Multiple myeloma (MM) accounts for over twenty percent of hematological cancer-related death worldwide. Long noncoding RNA (lncRNA) H19 is associated with multiple tumorigenesis and is increased in MM, but the underlying mechanism of H19 in MM is unclear. In this study, the expression of H19, microRNA 152-3p (miR-152-3p), and BRD4 in MM patients was evaluated by quantitative real-time PCR (qRT-PCR) and Western blotting.
Keywords: multiple myeloma, lncRNA-H19, miR-152-3p, BRD4, cell proliferation, apoptosis
ABSTRACT
Multiple myeloma (MM) accounts for over twenty percent of hematological cancer-related death worldwide. Long noncoding RNA (lncRNA) H19 is associated with multiple tumorigenesis and is increased in MM, but the underlying mechanism of H19 in MM is unclear. In this study, the expression of H19, microRNA 152-3p (miR-152-3p), and BRD4 in MM patients was evaluated by quantitative real-time PCR (qRT-PCR) and Western blotting. Colony formation and flow cytometry analysis were used to determine the effects of H19 and miR-152-3p on MM cell proliferation, apoptosis, and cell cycle. A luciferase reporter assay was conducted to confirm the interaction among H19, miR-152-3p, and BRD4. A nude mouse xenograft model was established, and the cell proliferation and apoptosis were evaluated by immunohistochemistry (IHC) staining and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling assay. We found that levels of H19 and BRD4 were upregulated and the expression of miR-152-3p was downregulated in MM patients. Dual luciferase reporter assay showed H19 targeted miR-152-3p to promote BRD4 expression. Knockdown of H19 repressed proliferation and enhanced apoptosis and cell cycle G1 arrest by upregulating miR-152-3p in MM cells. Furthermore, H19 knockdown suppressed the growth of xenograft tumor, reduced Ki-67 and BRD4 levels, and increased cell apoptosis in xenograft tumor tissues. Taking these results together, H19 knockdown suppresses MM tumorigenesis via inhibiting BRD4-mediated cell proliferation through targeting miR-152-3p, implying that H19 is a promising biomarker and drug target for MM.
INTRODUCTION
Multiple myeloma (MM), which is a result of bone marrow-resident plasma cells (PCs) malignantly expanding, is a deadly hematopoietic disease with no cure (1, 2). It has been estimated that over thirty thousand new MM cases were diagnosed in the United States in 2017, causing around twelve thousand deaths (3). With the development of society and the advancement of science and technology, the rate of median survival of MM has been significantly improved, with an overall 5-year survival rate of above 50%, which is even higher in younger patients (4, 5). However, treatments of MM always induce various adverse effects, including neutropenia, myelosuppression, thrombocytopenia, serious infection, and drug resistance, which need to be solved immediately (6). Thus, the improvement of therapeutic methods for MM, including new agents or innovative combination therapies, is urgent.
The bromodomain and extraterminal domain (BET) family is composed of BRD2, BRD3, BRD4, and BRDT (7). By regulating chromatin modification and genetic transcription, these proteins play important roles in cellular processes, such as cell proliferation, cell cycle, and cell apoptosis (8, 9). The BRDs exert their functions by recognizing and binding to the N-acetylation of lysine residues on the histone tail, releasing DNA sequences, recruiting transcription factors, and thereby enhancing gene expression (10). It was reported that BRD4 was indispensable for the expression of “tumor-driving” oncogenes such as the c-Myc gene during the development of hematologic cancers, including MM, acute lymphoblastic leukemia, and acute myelogenous leukemia (11), implying that BRD4 is a promising target for cancer therapy. Previous studies proved that BRD4 was overexpressed in the samples derived from MM patients. BRD4 inhibitor showed promising therapeutic effects in MM, reflecting inhibiting the proliferation of MM cells and promoting cell apoptosis (12). Nevertheless, the role and molecular mechanisms of BRD4 in the progression of MM are still largely unknown.
