor 1 (RUNX1) is another protein that plays critical roles in development of hematopoietic cells, and can positively regulate the expression of PU.1 (Ran et al., 2013). A comparative analysis of miRNAs has shown that overexpression of miR-129 can promote granulopoiesis and inhibit monocytopoiesis by repressing the expression of RUNX1 (Zhao et al., 2017).
Macrophages are heterogenic and can be generated from embryonic yolk sac, fetal liver, or bone marrow. Monocytes derived from the embryonic yolk sac are lymphocyte antigen 6 complex (Ly6C)negative, whereas those derived from bone marrow are Ly6Chigh or Ly6Cnegative/low (Guerriero, 2018). Studies using mice models have revealed that the progenitors of most TAMs comprise a Ly6Chigh subset of circulating monocytes (Movahedi et al., 2010; Franklin et al., 2014). Comparison of the expression of miRNAs in different subsets of monocytes indicated that miR-146a can be stably expressed in the Ly6Clow group without affecting its response to inflammatory signals, while in the Ly6Chigh group, its expression hampered the proliferation and trafficking of monocytes under inflammatory conditions (Etzrodt et al., 2012). In chronic myelo-monocytic leukemia (CMML) patients, the amount of non-classical monocytes (Ly6Clow group) is notably decreased compared with that in healthy people, which is also observed in miR-150 knock-out mouse models. Pulldown experiments proved that ten-eleven-translocation-3 (TET3) mRNA is one of the targets of miR-150 and is involved in the generation of non-classical monocytes (Selimoglu-Buet et al., 2018).
In the model of THP-1 cells transfected with miR-223 antagonist, under treatment with phorbol-12-myristate-13-acetate (PMA) or granulocyte-macrophage colony-stimulating factor (GM-CSF), monocytes do not become adherent, which is considered a phenotype indicative of macrophage differentiation. miR-223 can be transported through macrophage-derived microvesicles (MVs) to mediate the differentiation of other monocytes (Ismail et al., 2013). It is believed that a Notch signal is critical for the differentiation and polarization of macrophages. miR-148a-3p is regulated by the Notch signal pathway under GM-CSF stimulation, and can enhance differentiation of monocytes to macrophages (Huang et al., 2017). miRNAs involved in the differentiation of myeloid cells to macrophages are summarized in Table 1 and Fig. 1.
Table 1.
miRNAs involved in differentiation of myeloid cells to macrophages
| miRNA | Cell type | Target | Phenotype | Reference |
| miR-146a | HSC | Differentiation to macrophage↑ | Ghani et al., 2011 | |
| Monocyte | Relb | Proliferation and trafficking↓ | Etzrodt et al., 2012 | |
| miR-424 | Myeloid cell | NFIA | Differentiation to monocyte/macrophage↑ | Rosa et al., 2007 |
| miR-129 | Myeloid cell | RUNX1 | Granulopoiesis↑, monocytopoiesis↓ | Zhao et al., 2017 |
| miR-150 | Monocyte | TET3 | Generation↑ | Selimoglu-Buet et al., 2018 |
| miR-223 | Monocyte | Differentiation to macrophage↑ | Ismail et al., 2013 | |
| miR-148a | Monocyte | PTEN | Differentiation to macrophage↑ | Huang et al., 2017 |
HSC: hematopoietic stem cell; NFIA: nuclear factor 1 A; RUNX1: Runt-related transcription factor 1; TET3: ten-eleven-translocation-3; PTEN: phosphatase and tensin homologue
Fig. 1.
miRNAs involved in the differentiation and polarization of macrophages and tumor-associated macrophages (TAMs)
The development of macrophages from hematopoietic stem cells (HSCs) involves successive steps regulated by different miRNAs, and large numbers of miRNAs are engaged in the polarization of macrophages between M1 and M2
3. miRNAs involved in functional regulation of macrophages and TAMs
M1 (classically activated) and M2 (alternatively activated) macrophages represent the two extremes of the spectrum of macrophage phenotypes in tissue. M1, involved mainly in anti-microbial response and presenting a pro-inflammatory phenotype, is normally stimulated by interferon-γ (IFN-γ) and lipopolysaccharide (LPS), whereas M2, believed to be the main phenotype of TAMs, is normally stimulated by interleukin 4 (IL-4), IL-10, IL-13, and glucocorticoids (Curtale, 2018). Large numbers of miRNAs have been found to engage in the polarization of macrophages between M1 and M2, and subsequently inhibit or promote the initiation and progression of tumors. miRNAs involved in functional regulation of macrophages and TAMs are summarized in Table 2 and Fig. 1.
Table 2.
miRNAs involved in functional regulation of macrophages and tumor-associated macrophages (TAMs)
| miRNA | Cell type | Target | Phenotype | Reference |
| let-7a | Macrophagea | Proliferation↑, pro-inflammatory cytokine production↑ | Chafin et al., 2014 | |
| TAM | Pro-inflammatory cytokines secretion↑, tumor progression↓ | Schmid et al., 2018 | ||
| let-7f | Macrophage | A20 | Pro-inflammatory cytokines (TNF and IL-1β) secretion↑ | Kumar et al., 2015 |
| let-7c | Macrophage | PAK1/NF-κB | M1 polarization↓, M2 polarization↑ | Zhang et al., 2015 |
| Macrophage | C/EBP-δ | M1 polarization↓, M2 polarization↑ | Banerjee et al., 2013b | |
| let-7d | TAM | M2 polarization↑, amount of tumor-infiltrating CTLs↓ | Baer et al., 2016 | |
| miR-21 | TAM | Angiogenesis and tumor growth↑ | Mathsyaraja et al., 2015 | |
| Macrophage | PDCD4 | IL-10 expression↑ | Sheedy et al., 2010 | |
| TAM | PTEN, miR-200c | M2 polarization↑, tumor cell migration↑ | Li N et al., 2018 | |
| TAM | IFN-γ/STAT1 | PD-L1 expression↓, M1 polarization↓ | Xi et al., 2018 | |
| Macrophage | STAT3 | M2 genes expression↓ | Wang et al., 2015 | |
| miR-33 | Macrophage | AMPK | M2 polarization↓ | Ouimet et al., 2015 |
| Macrophage | ABCA1 | Inflammatory response↑ | Price et al., 2019 | |
| miR-125a | TAM | Phagocytic activity↑, tumor growth↓ | Zhao et al., 2016 | |
| Macrophage | KLF13 | M1 polarization↓, M2 polarization↑ | Banerjee et al., 2013a | |
| miR-125b | TAM | IRF4 | Costimulatory molecules expression↑, responsiveness to IFN-γ↑ | Chaudhuri et al., 2011 |
| TAM | M1 polarization↑ | Parayath et al., 2018 | ||
| Macrophage | TNF-α | TNF-α production↓, endotoxin tolerance↑ | Tili et al., 2007 | |
| miR-142 | TAM | gp130, C/EBP-β | M2 polarization↓, survival after tumor-specific T cell therapy↑ | Sonda et al., 2013 |
| Macrophage | SOCS1 | M2 polarization↑ | Su et al., 2015 | |
| miR-146a | Macrophage | Relb | TNF-α expression↓ | el Gazzar et al., 2011 |
| Macrophage | Notch1 | Endotoxin tolerance↑ | Bai et al., 2018; Funahashi et al., 2019 | |
| Macrophage | INHBA | M1 polarization↓, M2 polarization↑ | Li et al., 2016 | |
| TAM | Oncogenic transformation↓ | Boldin et al., 2011 | ||
| miR-146b | Macrophage | TLR-4 | IL-6, TNF-α, and IL-8 expression↓ | Curtale et al., 2013 |
| miR-155 | Macrophage | TNF-α secretion↑ | Bala et al., 2011 | |
| Macrophage | SOCS1 | IFN-mediated antiviral response↑ | Wang et al., 2010 | |
| Macrophage | SOCS1 | IL-6 and TNF-α expression↑ | Ye et al., 2016 | |
| Macrophage | BCL6 | CCL2 secretion↑, plaque formation↑ | Nazari-Jahantigh et al., 2012 | |
| TAM | FGF2 | TNF-α, IL-12, and iNOS expression↑, tumor cell survival, migration, and invasion↓ | Wang P et al., 2018 | |
| Macrophage | TAB2 | IFN-α/β expression↓ | Zhou et al., 2010 | |
| Macrophage | SHIP1, C/EBP-β | Inflammatory cytokine production↓ | He et al., 2009; O'Connell et al., 2009 | |
| miR-324 | TAM | CUEDC2 | Pro-inflammatory cytokines production↑, risk of colon tumorigenesis↑ | Chen et al., 2014 |
| miR-511 | TAM | Pro-tumoral gene expression↓, tumor growth↓ | Squadrito et al., 2012 | |
| miR-195 | TAM | Notch2 | EMT↓, M2 polarization↓, overall survival of colorectal cancer patients↑ | Lin et al., 2019 |
| miR-320a | Macrophage | STAT4 | M2 polarization↑ | Fortunato et al., 2019 |
| miR-98 | TAM | IL-10 | IL-10 expression↓, M1 polarization↑, tumor cell migration and invasion↓ | Li L et al., 2018 |
These functions need verification in tumor microenvironment (TME). PAK1: p21-activated kinase 1; NF-κB: nuclear factor κB; C/EBP: CCAAT/enhancer-binding protein; PDCD4: programmed cell death 4; PTEN: phosphatase and tensin homologue; IFN-γ: interferon-γ; STAT: signal transducer and activator of transcription; AMPK: adenosine 5'-monophosphate (AMP)-activated protein kinase; ABCA1: adenosine triphosphate (ATP)-binding cassette sub-family A member 1; KLF13: Kruppel-like factor 13; IRF4: IFN regulatory factor 4; TNF: tumor necrosis factor; SOCS1: suppressor of cytokine signaling 1; INHBA: inhibin subunit β A; TLR: Toll-like receptor; BCL6: B-cell lymphoma 6; FGF2: fibroblast growth factor-2; TAB2: transforming growth factor β-activated kinase 1 (TAK1)-binding protein 2; SHIP1: Src homology 2 domain-containing inositol 5-phosphatase; CUEDC2: CUE domain-containing protein 2; IL: interleukin; CTL: cytotoxic T cell; PD-L1: programmed death ligand 1; CCL2: C-C motif chemokine ligand 2; iNOS: inducible nitric oxide synthase; EMT: epithelial-mesenchymal transition
3.1. let-7
The let-7 miRNA family in humans includes 13 members expressed from nine chromosomes and is highly conserved in sequence and function among other vertebrates (Roush and Slack, 2008). In immune-stimulated cells, let-7a overexpression induces the expression of the cell cycle activator E2F2, promoting G1/S transition, and reduces the expression of the cell cycle inhibitor E2F5. In this way, let-7a is supposed to enhance the proliferation and pro-inflammatory cytokine production of macrophages (Chafin et al., 2014). The expression of let-7a is stimulated by CD11b, and the activation of CD11b promotes pro-inflammatory cytokine secretion and macrophage polarization. In contrast, downregulated let-7a in macrophages suppresses the inflammatory response and leads to tumor progression (Schmid et al., 2018). Another member of the let-7 family, let-7f, shows reduced expression in macrophages under tuberculosis infection. Re-expression of let-7f in macrophages can promote the production of pro-inflammatory cytokines, including tumor necrosis factor (TNF) and IL-1β, by inhibiting the expression of A20, an inhibitor of the NF-κB pathway (Kumar et al., 2015).
In contrast to let-7a and let-7f, let-7b/c/d/e seems to have greater effects on alternative activation of macrophages. Let-7c can prevent M1 polarization and promote M2 polarization by targeting CCAAT/enhancer-binding protein (C/EBP)-δ or p21-activated kinase 1 (PAK1)-NF-κB (Banerjee et al., 2013a; Zhang et al., 2015). Dicer is an RNAse-III enzyme involved in processing hairpin-shaped precursor miRNAs into mature miRNAs (Ha and Kim, 2014). Deletion of Dicer in macrophages promotes M1 polarization. However, in Dicer1-deficient TAMs, re-expression of let-7d-5p can reverse the polarization, restore the M2 phenotype, and decrease the amount of tumor-infiltrating cytotoxic T cells (CTLs) (Baer et al., 2016).
3.2. miR-21
In breast cancer and melanoma metastasis models, the expression of miR-21 is upregulated during tumor progression. The CSF1-ETS2 (V-ets erythroblastosis virus E26 oncogene homolog 2) pathway is responsible for the regulation of those miRNAs, and enforced expression of miR-21 in macrophages can promote angiogenesis and tumor growth (Mathsyaraja et al., 2015). One of the downstream effectors of the pro-tumor function of miR-21 is programmed cell death 4 (PDCD4), which is a tumor suppressor and pro-inflammatory protein that activates the NF-κB pathway and suppresses IL-10 expression (Sheedy et al., 2010). A recent study has proved that the tumor suppressor genes phosphatase and tensin homologue (PTEN) and miR-200c are also targets of miR-21a. Upregulation of miR-21a in macrophages can promote transformation to the anti-inflammatory phenotype through downregulation of PTEN and enhance the migratory ability of breast cancer cells (Li N et al., 2018). Consistent with these findings, a genetic deficiency of miR-21 can promote the polarization of macrophage to the M1 phenotype in the presence of tumor cells through the IFN-γ/signal transducer and activator of transcription 1 (STAT1) pathway. Augmented STAT1 signal caused by downregulation of miR-21 can also enhance the expression of programmed death ligand 1 (PD-L1) in macrophages, which consequently inhibits the anti-tumor ability of macrophages (Xi et al., 2018). Although miR-21 is regarded mainly as an anti-inflammatory mediator, there is also evidence showing that it is able to abolish the expression of M2 genes through targeting STAT3. Prostaglandin E2 (PGE2), a determining molecule of M2 polarization, can inhibit miR-21 expression and induce an anti-inflammatory phenotype (Wang et al., 2015).
3.3. miR-33
Cellular metabolism is a critical factor during the process of macrophage activation. The inhibition of miR-33 expression is responsible for M2 polarization through the targeting of adenosine 5'-monophosphate (AMP)-activated protein kinase (AMPK). Downregulation of miR-33 can increase the expression of aldehyde dehydrogenase 1 family member A2 (ALDH1A2) and activate retinal dehydrogenase, which in turn increases the production of retinal acids from macrophages to induce the differentiation of regulatory T cells (Tregs) (Ouimet et al., 2015). The adenosine triphosphate (ATP)-binding cassette subfamily A member 1 (Abca1) is another target of miR-33. In Abca1-binding site mutant mouse models, the re-expression of ABCA1 can repress the inflammatory response of macrophages during atherosclerotic plaque formation (Price et al., 2019). Thus, miR-33 could play a role in the maintenance of the pro-inflammatory microenvironment.
3.4. miR-125
Macrophage miRNA profiling shows that miR-125a is a downstream mediator of the Notch signaling pathway and regulates polarization of M1 and M2 macrophages. Overexpression of miR-125a in macrophages by transfection notably enhances their phagocytic activity and represses tumor growth (Zhao et al., 2016). When miR-125b is overexpressed in macrophages, it can induce the expression of co-stimulatory molecules and make macrophages more responsive to IFN-γ. miR-125b represses the expression of IFN regulatory factor 4 (IRF4), and in turn, activates macrophages and endows them with the ability to kill EL4 tumor cells more effectively (Chaudhuri et al., 2011). Recently, using nanoparticles containing miR-125b, Parayath et al. (2018) found that transfected TAMs showed a remarkable increase in the M1 to M2 ratio, which was proved by a 300-fold increase in the inducible nitric oxide synthase (iNOS)/arginase-1(Arg1) ratio.
miR-125a and miR-125b are able to promote the M1 phenotype of macrophages, but they also mediate the polarization of the anti-inflammatory M2 phenotype. M2 macrophages express a higher level of miR-125a-5p than M1 macrophages, mediated by activation of Toll-like receptor (TLR)-2 or TLR-4 and downstream myeloid differentiation factor (MyD88). The target of miR-125a-5p is transcription factor Kruppel-like factor 13 (KLF13), whose downregulation diminishes the M1 phenotype induced by LPS, and enhances the M2 phenotype induced by IL-4 (Banerjee et al., 2013b). miR-125b directly targets the 3'-untranslated region (3'-UTR) of TNF-α mRNA to inhibit its production in response to LPS stimulation, and might also be involved in the formation of endotoxin tolerance (Tili et al., 2007).
3.5. miR-142
The effect of miR-142 on macrophage polarization might also be bidirectional. Downregulation of miR-142-3p promotes macrophage differentiation into an immunosuppressive phenotype through binding to mRNA of gp130 and C/EBP-β. This impairs the differentiation process, and may increase survival after tumor-specific T cell therapy when constitutively expressed in bone marrow of mice (Sonda et al., 2013). miR-142-5p is induced in macrophages treated with IL-4 and IL-13, and transduction of anti-sense oligonucleotides of miR-142-5p in macrophages notably downregulates the secretion of M2 cytokines and expression of M2 surface markers including C-C motif chemokine ligand 13 (CCL13), CCL17, CCL18, TGF-β1, CD206, and CD36 (Su et al., 2015).