MicroRNAs (miRNAs) are a group of small noncoding RNAs of 19 to 25 nucleotides in length (13). These molecules mediate mRNA degradation or translational inhibition by directly binding to the complementary sequences within the 3′ untranslated region (3′-UTR) of these mRNAs (14). A variety of cellular and physiological processes, including cell proliferation, differentiation, apoptosis, and tumor metastasis, are regulated by posttranscriptional processing of miRNAs (15). miRNAs are deregulated in the vast majority of tumors, suggesting their important roles in the development of tumors (16). miRNA 152-3p (miR-152-3p) is a member of the miR-148/152 family (miR-148a, miR-148b, and miR-152-3p) and has a relatively conservative sequence (17). Research on miR-152-3p focused mainly on its roles in tumor suppression (18). It was reported that the expression of miR-152-3p was greatly suppressed in MM, and its overexpression could result in suppression of cell cycle arrest and apoptosis in MM cells (19). However, the downstream targets of miR-152-3p have not been well identified yet.
The transcripts of long noncoding RNAs (lncRNAs) are over 200 nucleotides in length and are often abnormally expressed in human cancers (20). lncRNAs have been found to play critical roles during tumor initiation and progression, including survival, proliferation, invasion, metastasis, and angiogenesis (21). H19, a 2.3-kb capped, spliced, and polyadenylated lncRNA, is the first identified lncRNA derived from the H19/igf2 gene cluster (22). Evidence indicated that H19 was overexpressed in MM and acted as a biomarker for cancer monitoring (23). By using bioinformatics methods, we found that miR-152-3p bound with H19 and BRD4 mRNA. Thus, we speculated that H19 targets miR-152-3p and promotes tumorigenesis of MM by activating the BRD4 signaling pathway.
In this study, our findings demonstrated that H19 could bind to miR-152-3p and could decrease the expression of miR-152-3p, leading to activation of BRD4 signaling and thereby promoting the proliferation of MM cells. These results provide new potential implications for prognosis and therapeutic interventions in the treatment of MM.
RESULTS
H19 and BRD4 were upregulated in MM tissues, along with miR-152-3p downregulation.
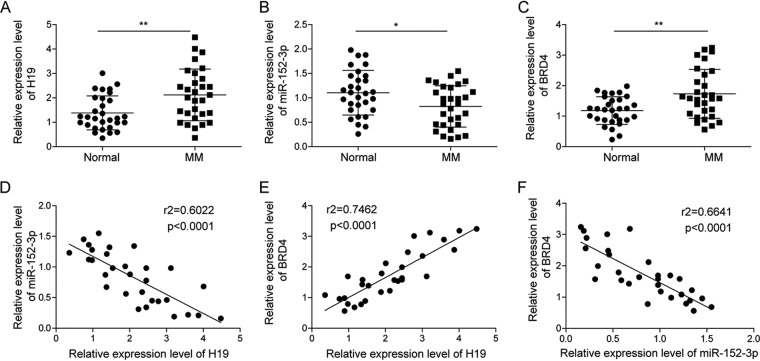
To study the relationship of H19, BRD4, and miR-152-3p to the progression of MM, we tested the expression of H19, BRD4, and miR-152-3p in our collection of bone marrow samples from 30 healthy controls and MM patients. The results showed that compared to the healthy controls, H19 and BRD4 were significantly upregulated in samples from patients with MM (Fig. 1A and C), while the expression of miR-152-3p was decreased in MM patients (Fig. 1B). We then analyzed the relationship of H19/miR-152-3p, H9/BRD4, and miR-152-3p/BRD4 using Spearman’s correlation analysis. We found that the level of H19 was positively correlated with BRD4, while H19 was negatively correlated with miR-152-3p and miR-152-3p was negatively correlated with BRD4 (Fig. 1D to F). These results suggested that H19 increased the expression of BRD4 by regulating miR-152-3p. In addition, the correlation analysis between H19 expression and clinicopathological features in MM patients was performed, and the results demonstrated that H19 expression was not related to gender, age, ISS stage, and renal insufficiency in MM patients but was associated with bone diseases (P = 0.044) (data not shown).
FIG 1.
Expression level of H19, miR-152-3p, and BRD4 in MM tissues. Expression of H19 (A), miR-152-3p (B), and BRD4 (C) was detected by qRT-PCR in plasma of 30 patients with or without MM. (D) Spearman’s correlation analysis between H19 and miR-152-3p expression level. (E) Spearman’s correlation analysis between H19 and BRD4 expression level. (F) Spearman’s correlation analysis between miR-152-3p and BRD4 expression level. Each data point represents fold change for real-time PCR. *, P < 0.05; **, P < 0.01.