3.6. miR-146
miR-146a has long been regarded as an anti-inflammatory miRNA. Compared with M1 macrophages, miR-146a is highly expressed in M2 macrophages and targets inhibin subunit β A (INHBA). Downregulation of these downstream molecules can effectively reduce the expression of several cytokines related to the M1 phenotype, such as IL-6, IL-12, TNF-α, and iNOS, and increase the levels of M2 markers, including Arg1, CD206, CCL17, and CCL22 (Li et al., 2016). Consistent with the functions in vitro, miR-146a-null mice are hypersensitive to LPS stimulation and present a higher risk of oncogenic transformation (Boldin et al., 2011).
In addition, miR-146a and miR-146b are involved in the regulation of endotoxin tolerance, which seems to be a mechanism to protect macrophages from overwhelming activation. In general, macrophages become polarized to the M1 phenotype in response to the stimulation of LPS/endotoxin, but when the exposure time is long enough, they can become tolerant to the stimulation and alter their response mode (West and Heagy, 2002). Through kinetic analysis of miRNA expression, the level of miR-146a in THP-1 cells was observed to increase after 4 h of LPS stimulation and continue rising, which is likely to correlate negatively with the expression level of TNF-α (Nahid et al., 2009). This function of miR-146a is accomplished by regulating the transcription repressor Relb, which binds to the TNF-α promoter, and by interfering with the interaction between argonaute 2 (Ago2) and RNA-binding motif protein 4 (RBM4), which in turn assists the assembly of the translation repressor complex of TNF-α (el Gazzar et al., 2011). Recent studies have revealed that Notch1 could be another target of miR-146a. Overexpression of miR-146a leads to attenuated activation of the NF-κB pathway, which protects against organ damage from LPS-induced inflammation (Bai et al., 2018; Funahashi et al., 2019). Through directly targeting TLR-4, which is the receptor of LPS, and critical adaptor/signaling proteins including MyD88, IL-1 receptor-associated kinase 1 (IRAK-1), and TNF receptor-associated factor 6 (TRAF6), enforced expression of miR-146b can notably repress the expression of IL-6, TNF-α, IL-8, and some other cytokines in macrophages (Curtale et al., 2013).
3.7. miR-155
miR-155 is characteristically expressed at a high level in M1 macrophages under stimulation from LPS, IFN-γ, and mildly oxidized low-density lipoprotein (moxLDL) (Wei and Schober, 2016). Various mechanisms underlie its pro-inflammatory effects. In mice models of alcoholic liver disease, expression of miR-155 via the NF-κB pathway can increase the half-life of TNF-α mRNA and the secretion of TNF-α from macrophages (Bala et al., 2011). Suppressor of cytokine signaling 1 (SOCS1) and B-cell lymphoma 6 (BCL6) are important downstream mediators of its function. SOCS1 is a canonical negative regulator of IFN signaling, and its downregulation by miR-155 activates the IFN-mediated antiviral response (Wang et al., 2010). In an atherosclerotic mice model, inhibition of miR-155 expression remarkably decreased the levels of IL-6 and TNF-α through upregulation of SOCS1 and downregulation of STAT3 and PDCD4 (Ye et al., 2016). BCL6 is a transcription factor that hampers NF-κB signaling. Silencing of BCL6 expression by miR-155 induced the secretion of chemokine CCL2 and enhanced plaque formation in an atherosclerosis model (Nazari-Jahantigh et al., 2012). Studies focusing on esophageal cancer (EC) found that upregulation of miR-155 in TAMs could enhance the expression of TNF-α, IL-12, and iNOS, and consequently inhibit the ability of EC cells to survive, migrate, and invade. This effect was proved to be mediated by fibroblast growth factor-2 (FGF2) (Wang P et al., 2018).
However, there is some evidence showing that miR-155 is also engaged in the anti-inflammatory response of macrophages. Kinetic analysis revealed that miR-155 was upregulated mainly in the later stages of TLR7 activation, and targeted transforming growth factor β-activated kinase 1 (TAK1)-binding protein 2 (TAB2) to repress the expression of IFN-α/β (Zhou et al., 2010). Src homology 2 domain-containing inositol 5-phosphatase (SHIP1) and C/EBP-β could also be the downstream targets of miR-155, and play a role in the regulation of alternatively activated macrophages (He et al., 2009; O'Connell et al., 2009).
3.8. miR-324-5p
In colitis-associated tumorigenesis, CUE domain-containing protein 2 (CUEDC2) is a critical protein regulating macrophage function, and is also the target of miR-324-5p. Under stimulation of IL-4, expression of miR-324-5p is elevated and CUEDC2 is deficient, which results in excessive production of pro-inflammatory cytokines and increases the risk of colon tumorigenesis (Chen et al., 2014).
3.9. miR-511-3p
miR-511-3p is more likely to be a modulator that negatively regulates the alternative activation of macrophages or their pro-tumoral functions. In CD206+ M2-polarized TAMs, overexpression of miR-511-3p can downregulate the level of pro-tumoral gene signature and limit the growth of tumors (Squadrito et al., 2012).
3.10. miR-195-5p
Epithelial-mesenchymal transition (EMT) is considered to be a major mechanism of tumorigenesis and metastasis, and includes the interaction between cancer cells and TAMs (Suarez-Carmona et al., 2017). In colorectal cancer tissue, the level of miR-195-5p is notably decreased compared with normal tissue, and patients with a higher miR-195-5p expression level have longer overall survival. Mechanistically, Notch2 is the target of miR-195-5p. Downregulation of Notch2 in cancer cells inhibits the EMT process, leading to a decrease in IL-4 secretion and suppression of anti-inflammatory TAM polarization (Lin et al., 2019).
3.11. miR-320a
In recent years, tumor-circulating cell-free miRNAs have been regarded as potential biomarkers for early diagnosis of cancer and predictive factors for prognosis (Schwarzenbach et al., 2014). In heavy smokers, the level of circulating miR-320a secreted by neutrophils is upregulated. miR-320a can be shuttled into macrophages to promote an M2-like phenotype through targeting STAT4 (Fortunato et al., 2019). This finding suggests that miR-320a might have a causative role in the immunosuppressive microenvironment and tumorigenesis in lung cancer.
3.12. miR-98
miR-98 suppresses the progression of hepatocellular carcinoma (HCC), and IL-10 is the direct target. Overexpression of miR-98 in HCC-conditioned TAMs suppresses the expression of IL-10 and leads to a pro-inflammatory phenotype, thereby reducing the migration and invasion of HCC (Li L et al., 2018).
The regulation of macrophage polarization is a very complicated process in which dozens of cytokines, receptors, and transcription regulators are involved. The effects of miRNAs on those regulatory proteins add to the complexity. According to previous studies of macrophage polarization, the expression of several proteins, including IL-4R, STAT6, IRF4, Jumonji doman-containing protein 3 (JMJD3), peroxisome proliferator-activated receptor δ (PPARδ), and PPARγ, with normal function is essential for alternative activation of macrophages. As for classical activation, it seems that the NF-κB pathway and its activators (TLRs, TNF, and IL-1R) are important for M1 polarization (Murray, 2017). Among the miRNAs mentioned above, many target the mRNAs of critical regulatory molecules, such as IRF4 (by miR-125b), C/EBP (by miR-142, miR-155, and let-7c), TLRs (by miR-146b), IFN-γ (by miR-21), and TNF-α (by miR-125b and miR-146a). Some can influence the activation level of signaling pathways directly or indirectly, such as the Janus kinase (JAK)/STAT/SOCS pathway (by miR-21, miR-142, miR-155, and miR-320a), Notch pathway (by miR-146a and miR-195), and NF-κB pathway (by let-7c, let-7f, miR-21, miR-146a, and miR-155). Because of the complexity of the regulatory networks of polarization, in future studies it will be important to focus on how those miRNAs cooperate with each other under specific in vivo circumstances.
4. miRNAs involved in communication between macrophages and tumor cells
Microvesicles and exosomes, which contain proteins, lipids, and nucleic acids, are important extracellular vesicles transported between cells and have significant roles in intercellular communication (el Andaloussi et al., 2013; Yao et al., 2018). The difference between microvesicles and exosomes is that microvesicles are derived from the plasma membrane and are normally larger than 100 nm in diameter, while exosomes are derived from multi-vesicular bodies (MVBs) or late endosomes, and are usually between 50 and 100 nm in diameter (Théry et al., 2009). Secretion of extracellular vesicles containing functional mRNAs and non-coding RNAs by cancer cells or normal cells has been proved to be a mechanism for intercellular transportation of genetic information by docking with the target cells and releasing their contents into them (Robbins and Morelli, 2014). miRNAs involved in communication between macrophages and tumor cells are summarized in Table 3 and Fig. 2.
Table 3.
miRNAs involved in communication between macrophages and tumor cells
| miRNA | Cell type | Target | Phenotype | Reference |
| miR-21 | Tumor cell→TAMa | EMT↑ | Hsieh et al., 2018 | |
| Tumor cell→TAM | TLR-7/8 | Tumor growth and metastasis↑ | Fabbri et al., 2012 | |
| Tumor cell→TAM | TLR-8 | miR-155 expression↑, tumor resistance to cisplatin↑ | Challagundla et al., 2015 | |
| miR-29a | Tumor cell→TAM | TLR-7/8 | Tumor growth and metastasis↑ | Fabbri et al., 2012 |
| miR-222 | Tumor cell→TAM | SOCS3 | M2 polarization↑ | Ying et al., 2016 |
| miR-203 | Tumor cell→TAM | M2 polarization↑, tumor metastasis↑ | Takano et al., 2017 | |
| miR-16 | Tumor cell→TAM | NF-κB pathway | M2 polarization↓ | Jang et al., 2013 |
| miR-1246 | Tumor cell→TAM | M2 polarization↑, tumor progression and metastasis↑ | Cooks et al., 2018 | |
| miR-940 | Tumor cell→TAM | M2 polarization↑, tumor growth and migration↑ | Chen et al., 2017 | |
| let-7a | Tumor cell→TAM | Insulin/AKT/mTOR | M2 polarization↑, tumor progression↑ | Park et al., 2019 |
| miR-301a | Tumor cell→TAM | PTEN | M2 polarization↑, tumor TNM stage↑, prognosis↓ | Wang XF et al., 2018 |
| miR-103a | Tumor cell→TAM | PTEN | M2 polarization↑, tumor angiogenesis↑ | Hsu et al., 2018 |
| miR-21 | TAM→tumor cellb | PTEN | Tumor cell apoptosis↓, resistance to cisplatin↑ | Zheng et al., 2017 |
| miR-223 | TAM→tumor cell | PTEN | Resistance to cisplatin↑, progression-free survival of patients↓ | Zhu et al., 2019 |
| TAM→tumor cell | MEF2C | Tumor cell invasion↑ | Yang et al., 2011 | |
| miR-365 | TAM→tumor cell | Resistance to gemcitabine↑ | Binenbaum et al., 2018 | |
| miR-21 | TAM→tumor cell | BRG1 | Tumor metastasis↑ | Lan et al., 2019 |
| miR-155 | TAM→tumor cell | BRG1 | Tumor metastasis↑ | Lan et al., 2019 |
| miR-125a/b | TAM→tumor cell | CD90 | Tumor cell proliferation and migration↓ | Wang et al., 2019 |
| miR-375 | Tumor cell | CCL2, TNS3, PXN | Recruitment of macrophages↑ | Frank et al., 2019 |
| miR-342 | Tumor cell | CXCL12 | Recruitment of macrophages↓, tumor angiogenesis↓ | Tian et al., 2018 |
| miR-26a | Tumor cell | M-CSF | Recruitment of macrophages↓, tumor growth↓ | Chai et al., 2015 |
| miR-125b | Tumor cell | CSF1, CX3CL1 | Recruitment of macrophages↓ | Batool et al., 2018 |
miRNA is transported from tumor cells to tumor-associated macrophages (TAMs);
miRNA is transported from TAMs to tumor cells. TLR: Toll-like receptor; SOCS3: suppressor of cytokine signaling 3; NF-κB: nuclear factor κB; AKT: protein kinase B; mTOR: mammalian target of rapamycin; PTEN: phosphatase and tensin homologue; MEF2C: myocyte enhancer factor 2; BRG1: ATP-dependent chromatin remodeler SMARCA4; CCL2: C-C motif chemokine ligand 2; TNS3: tensin-3; PXN: paxillin; CXCL12: CXC chemokine ligand 12; M-CSF: macrophage colony-stimulating factor; CX3CL1: (C-X3-C motif) ligand 1; EMT: epithelial-mesenchymal transition; TNM: tumor node metastasis
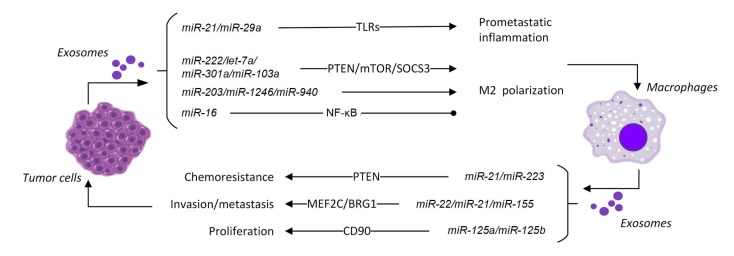
Fig. 2.
miRNAs involved in communication between macrophages and tumor cells
miRNAs can be transported between tumor cells and macrophages via microvesicles and exosomes. This will affect the functions of macrophages and the phenotypes of tumor cells
4.1. miRNAs transported from tumor cells to macrophages
miR-21 is engaged in the communication between tumor cells and macrophages in different ways. During the process of EMT, the transcription of miR-21 in tumor cells can be directly activated by Snail, a critical transcription factor of the EMT process. Subsequently, miR-21-abundant exosomes derived from tumor cells can be engulfed by macrophages (Hsieh et al., 2018). This differs from the traditional mechanisms in which miR-21 and miR-29a act as paracrine ligands secreted from tumor cells within exosomes and bind to TLR-7/8 in immune cells. These miRNAs can trigger an inflammatory response in macrophages mediated by TLRs and finally enhance tumor growth and metastasis (Fabbri et al., 2012). In addition, miR-21 from tumor-secreted exosomes is able to increase the expression of miR-155 in both macrophages and TAM-derived exosomes by binding with TLR-8. miR-155 can be transported back to tumor cells and enable them to become resistant to cisplatin by repressing telomerase inhibitor, telomeric repeat binding factor 1 (TERF1) (Challagundla et al., 2015), forming a regulatory circuit to enhance the drug resistance of tumor cells.
An important function of extracellular miRNAs derived from tumor cells is to regulate macrophage polarization. miR-222-3p is expressed in exosomes derived from epithelial ovarian cancer (EOC) and can induce M2 polarization. Studies of the mechanisms revealed that miR-222-3p can target SOCS3, and a low expression level of SOCS3 activates the STAT3 pathway (Ying et al., 2016). In metastatic colorectal cancer patients, miR-203 in serum exosomes is regarded as a poor independent prognostic factor related to distant metastasis. miR-203 in exosomes transported to macrophages induces the expression of M2 markers. In mouse models, colorectal cancer develops more liver metastatic sites when transfected with miR-203 (Takano et al., 2017). Epigallocatechin gallate (EGCG) is an anti-tumor molecule that can inhibit tumor growth and repress the infiltration of pro-tumoral M2 macrophages in tumor tissue. It is reported that miR-16 is upregulated in both tumor cells and tumor-derived exosomes when the tumor cells are treated with EGCG. It can then be delivered to macrophages and inhibit the NF-κB pathway, preventing the polarization of macrophages to the M2 phenotype (Jang et al., 2013). miR-1246-enriched exosomes are derived from p53-mutant cancer cells, and neighboring macrophages which take up these exosomes are reprogrammed to an anti-inflammatory state, promoting cancer progression and metastasis (Cooks et al., 2018). A hypoxic microenvironment is a common feature of solid tumors, which contributes to different characteristics of cancers, including chemoresistance, radioresistance, angiogenesis, and invasiveness (Wilson and Hay, 2011). The expression of miR-940 in exosomes secreted from EOC cells is highly induced by hypoxia. miR-940 in exosomes can be delivered into macrophages to promote M2 polarization, thus stimulating the growth and migration of EOC (Chen et al., 2017). Exosomes derived from hypoxic tumor cells are enriched with immunomodulatory molecules, including let-7a. The transfer of let-7a to macrophages suppresses the insulin-protein kinase B (AKT)-mammalian target of rapamycin (mTOR) pathway and induces the expression of TAM-associated genes, leading to host immunosuppression and tumor progression (Park et al., 2019). In pancreatic cancer patients, the TNM stage (a stage in a classification system that describes tumor development based on the extent of the primary tumor, the spread to lymph nodes, and the presence of distant metastasis (Amin et al., 2017)) and poor prognosis are positively associated with the level of circulating miR-301a-3p. Studies focusing on mechanisms revealed that miR-301a-3p is enriched in tumor-derived exosomes in hypoxic microenvironments and can induce M2 polarization of macrophages through the PTEN/phosphatidylinositol 3-kinase γ (PI3Kγ) pathway (Wang XF et al., 2018). A similar phenomenon is also observed in lung cancer, where miR-103a is transferred by extracellular vesicles produced by hypoxic lung cancer cells and leads to differentiation of macrophages into the M2 phenotype. Macrophages that receive miR-103a decrease the PTEN level and increase production of several cytokines that are immunosuppressive and pro-angiogenic (Hsu et al., 2018).