H19 promoted the expression of BRD4 by targeting miR-152-3p.
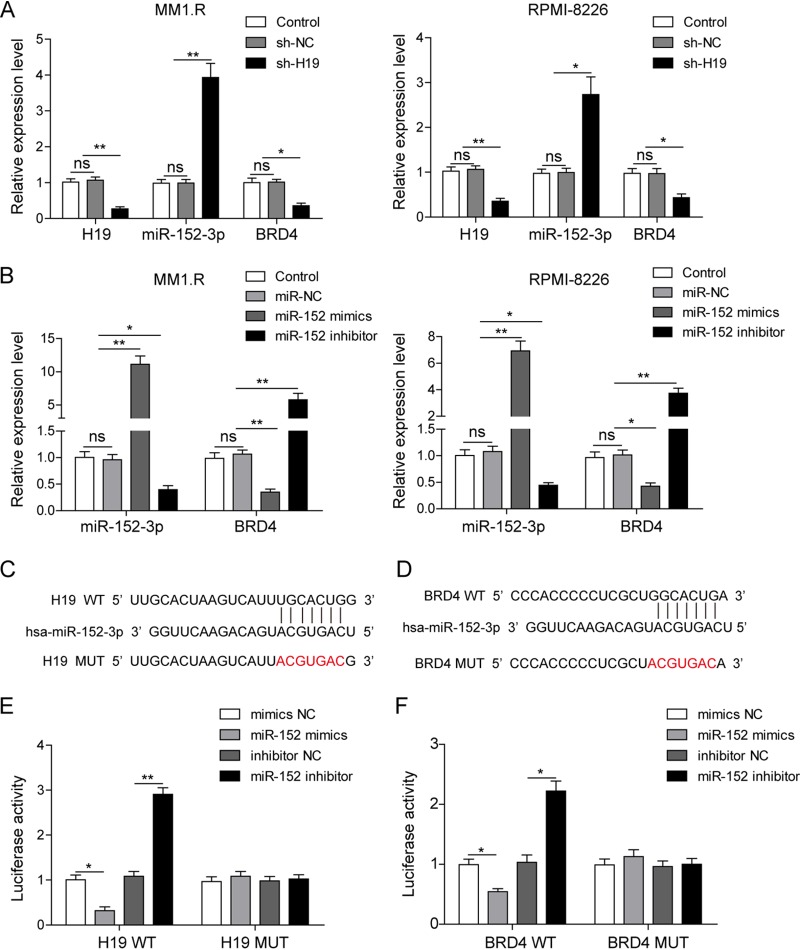
In order to study the relationship of H19, BRD4, and miR-152-3p to the progression of MM, H19 was inhibited in MM1.R and RPMI-8226 cells by transfection with sh-H19, and then the expression of H19, BRD4, and miR-152-3p was detected by qRT-PCR. As confirmed by qRT-PCR, the expression of H19 was significantly silenced in cells transfected with H19 short hairpin RNA (shRNA) (Fig. 2A). The expression of miR-152-3p was significantly increased, and the expression of BRD4 was greatly suppressed by knockdown of H19 (Fig. 2A). These results indicated that H19 could suppress the expression of miR-152-3p and activate the expression of BRD4.
FIG 2.
H19 promoted the expression of BRD4 by binding to miR-152-3p. (A) The expression levels of H19, miR-152-3p, and BRD4 were detected by qRT-PCR in MM1.R and RPMI-8226 cells transfected with scramble vector (shNC) or sh-H19 vector. (B) The expression of miR-152-3p and BRD4 was detected by qRT-PCR in MM1.R and RPMI-8226 cells transfected with miR-152-3p mimics and inhibitor. The potential binding site in the sequences of H19 with miR-152-3p (C) and miR-152-3p with the 3′-UTR of BRD4 (D) was analyzed through bioinformatics. A dual luciferase activity assay was implemented to confirm the binding of H19/miR-152-3p (E) and miR-152-3p/BRD4 mRNA (F). All data are representative of 3 independent experiments. *, P < 0.05; **, P < 0.01.
To prove the relationship between miR-152-3p and BRD4, miR-152-3p was overexpressed or inhibited in MM1.R and RPMI-8226 cells by transfection with miR-152-3p mimics or miR-152-3p inhibitor. As confirmed by qRT-PCR, the expression of miR-152-3p was significantly upregulated by transfection of miR-152-3p mimics and was markedly decreased by transfection with miR-152-3p inhibitor (Fig. 2B). Moreover, the expression of BRD4 was significantly changed on the reverse trend of miR-152-3p expression (Fig. 2B). As predicted by bioinformatics, miR-152-3p could bind with H19 and the 3′-UTR of BRD4 mRNA. In order to confirm this interaction, we introduced two point mutations in the binding sequences of H19 or 3′-UTR of BRD4 mRNA (Fig. 2C and D). Dual luciferase reporter assays then were performed to further corroborate the specific interaction among H19, miR-152-3p, and 3′-UTR of BRD4. It was shown that cotransfection of WT-H19 and miR-152-3p mimics distinctly diminished the relative luciferase activity compared to mimics of the negative-control (NC) group, while the relative luciferase activity showed no significant change with the cotransfection of MUT-H19 and miR-152-3p mimics or miR-152-3p inhibitor (Fig. 2E). Cotransfection of WT-3′-UTR of BRD4 and miR-152-3p mimics significantly reduced the relative luciferase activity compared to that of the inhibitor NC group, while the relative luciferase activity showed no significant difference with the cotransfection of MUT-3′-UTR of BRD4 and miR-152-3p mimics or miR-152-3p inhibitor (Fig. 2F). These results suggested that miR-152-3p was regulated by H19 and targeted BRD4.
H19 knockdown inhibited MM cell proliferation and enhanced cell apoptosis by stimulating the expression of miR-152-3p.
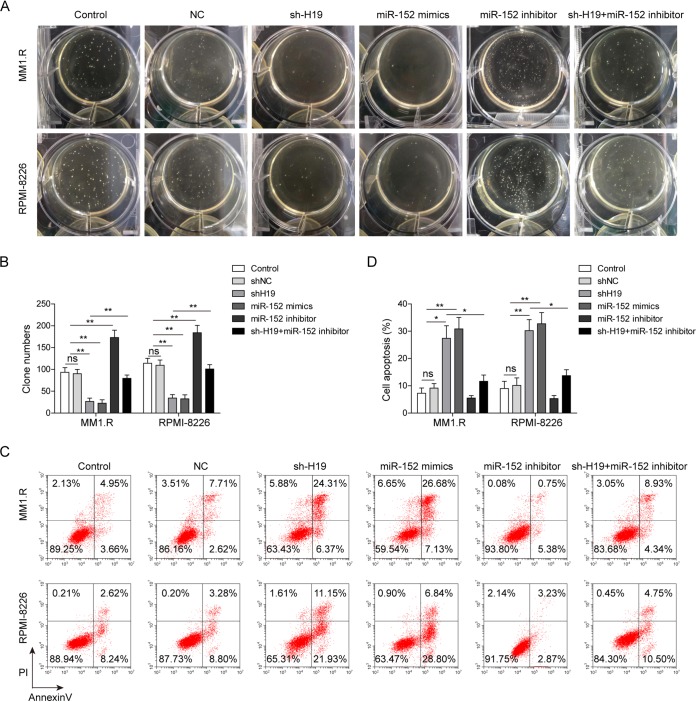
To further study the effects of H19 and miR-152-3p on cellular biological function, we knocked down the expression of H19 by transfection with sh-H19 in MM1.S and RPMI-8226 cells and inhibited miR-152-3p by cotransfection with miR-152-3p inhibitor. The soft-agar colony formation assay indicated that the proliferation of MM cells was significantly inhibited by H19 knockdown and miR-152-3p upregulation, while the inhibition of miR-152-3p by miR-152-3p inhibitor remarkably promoted the proliferation of MM cells and reversed the effect induced by H19 knockdown (Fig. 3A and B). Additionally, overexpression of H19 significantly increased the colony numbers of MM cells, and overexpression of miR-152-3p remarkably reversed the effects (data not shown), indicating that H19 promoted MM proliferation by regulating miR-152-3p. Cell apoptotic assay indicated that the process of cell apoptosis was significantly promoted by H19 knockdown or miR-152-3p overexpression (Fig. 3C and D), and the effects of H19 knockdown were also alleviated by miR-152-3p inhibitor (Fig. 3C and D). Taken together, these results indicated that the cellular biological functions of H19 were mainly mediated by the suppression of miR-152-3p.