4.2. miRNAs transported from macrophages to tumor cells
Drug resistance is a major factor that influences the treatment and prognosis of cancer, and is thought to be one of the characteristics of cancer stem cells (Dean et al., 2005). Recent research has revealed that exosomal miRNAs derived from macrophages could be responsible for the formation or maintenance of drug resistance of cancer cells. Analysis of the miRNA expression profile of exosomes derived from M2 macrophages demonstrates that miR-21 is highly expressed, which is consistent with the knowledge that miR-21 is upregulated in M2 macrophages, as mentioned above. miR-21 in macrophage-derived exosomes can be delivered to gastric cancer cells and target PTEN to activate the PI3K/AKT pathway, thereby preventing cell apoptosis and making tumor cells resistant to cisplatin (Zheng et al., 2017). In EOC, miR-223 is abundant in macrophage-derived exosomes under hypoxia and, similar to the mechanism of miR-21, miR-223 can also be transferred to cancer cells and promote resistance to cisplatin via the PTEN-PI3K/AKT signaling pathway. As for EOC patients, a lower expression level of miR-223 is associated with longer progression-free survival (Zhu et al., 2019). Using a mouse model of pancreatic ductal adenocarcinoma (PDAC), Binenbaum et al. (2018) demonstrated that macrophage-derived miR-365 can be transferred to cancer cells and induce the activity of cytidine deaminase. This would inactivate gemcitabine, thus making cancer cells resistant to chemotherapy.
miRNAs in exosomes derived from macrophages also contribute to the invasion and metastasis of cancer cells. Using fluorescence labels, it was found that miR-223 can be transferred from IL-4-activated macrophages to co-cultured breast cancer cells through exosomes. Functional assays have discovered that miR-223 in cancer cells can downregulate myocyte enhancer factor 2 (MEF2C) and enhance the translocation of β-catenin from the cytosol into the nucleus to promote breast cancer invasion (Yang et al., 2011). Recently, it has been discovered that miR-21-5p and miR-155-5p are enriched in M2 macrophage-derived exosomes in colorectal cancer, and BRG1 (also known as ATP-dependent chromatin remodeler SMARCA4) has been shown to be the target of these two miRNAs. Downregulation of BRG1, in turn, promotes colon cancer metastasis through activating the Wnt/β-catenin pathway (Lan et al., 2019). Using a HCC cell line, Wang et al. (2019) found that miR-125a and miR-125b were downregulated in exosomes from TAMs, and transfection of miR-125a/125b effectively suppressed the proliferation and migration of HCC cells via downregulation of CD90.
4.3. miRNAs involved in recruitment of macrophages
Extracellular vesicles are not the only way for miRNAs to regulate the TME. miRNAs in tumor cells can also alter the expression of several cytokines responsible for the recruitment of macrophages in the tumor site. miR-375 is highly expressed in breast cancer cells, and can regulate the expression of CCL2 to facilitate the recruitment of macrophages into the tumor site and the development of a pro-tumoral microenvironment. miR-375 can also be released by apoptotic breast cancer cells and engulfed by macrophages in a non-exosome manner, and in macrophages, miR-375 can enhance migration and infiltration into tumor tissue through targeting tensin-3 (TNS3) and paxillin (PXN) (Frank et al., 2019).
In contrast to the function of miR-375, some other miRNAs are believed to be tumor-suppressive. In murine MS-K cell lines, increased expression of miR-342 interferes with angiogenesis and inhibits accumulation of macrophages. CXC chemokine ligand 12 (CXCL12) is thought to be the target of miR-342 responsible for its tumor-suppressing function (Tian et al., 2018). In HCC cell lines, ectopic expression of miR-26a effectively reduced the expression of M-CSF and infiltration of macrophages into tumors. The growth rate of tumors was also inhibited (Chai et al., 2015). In addition, the recruitment of macrophages could also be impaired by miR-125b, which targets the chemokines CSF1 and (C-X3-C motif) ligand 1 (CX3CL1) (Batool et al., 2018).
5. Macrophage-related miRNAs (MRMs) involved in clinical cancer applications
5.1. MRMs in clinical cancer diagnosis
Abnormal expression of miRNAs has been proved to be the significant feature of many types of cancer. More and more evidence supports the idea that miRNAs have the potential to help clinical diagnosis and prognosis analysis in cancer. For instance, our recent research revealed that serum miR-1915-3p and miR-455-3p could serve as diagnostic and predictive biomarkers for breast cancer (Guo et al., 2018). Among those biomarkers, MRMs are increasingly compelling in recent years. miR-130a expression is related to the metastasis of lung cancer, and its downregulation is a predictive factor of poor prognosis and the TNM stage. Mechanistically, miR-130a could suppress PPARγ protein expression and regulate the polarization of macrophages (Lin et al., 2015). Using in situ hybridization, the expression of miR-92a was evaluated in invasive breast cancer samples. The result revealed that the level of miR-92a was inversely correlated with the infiltration grade of macrophages in tumors and the stage of the tumor itself. The miR-92a level was also found to be an independent prognostic factor associated with recurrence-free survival of breast cancer patients (Nilsson et al., 2012). Upregulation of miR-195-5p in colorectal cancer cells can suppress IL-4 secretion and M2-like TAM polarization, and a low level of miR-195-5p is correlated with a shortened overall survival of colorectal cancer patients. In colorectal cancer patients, chemoresistance is associated with macrophage infiltration. miR-155 is downregulated in TAMs and results in increased production of IL-6, which can then induce chemoresistance in colorectal cancer cells by inhibiting the tumor suppressor miR-204-5p (Yin et al., 2017). miR-28-5p deficiency in HCC cells can upregulate the expression of IL-34, thereby promoting TAM infiltration into the tumor site. Patients with low miR-28-5p expression have a poor prognosis and a higher rate of recurrence (Zhou et al., 2016). In addition, miR-23a-3p is highly upregulated in exosomes derived from HCC cells treated with tunicamycin, and the expression of miR-23a-3p in tumor tissue is negatively associated with overall survival (Liu et al., 2019). Based on the TCGA database, a higher level of miR-1246 is associated with a worse overall prognosis for ovarian cancer patients. Research on the mechanism suggested that miR-1246 could be transferred to M2-type TAMs from tumor cells. These cells might participate in the induction of chemoresistance to paclitaxel. Consistent with this finding, treatments that combine chemotherapy and miR-1246 inhibitor can effectively reduce the tumor burden (Kanlikilicer et al., 2018). In a recent study, it was shown that miR-29a-3p and miR-21-5p are enriched in M2 macrophage-derived exosomes and can be shuttled into T lymphocytes to induce an increase in the Treg/Th17 ratio. The Treg/Th17 ratio in ovarian cancer tissue is positively correlated with histologic grade and poor prognosis of patients, which suggests that these miRNAs could possibly be the target of novel treatments for ovarian cancer (Zhou et al., 2018).
5.2. MRMs in cancer clinical therapy
Considering the pro-inflammatory function of miR-125b, the miRNA can be transfected along with wild-type p53 in lung cancer cells, delivered by dual CD44/epidermal growth factor receptor (EGFR)-targeted hyaluronic acid-based nanoparticles. The transfection of miR-125b successfully repolarizes the co-cultured macrophages into the M1 phenotype and effectively inhibits tumor growth, which reveals the potential function of miR-125b in treatment of lung cancer (Talekar et al., 2016). In a mouse breast cancermodel, the expression of miR-100 in TAMs is positively associated with the secretion of IL-1 receptor antagonist (IL-1ra), which can promote tumor cell stemness. Injection of miR-100 antagonist can effectively suppress the capacity of tumor invasion and metastasis (Wang W et al., 2018). miR-19a-3p is another miRNA with an anti-tumor function, and a mechanistic study revealed that ectopic expression of miR-19a-3p in TAMs could notably downregulate Fos-related antigen-1 (Fra-1), a pro-oncogene that can facilitate breast tumor progression and invasion. Intratumoral injection of miR-19a-3p can effectively impair the migration and invasion capacity of breast cancer, which makes miR-19a-3p a potential therapeutic molecule (Yang et al., 2014). There are some other anti-cancer drugs that exert their function through regulating miRNAs. Norcantharidin (NCTD) is a drug used for treatment of primary liver cancer. Injection of NCTD into tumor-bearing mice effectively induces the expression of miR-214 in macrophages, which promotes M1-polarization of TAMs and impairs tumor growth (Lu et al., 2014). Propofol is a widely used anesthetic, but also has the ability to inhibit the proliferation and invasiveness of HCC cells. Investigation of the mechanism revealed that propofol can stimulate the secretion of exosomes from TAMs which deliver miR-142-3p to recipient HCC cells and cause the suppression of tumor growth and invasion (Zhang et al., 2014). miRNAs involved in cancer prognosis and treatment are summarized in Table 4.
Table 4.
Macrophage-related miRNAs (MRMs) with potential value in prognosis and treatment of cancer
| miRNA | Cell type | Target | Clinical significance | Reference |
| miR-130a | TAM | PPARγ | TNM stage↓, tumor metastasis↓, prognosis↑ | Lin et al., 2015 |
| miR-92a | Tumor cell | Recurrence-free survival↑ | Nilsson et al., 2012 | |
| miR-155 | TAM | C/EBP-β | Tumor chemoresistance↓ | Yin et al., 2017 |
| miR-28 | Tumor cell | IL-34 | Prognosis↑ | Zhou et al., 2016 |
| miR-23a | Tumor cell→TAM | PTEN | PD-L1 expression↑, overall survival↓ | Liu et al., 2019 |
| miR-1246 | Tumor cell→TAM | Cav1 | Chemoresistance to paclitaxel↑, prognosis↓ | Kanlikilicer et al., 2018 |
| miR-29a | TAM→T cell | STAT3 | Treg/Th17 ratio↑, prognosis↓ | Zhou et al., 2018 |
| miR-21 | TAM→T cell | STAT3 | Treg/Th17 ratio↑, prognosis↓ | Zhou et al., 2018 |
| miR-125b | TAM | Tumor growth↓ | Talekar et al., 2016 | |
| miR-100 | TAM | mTOR pathway | Tumor cell stemness↑ | Wang W et al., 2018 |
| miR-19a | TAM | Fra-1 | Tumor cell migration and invasion↓ | Yang et al., 2014 |
| miR-214 | TAM | Tumor growth↓ | Lu et al., 2014 | |
| miR-142 | TAM→tumor cell | Tumor growth and invasion↓ | Zhang et al., 2014 |
TAM: tumor-associated macrophage; PPARγ: peroxisome proliferator-activated receptor γ; C/EBP-β: CCAAT/enhancer-binding protein β; IL-34: interleukin 34; PTEN: phosphatase and tensin homologue; Cav1: caveolin-1; STAT3: signal transducer and activator of transcription 3; mTOR: mammalian target of rapamycin; Fra-1: Fos-related antigen-1; TNM: tumor node metastasis; PD-L1: programmed death ligand 1; Treg: regulatory T cell
6. Conclusions
In their regulatory roles, a number of miRNAs participate in the process of macrophage generation, differentiation, and polarization. In solid tumors, macrophages are critical components in the TME, and their abilities to recognize tumor cells and regulate the immune system have significant impacts on tumor generation and progression. The activation or silencing of the expression of different miRNAs can have a marked effect on the phenotype of TAMs through various targets and signaling pathways, which influences other cell components and tumor progression.
In recent years, considering the significant effects of miRNAs on the TAM phenotype and tumor cell behavior, researchers have started to pay attention to the treatment of cancer through altering the expression and distribution pattern of MRMs in the TME. It is hoped that by analyzing the expression of MRMs, we can screen cancers in their early stages, make better clinical decisions, and evaluate prognosis after treatment. For now, most studies of MRMs are limited to individual miRNAs, but the regulatory network is extremely complex. Therefore, future studies should pay more attention to miRNA networks, which may be a more valuable approach for practical application in complicated clinical situations.
Footnotes
Project supported by the National Natural Science Foundation of China (Nos. 81972795 and 1672914)
Contributors: Chong CHEN and Jia-ming LIU wrote and edited the manuscript. Yun-ping LUO edited and checked the final version. All authors read and approved the final manuscript.
Compliance with ethics guidelines: Chong CHEN, Jia-ming LIU, and Yun-ping LUO declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Amin MB, Greene FL, Edge SB, et al. The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67(2):93–99. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 2.Baer C, Squadrito ML, Laoui D, et al. Suppression of microRNA activity amplifies IFN-γ-induced macrophage activation and promotes anti-tumour immunity. Nat Cell Biol. 2016;18(7):790–802. doi: 10.1038/ncb3371. [DOI] [PubMed] [Google Scholar]
- 3.Bai XZ, Zhang JL, Cao MY, et al. MicroRNA-146a protects against LPS-induced organ damage by inhibiting Notch1 in macrophage. Int Immunopharmacol. 2018;63:220–226. doi: 10.1016/j.intimp.2018.07.040. [DOI] [PubMed] [Google Scholar]
- 4.Bala S, Marcos M, Kodys K, et al. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor α (TNFα) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. 2011;286(2):1436–1444. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee S, Xie N, Cui HC, et al. MicroRNA let-7c regulates macrophage polarization. J Immunol. 2013;190(12):6542–6549. doi: 10.4049/jimmunol.1202496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee S, Cui HC, Xie N, et al. miR-125a-5p regulates differential activation of macrophages and inflammation. J Biol Chem. 2013;288(49):35428–35436. doi: 10.1074/jbc.M112.426866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batool A, Wang YQ, Hao XX, et al. A miR-125b/CSF1-CX3CL1/tumor-associated macrophage recruitment axis controls testicular germ cell tumor growth. Cell Death Dis. 2018;9(10):962. doi: 10.1038/s41419-018-1021-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binenbaum Y, Fridman E, Yaari Z, et al. Transfer of miRNA in macrophage-derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Res. 2018;78(18):5287–5299. doi: 10.1158/0008-5472.CAN-18-0124. [DOI] [PubMed] [Google Scholar]
- 9.Boldin MP, Taganov KD, Rao DS, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208(6):1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chafin CB, Regna NL, Caudell DL, et al. MicroRNA-let-7a promotes E2F-mediated cell proliferation and NFκB activation in vitro . Cell Mol Immunol. 2014;11(1):79–83. doi: 10.1038/cmi.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chai ZT, Zhu XD, Ao JY, et al. MicroRNA-26a suppresses recruitment of macrophages by down-regulating macrophage colony-stimulating factor expression through the PI3K/Akt pathway in hepatocellular carcinoma. J Hematol Oncol, 8:56. 2015 doi: 10.1186/s13045-015-0150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Challagundla KB, Wise PM, Neviani P, et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J Natl Cancer Inst. 2015;107(7):djv135. doi: 10.1093/jnci/djv135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhuri AA, So AYL, Sinha N, et al. MicroRNA-125b potentiates macrophage activation. J Immunol. 2011;187(10):5062–5068. doi: 10.4049/jimmunol.1102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Ying X, Wang XJ, et al. Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol Rep. 2017;38(1):522–528. doi: 10.3892/or.2017.5697. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Wang SX, Mu R, et al. Dysregulation of the miR-324-5p-CUEDC2 axis leads to macrophage dysfunction and is associated with colon cancer. Cell Rep. 2014;7(6):1982–1993. doi: 10.1016/j.celrep.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Cooks T, Pateras IS, Jenkins LM, et al. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun. 2018;9(1):771. doi: 10.1038/s41467-018-03224-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortez-Retamozo V, Etzrodt M, Newton A, et al. Origins of tumor-associated macrophages and neutrophils. Proc Natl Acad Sci USA. 2012;109(7):2491–2496. doi: 10.1073/pnas.1113744109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curtale G. MiRNAs at the crossroads between innate immunity and cancer: focus on macrophages. Cells. 2018;7(2):12. doi: 10.3390/cells7020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curtale G, Mirolo M, Renzi TA, et al. Negative regulation of Toll-like receptor 4 signaling by IL-10-dependent microRNA-146b. Proc Natl Acad Sci USA. 2013;110(28):11499–11504. doi: 10.1073/pnas.1219852110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5(4):275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 21.el Andaloussi S, Mäger I, Breakefield XO, et al. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12(5):347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 22.el Gazzar M, Church A, Liu TF, et al. MicroRNA-146a regulates both transcription silencing and translation disruption of TNF-α during TLR4-induced gene reprogramming. J Leukoc Biol. 2011;90(3):509–519. doi: 10.1189/jlb.0211074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Etzrodt M, Cortez-Retamozo V, Newton A, et al. Regulation of monocyte functional heterogeneity by miR-146a and Relb. Cell Rep. 2012;1(4):317–324. doi: 10.1016/j.celrep.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabbri M, Paone A, Calore F, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109(31):E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fortunato O, Borzi C, Milione M, et al. Circulating miR-320a promotes immunosuppressive macrophages M2 phenotype associated with lung cancer risk. Int J Cancer. 2019;144(11):2746–2761. doi: 10.1002/ijc.31988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank AC, Ebersberger S, Fink AF, et al. Apoptotic tumor cell-derived microRNA-375 uses CD36 to alter the tumor-associated macrophage phenotype. Nat Commun. 2019;10(1):1135. doi: 10.1038/s41467-019-08989-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franklin RA, Liao W, Sarkar A, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344(6186):921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Funahashi Y, Kato N, Masuda T, et al. miR-146a targeted to splenic macrophages prevents sepsis-induced multiple organ injury. Lab Invest. 2019;99(8):1130–1142. doi: 10.1038/s41374-019-0190-4. [DOI] [PubMed] [Google Scholar]