FIG 3.
H19 knockdown inhibited MM cell proliferation and promoted cell apoptosis via stimulating the expression of miR-152-3p. The soft-agar colony formation assay was performed (A), and cell numbers were counted in MM1.R and RPMI-8226 cells transfected with sh-H19, miR-152-3p mimics, or miR-152-3p inhibitor (B). Cell apoptosis was analyzed by flow cytometry (C), and the proportions of apoptotic cells are shown for MM1.R and RPMI-8226 cells transfected with sh-H19, miR-152-3p mimics, or miR-152-3p inhibitor (D). All data are representative of 3 independent experiments. *, P < 0.05; **, P < 0.01.
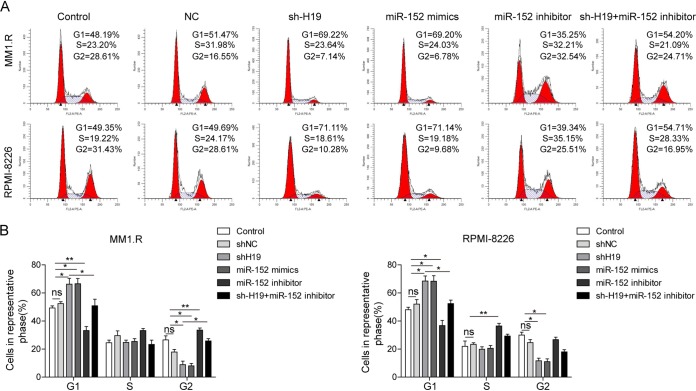
H19 knockdown induced cell phase G1 arrest by upregulating miR-152-3p in MM cells.
We measured the effect of H19 knockdown on cell cycle arrest. H19 shRNA-transfected MM cells were cotransfected with miR-152-3p mimics or miR-152-3p inhibitor, and then these cells were subjected to propidium iodide (PI) staining. As shown in Fig. 4, cells transfected with shH19 and cells transfected with miR-152-3p mimics exhibited higher distribution of G1 phase, while cells transfected with miR-152-3p inhibitor showed higher S/G2 cell numbers. Furthermore, the cell phase G1 arrest induced by H19 knockdown in MM cells also could be significantly alleviated by miR-152-3p inhibitor (Fig. 4). Together, these results indicated that the function of H19 on regulation of the cell cycle was also closely related to miR-152-3p.
FIG 4.
H19 knockdown induces cell phase G1 arrest by upregulating miR-152-3p in MM cells. The cells were stained with PI and analyzed by flow cytometry (A) to determine cell cycle distribution in MM1.R and RPMI-8226 cells transfected with sh-H19, miR-152-3p mimics, or miR-152-3p inhibitor (B). All data are representative of 3 independent experiments. *, P < 0.05; **, P < 0.01.
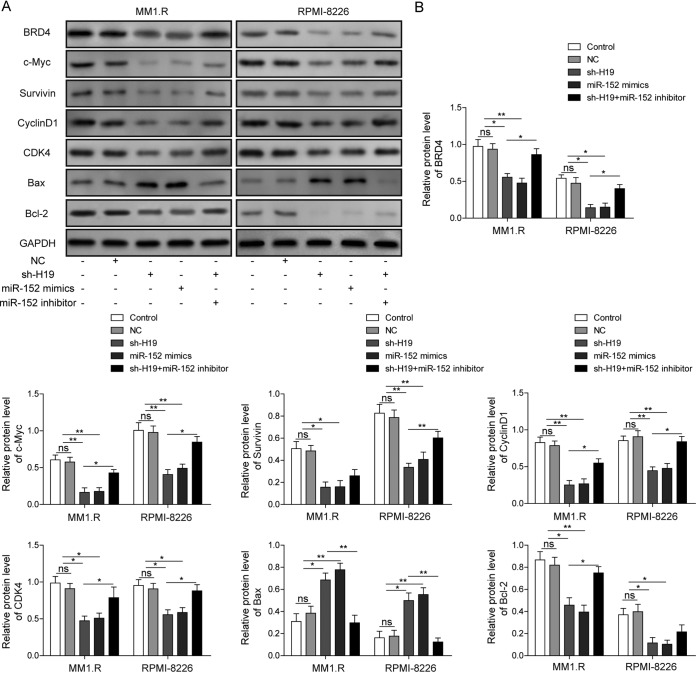
H19 knockdown suppressed the expression of proliferation-associated genes by inhibiting BRD4 signaling mediated by miR-152-3p upregulation.