- 29.Geissmann F, Manz MG, Jung S, et al. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327(5966):656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghani S, Riemke P, Schönheit J, et al. Macrophage development from HSCs requires PU.1-coordinated microRNA expression. Blood. 2011;118(8):2275–2284. doi: 10.1182/blood-2011-02-335141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerriero JL. Macrophages: the road less traveled, changing anticancer therapy. Trends Mol Med. 2018;24(5):472–489. doi: 10.1016/j.molmed.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo J, Liu C, Wang W, et al. Identification of serum miR-1915-3p and miR-455-3p as biomarkers for breast cancer. PLoS ONE. 2018;13(7):e0200716. doi: 10.1371/journal.pone.0200716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ha MJ, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 34.He M, Xu ZQ, Ding T, et al. MicroRNA-155 regulates inflammatory cytokine production in tumor-associated macrophages via targeting C/EBPβ. Cell Mol Immunol. 2009;6(5):343–352. doi: 10.1038/cmi.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh CH, Tai SK, Yang MH. Snail-overexpressing cancer cells promote M2-like polarization of tumor-associated macrophages by delivering miR-21-abundant exosomes. Neoplasia. 2018;20(8):775–788. doi: 10.1016/j.neo.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu YL, Hung JY, Chang WA, et al. Hypoxic lung-cancer-derived extracellular vesicle microRNA-103a increases the oncogenic effects of macrophages by targeting PTEN. Mol Ther. 2018;26(2):568–581. doi: 10.1016/j.ymthe.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang F, Zhao JL, Wang L, et al. miR-148a-3p mediates notch signaling to promote the differentiation and M1 activation of macrophages. Front Immunol, 8:1327. 2017 doi: 10.3389/fimmu.2017.01327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ismail N, Wang YJ, Dakhlallah D, et al. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood. 2013;121(6):984–995. doi: 10.1182/blood-2011-08-374793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang JY, Lee JK, Jeon YK, et al. Exosome derived from epigallocatechin gallate treated breast cancer cells suppresses tumor growth by inhibiting tumor-associated macrophage infiltration and M2 polarization. BMC Cancer, 13:421. 2013 doi: 10.1186/1471-2407-13-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanlikilicer P, Bayraktar R, Denizli M, et al. Exosomal miRNA confers chemo resistance via targeting Cav1/p-gp/M2-type macrophage axis in ovarian cancer. EBioMedicine. 2018;38:100–112. doi: 10.1016/j.ebiom.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar M, Sahu SK, Kumar R, et al. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-κB pathway. Cell Host Microbe. 2015;17(3):345–356. doi: 10.1016/j.chom.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 42.Lan JQ, Sun L, Xu F, et al. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2019;79(1):146–158. doi: 10.1158/0008-5472.CAN-18-0014. [DOI] [PubMed] [Google Scholar]
- 43.Li D, Duan MY, Feng Y, et al. MiR-146a modulates macrophage polarization in systemic juvenile idiopathic arthritis by targeting INHBA. Mol Immunol. 2016;77:205–212. doi: 10.1016/j.molimm.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 44.Li L, Sun PF, Zhang CS, et al. MiR-98 suppresses the effects of tumor-associated macrophages on promoting migration and invasion of hepatocellular carcinoma cells by regulating IL-10. Biochimie. 2018;150:23–30. doi: 10.1016/j.biochi.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 45.Li N, Qin JF, Han X, et al. miR-21a negatively modulates tumor suppressor genes PTEN and miR-200c and further promotes the transformation of M2 macrophages. Immunol Cell Biol. 2018;96(1):68–80. doi: 10.1111/imcb.1016. [DOI] [PubMed] [Google Scholar]
- 46.Lin L, Lin HB, Wang L, et al. miR-130a regulates macrophage polarization and is associated with non-small cell lung cancer. Oncol Rep. 2015;34(6):3088–3096. doi: 10.3892/or.2015.4301. [DOI] [PubMed] [Google Scholar]
- 47.Lin XB, Wang SY, Sun M, et al. miR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J Hematol Oncol. 2019;12(1):20. doi: 10.1186/s13045-019-0708-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Liu JT, Fan LL, Yu HQ, et al. Endoplasmic reticulum stress causes liver cancer cells to release exosomal miR-23a-3p and up-regulate programmed death ligand 1 expression in macrophages. Hepatology. 2019;70(1):241–258. doi: 10.1002/hep.30607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu S, Gao Y, Huang XL, et al. Cantharidin exerts anti-hepatocellular carcinoma by miR-214 modulating macrophage polarization. Int J Biol Sci. 2014;10(4):415–425. doi: 10.7150/ijbs.8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep, 6:13. 2014 doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathsyaraja H, Thies K, Taffany DA, et al. CSF1-ETS2-induced microRNA in myeloid cells promote metastatic tumor growth. Oncogene. 2015;34(28):3651–3661. doi: 10.1038/onc.2014.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Movahedi K, Laoui D, Gysemans C, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010;70(14):5728–5739. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 53.Murray PJ. Macrophage polarization. Annu Rev Physiol. 2017;79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 54.Nahid MA, Pauley KM, Satoh M, et al. miR-146a is critical for endotoxin-induced tolerance: implication in innate immunity. J Biol Chem. 2009;284(50):34590–34599. doi: 10.1074/jbc.M109.056317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nazari-Jahantigh M, Wei YY, Noels H, et al. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J Clin Invest. 2012;122(11):4190–4202. doi: 10.1172/JCI61716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nilsson S, Moller C, Jirström K, et al. Downregulation of miR-92a is associated with aggressive breast cancer features and increased tumour macrophage infiltration. PLoS ONE. 2012;7(4):e36051. doi: 10.1371/journal.pone.0036051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41(1):49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O'Connell RM, Chaudhuri AA, Rao DS, et al. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106(17):7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ouimet M, Ediriweera HN, Gundra UM, et al. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J Clin Invest. 2015;125(12):4334–4348. doi: 10.1172/JCI81676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parayath NN, Parikh A, Amiji MM. Repolarization of tumor-associated macrophages in a genetically engineered nonsmall cell lung cancer model by intraperitoneal administration of hyaluronic acid-based nanoparticles encapsulating microRNA-125b. Nano Lett. 2018;18(6):3571–3579. doi: 10.1021/acs.nanolett.8b00689. [DOI] [PubMed] [Google Scholar]
- 61.Park JE, Dutta B, Tse SW, et al. Hypoxia-induced tumor exosomes promote M2-like macrophage polarization of infiltrating myeloid cells and microRNA-mediated metabolic shift. Oncogene. 2019;38(26):5158–5173. doi: 10.1038/s41388-019-0782-x. [DOI] [PubMed] [Google Scholar]