As we proved that H19 knockdown could enhance cell apoptosis and suppress the expression of BRD4, we next detected the expression of proliferation-associated genes and the BRD4 signaling pathway in MM cells after H19 knockdown. It was observed that the expression of BRD4 was markedly decreased after H19 silencing or transfection of miR-152-3p mimics (Fig. 5). As we expected, the expression of proliferation-associated genes, including the c-Myc, survivin, cyclin D1, and cyclin-dependent kinase 4 (CDK4) genes, was significantly decreased after H19 silencing or transfection of miR-152-3p mimics, and such effects could be markedly rescued by the transfection of miR-152-3p inhibitor (Fig. 5). Moreover, the expression of the proapoptotic Bax gene was greatly upregulated and the antiapoptotic Bcl-2 gene was significantly suppressed by H19 silencing or overexpression of miR-152-3p (Fig. 5). Consistent with these results, this effect could be markedly inhibited by the transfection of miR-152-3p inhibitor (Fig. 5). Taken together, these results indicated that H9 knockdown suppressed the proliferation and enhanced the apoptosis of MM cells mainly through promoting the expression of miR-152-3p, thereby activating BRD4 signaling.
FIG 5.
H19 knockdown suppressed the expression of proliferation-associated gene by inhibiting miR-152-3p-mediated BRD4 signaling pathway. (A) The expression of BRD4, c-Myc, survivin, cyclin D1, CDK4, Bax, and Bcl-2 was detected by Western blotting in MM1.R and RPMI-8226 cells transfected with sh-H19, miR-152-3p mimics, or miR-152-3p inhibitor. (B) Quantitative analysis of Western blotting. The levels of their expression were normalized to that of GAPDH. All data are representative of 3 independent experiments. *, P < 0.05; **, P < 0.01.
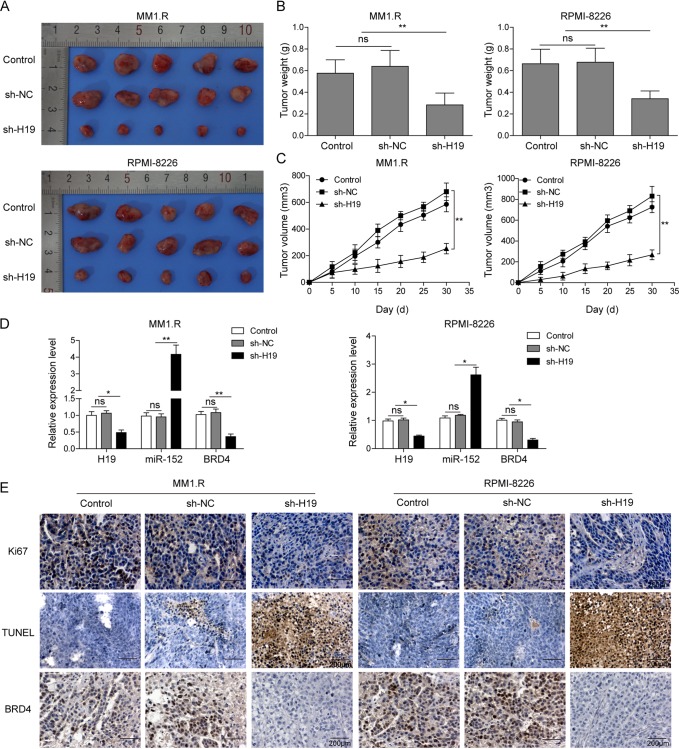
H19 knockdown repressed tumor growth by upregulating expression of miR-152-3p in vivo.
The effect of H19 on tumor progression then was tested in vivo using two xenograft mouse models generated by MM1.R and RPMI-8226 lines. At the end of the experiment, obvious differences in tumor size between mice injected with shNC or H19 knockdown MM cells were observed (Fig. 6A). The weight of tumors was much lighter, while the mean volume of the tumors was significantly lower in H19 knockdown mice than in the tumors in NC mice (Fig. 6B and C). Consistent with the results gathered from the in vitro experiments, the expression of BRD4 was greatly inhibited and the expression of miR-152-3p was significantly enhanced in vivo after H19 knockdown (Fig. 6D). Furthermore, the expression of cell proliferation marker Ki-67 and BRD4 in xenograft tumors was measured by immunohistochemistry (IHC) staining. As shown in Fig. 6E, the expression of Ki67 and BRD4 was largely suppressed by H19 silence in vivo. Consistent with this, the rate of apoptotic cells in these H19 silenced tumors was markedly increased as detected by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay (Fig. 6E). These results fully proved that H19 played an important role in the progression of MM, and these effects were mediated by the BRD4 signaling pathway.
FIG 6.
H19 knockdown inhibited tumor growth by upregulating the expression of miR-152-3p in vivo. BALB/c nude mice were subcutaneously injected with 5 × 106 MM1.R or RPMI-8226 cells stably transfected with scramble vector (shNC) or sh-H19 vector. The tumor was excised and weighed after 30 days. (A) The tumor specimen figure of nude mice. (B) The tumors were weighed. (C) Tumor volume was monitored every 5 days. (D) The expression levels of H19, miR-152-3p, and BRD4 in xenograft tumor tissues were detected by qRT-PCR. (E) The expression of Ki67 and BRD4 and apoptosis in xenograft tumor tissues was analyzed by IHC staining and TUNEL assay. All data are representative of 3 independent experiments. *, P < 0.05; **, P < 0.01.
DISCUSSION
MM is a type of cancer resulting from malignant expansion of plasma cells in bone marrow, leading to elevated levels of inflammatory cytokines, such as tumor necrosis factor alpha and the excessive formation of osteoclasts, along with bone destruction and loss (24). Despite extensive studies having been conducted to reveal accurate molecular mechanisms of this cancer, the treatment of MM is still a huge challenge (25). Our study essentially addresses a novel role for lncRNA H19 and its inverse relationship with miR-152-3p in MM development. We found that miR-152-3p was downregulated in the majority of MM patients, which was negatively related to H19 and BRD4. Knockdown of H19 inhibited the proliferation of MM cells, induced G1 cell cycle arrest, and enhanced cell apoptosis. Overexpression of miR-152-3p reversed the effects of H19 inhibition by targeting 3′-UTR of BRD4 in MM cell lines. miR-152-3p mimics induced apoptosis in MM cells. Furthermore, knockdown of H19 in xenograft mouse MM models led to attenuated tumor growth in vivo, providing evidence for the role of H19 in the regulation of MM.
H19 is a long noncoding RNA that has been reported to play central roles in the initiation of cancer and promotes the progression and invasion of various tumors (26). A previous study proved that the expression of H19 was significantly elevated in the serum derived from MM patients, suggesting that H19 could serve as a tumor biomarker for MM detection (23). It was also demonstrated that overexpression of H19 could induce the drug resistance in MM by targeting MCL-1 via miR-29b-3p (27). In addition, other studies reported that knockdown of H19 inhibited MM cell growth via inhibiting NF-κB signaling (28). Furthermore, H19 is primarily expressed in the embryo, where cells are proliferating very rapidly, and previous studies demonstrated that H19 could enhance cell proliferation in bladder cancer cells, gastric cancer cells, and fibroblasts (29–31), suggesting that H19 played a vital role in cell proliferation and promoted many kinds of cell proliferation. By analyzing patient samples, we found that the expression of H19 was significantly elevated in the majority of MM patients. H19 knockdown significantly inhibited the proliferation of MM cells, promoted cell apoptosis, and induced G1 cell cycle arrest, which was confirmed in nude mouse xenograft experiments. These results suggested that H19 plays a key role in MM.