- 62.Price NL, Rotllan N, Zhang XB, et al. Specific disruption of abca1 targeting largely mimics the effects of miR-33 knockout on macrophage cholesterol efflux and atherosclerotic plaque development. Circ Res. 2019;124(6):874–880. doi: 10.1161/CIRCRESAHA.118.314415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ran D, Shia WJ, Lo MC, et al. RUNX1a enhances hematopoietic lineage commitment from human embryonic stem cells and inducible pluripotent stem cells. Blood. 2013;121(15):2882–2890. doi: 10.1182/blood-2012-08-451641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14(3):195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosa A, Ballarino M, Sorrentino A, et al. The interplay between the master transcription factor PU. Proc Natl Acad Sci USA. 2007;104(50):19849–19854. doi: 10.1073/pnas.0706963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 67.Schmid MC, Khan SQ, Kaneda MM, et al. Integrin CD11b activation drives anti-tumor innate immunity. Nat Commun. 2018;9(1):5379. doi: 10.1038/s41467-018-07387-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwarzenbach H, Nishida N, Calin GA, et al. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11(3):145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 69.Selimoglu-Buet D, Rivière J, Ghamlouch H, et al. A miR-150/TET3 pathway regulates the generation of mouse and human non-classical monocyte subset. Nat Commun. 2018;9(1):5455. doi: 10.1038/s41467-018-07801-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11(2):141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 71.Shidal C, Singh NP, Nagarkatti P, et al. MicroRNA-92 expression in CD133+ melanoma stem cells regulates immunosuppression in the tumor microenvironment via integrin-dependent activation of TGFβ. Cancer Res. 2019;79(14):3622–3635. doi: 10.1158/0008-5472.CAN-18-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sonda N, Simonato F, Peranzoni E, et al. miR-142-3p prevents macrophage differentiation during cancer-induced myelopoiesis. Immunity. 2013;38(6):1236–1249. doi: 10.1016/j.immuni.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 73.Squadrito ML, Pucci F, Magri L, et al. miR-511-3p modulates genetic programs of tumor-associated macrophages. Cell Rep. 2012;1(2):141–154. doi: 10.1016/j.celrep.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 74.Su SC, Zhao QY, He CH, et al. miR-142-5p and miR-130a-3p are regulated by IL-4 and IL-13 and control profibrogenic macrophage program. Nat Commun, 6:8523. 2015 doi: 10.1038/ncomms9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suarez-Carmona M, Lesage J, Cataldo D, et al. EMT and inflammation: inseparable actors of cancer progression. Mol Oncol. 2017;11(7):805–823. doi: 10.1002/1878-0261.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takano Y, Masuda T, Iinuma H, et al. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget. 2017;8(45):78598–78613. doi: 10.18632/oncotarget.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Talekar M, Trivedi M, Shah P, et al. Combination wt-p53 and microRNA-125b transfection in a genetically engineered lung cancer model using dual CD44/EGFR-targeting nanoparticles. Mol Ther. 2016;24(4):759–769. doi: 10.1038/mt.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 79.Tian YJ, Matsui S, Touma M, et al. MicroRNA-342 inhibits tumor growth via targeting chemokine CXCL12 involved in macrophages recruitment/activation. Genes Cells. 2018;23(12):1009–1022. doi: 10.1111/gtc.12650. [DOI] [PubMed] [Google Scholar]
- 80.Tili E, Michaille JJ, Cimino A, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179(8):5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 81.Wang P, Hou J, Lin L, et al. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol. 2010;185(10):6226–6233. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- 82.Wang P, Xu LJ, Qin JJ, et al. MicroRNA-155 inversely correlates with esophageal cancer progression through regulating tumor-associated macrophage FGF2 expression. Biochem Biophys Res Commun. 2018;503(2):452–458. doi: 10.1016/j.bbrc.2018.04.094. [DOI] [PubMed] [Google Scholar]
- 83.Wang W, Liu Y, Guo J, et al. miR-100 maintains phenotype of tumor-associated macrophages by targeting mTOR to promote tumor metastasis via Stat5a/IL-1ra pathway in mouse breast cancer. Oncogenesis. 2018;7(12):97. doi: 10.1038/s41389-018-0106-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang XF, Luo GT, Zhang KD, et al. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kγ to promote pancreatic cancer metastasis. Cancer Res. 2018;78(16):4586–4598. doi: 10.1158/0008-5472.CAN-17-3841. [DOI] [PubMed] [Google Scholar]
- 85.Wang YF, Wang BY, Xiao S, et al. miR-125a/b inhibits tumor-associated macrophages mediated in cancer stem cells of hepatocellular carcinoma by targeting CD90. J Cell Biochem. 2019;120(3):3046–3055. doi: 10.1002/jcb.27436. [DOI] [PubMed] [Google Scholar]
- 86.Wang Z, Brandt S, Medeiros A, et al. MicroRNA 21 is a homeostatic regulator of macrophage polarization and prevents prostaglandin E2-mediated M2 generation. PLoS ONE. 2015;10(2):e0115855. doi: 10.1371/journal.pone.0115855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wei YY, Schober A. MicroRNA regulation of macrophages in human pathologies. Cell Mol Life Sci. 2016;73(18):3473–3495. doi: 10.1007/s00018-016-2254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.West MA, Heagy W. Endotoxin tolerance: a review. Crit Care Med. 2002;30(1):S64–S73. [PubMed] [Google Scholar]
- 89.Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11(6):393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 90.Xi JJ, Huang Q, Wang L, et al. miR-21 depletion in macrophages promotes tumoricidal polarization and enhances PD-1 immunotherapy. Oncogene. 2018;37(23):3151–3165. doi: 10.1038/s41388-018-0178-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang J, Zhang Z, Chen C, et al. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene. 2014;33(23):3014–3023. doi: 10.1038/onc.2013.258. [DOI] [PubMed] [Google Scholar]
- 92.Yang M, Chen JQ, Su F, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer, 10:117. 2011 doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yao ZY, Chen WB, Shao SS, et al. Role of exosome-associated microRNA in diagnostic and therapeutic applications to metabolic disorders. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2018;19(3):183–198. doi: 10.1631/jzus.B1600490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ye JS, Guo RW, Shi YK, et al. miR-155 regulated inflammation response by the SOCS1-STAT3-PDCD4 axis in atherogenesis. Mediators Inflamm, 2016:8060182. 2016 doi: 10.1155/2016/8060182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yin Y, Yao SR, Hu YL, et al. The immune-microenvironment confers chemoresistance of colorectal cancer through macrophage-derived IL6. Clin Cancer Res. 2017;23(23):7375–7387. doi: 10.1158/1078-0432.CCR-17-1283. [DOI] [PubMed] [Google Scholar]
- 96.Ying X, Wu QF, Wu XL, et al. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget. 2016;7(28):43076–43087. doi: 10.18632/oncotarget.9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yona S, Kim KW, Wolf Y, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38(1):79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang J, Shan WF, Jin TT, et al. Propofol exerts anti-hepatocellular carcinoma by microvesicle-mediated transfer of miR-142-3p from macrophage to cancer cells. J Transl Med, 12:279. 2014 doi: 10.1186/s12967-014-0279-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang W, Liu H, Liu W, et al. Polycomb-mediated loss of microRNA let-7c determines inflammatory macrophage polarization via PAK1-dependent NF-κB pathway. Cell Death Differ. 2015;22(2):287–297. doi: 10.1038/cdd.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao HM, Wang XS, Yi P, et al. KSRP specifies monocytic and granulocytic differentiation through regulating miR-129 biogenesis and RUNX1 expression. Nat Commun. 2017;8(1):1428. doi: 10.1038/s41467-017-01425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhao JL, Huang F, He F, et al. Forced activation of Notch in macrophages represses tumor growth by upregulating miR-125a and disabling tumor-associated macrophages. Cancer Res. 2016;76(6):1403–1415. doi: 10.1158/0008-5472.CAN-15-2019. [DOI] [PubMed] [Google Scholar]
- 102.Zhao Y, Zou WL, Du JF, et al. The origins and homeostasis of monocytes and tissue-resident macrophages in physiological situation. J Cell Physiol. 2018;233(10):6425–6439. doi: 10.1002/jcp.26461. [DOI] [PubMed] [Google Scholar]
- 103.Zheng PM, Chen L, Yuan XL, et al. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J Exp Clin Cancer Res. 2017;36(1):53. doi: 10.1186/s13046-017-0528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou HB, Huang XF, Cui HJ, et al. miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells. Blood. 2010;116(26):5885–5894. doi: 10.1182/blood-2010-04-280156. [DOI] [PubMed] [Google Scholar]
- 105.Zhou JR, Li XD, Wu XL, et al. Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol Res. 2018;6(12):1578–1592. doi: 10.1158/2326-6066.CIR-17-0479. [DOI] [PubMed] [Google Scholar]
- 106.Zhou SL, Hu ZQ, Zhou ZJ, et al. miR-28-5p-IL-34-macrophage feedback loop modulates hepatocellular carcinoma metastasis. Hepatology. 2016;63(5):1560–1575. doi: 10.1002/hep.28445. [DOI] [PubMed] [Google Scholar]
- 107.Zhu XL, Shen HL, Yin XM, et al. Macrophages derived exosomes deliver miR-223 to epithelial ovarian cancer cells to elicit a chemoresistant phenotype. J Exp Clin Cancer Res. 2019;38(1):81. doi: 10.1186/s13046-019-1095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]