miR-152-3p is a member of the miR-148/152 family, 21 nucleotides in length, and is significantly suppressed in MM patients (32). A previous study found that miR-152-3p inhibited glioma cell proliferation and invasion activities by targeting DNMT1 (33). However, the role of miR-152-3p in MM is still unclear. miRNAs exert their gene-silencing function mainly by binding to the 3′-UTR of mRNAs. Our results suggested that H19 could directly target miR-152-3p and repressed the expression of miR-152-3p. Many reports about the mechanism of lncRNA sponge showed that the expression of miRNA was negatively regulated after overexpression or knockdown of lncRNA (34–36), but the specific mechanism of reducing the level of miRNA by the lncRNA sponge was not described in detail. However, a previous study demonstrated that HSURs, as a noncoding RNA, could interact with miR-27 in virally transformed T cells and dramatically decreased the level of miR-27 by directing degradation of mature miR-27 in a binding-dependent manner (37). Therefore, in MM cells, H19 may also reduce the level of miR-152-3p by directing the degradation of miR-152-3p in a binding-dependent manner. Additionally, overexpression of miR-152-3p in MM cells markedly inhibited cell proliferation, enhanced cell apoptosis, and resulted in G1 phase cell cycle arrest. Altogether, these results suggested that H19 regulated cell cycle, proliferation, and apoptosis by targeting miR-152-3p in MM.
BRD4 is one of the most important epigenetic proteins that specifically recognizes acetylated lysine residues in nucleosomal histones, interacting with other transcriptional proteins in chromatin to promote the expression of oncogenetic proteins, which are critical for cancer cell growth and division (38). However, the exact molecular mechanisms for BRD4 expression in MM have not been illustrated yet. Preclinical evidence showed that BRD4 inhibitors, including I-BET151 and I-BET762, displayed a promising antimyeloma activity both in vitro and in vivo (39). BRD4 is responsible for the expression of oncogenes, such as those for cyclin D1, c-Myc, and CDK4, in skin squamous cell carcinoma (40). However, the exact mechanism of BRD4 in regulating cancer cell proliferation is still unclear. Our study showed that H19 silencing or miR-152-3p overexpression significantly inhibited the expression of BRD4 and the expression of proliferation-associated proteins, such as c-Myc, survivin, cyclin D1, and CDK4 in MM cells. H19 could increase the expression of BRD4 by binding to miR-152-3p. These results fully demonstrated that H19 could regulate the tumorigenesis of MM by promoting BRD4-mediated survival signaling by directly targeting miR-152-3p.
In conclusion, we confirmed the oncogenic effects of H19 in MM and illustrated a novel mechanism for the role of H19 in MM. It was found that knockdown of H19 could elevate the expression of miR-152-3p via sponging miR-152-3p through targeting BRD4 gene and further activated BRD4-mediated proliferation related signals, thereby resulting in inhibition of proliferation, promotion of cell apoptosis, and induction of cell cycle G1 arrest. Our study discovered a novel potential implication for prognosis and therapeutic intervention of MM.
MATERIALS AND METHODS
MM patients.
In this study, 30 patients newly diagnosed with MM were enrolled at the Second Affiliated Hospital of Nanchang University, and this study was approved by the Medical Ethics Committee of the Second Affiliated Hospital of Nanchang University. Additionally, 30 healthy donors were recruited as a control. All of the samples were kept in liquid nitrogen before use. All patients signed informed written consent in accordance with the Declaration of Helsinki.
Cell culture.
Multiple myeloma (MM) cell lines of MM1.S and RPMI-8226 cells were purchased from the American Type Culture Collection (ATCC, USA). The cells were cultured in RPMI 1640 containing 10% fetal bovine serum (Sigma-Aldrich, USA) supplemented with 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen, USA) at 37°C and 5% CO2.
Lentivirus transfection.
Recombinant vectors harboring sh-NC and sh-H19 were purchased from the GeneChem Company (Shanghai, China). The vectors were transfected into cells using Lipofectamine 2000 (Invitrogen). Lentivirus supernatants were collected and filtered through a 0.45-μm filter and were immediately used to infect MM cells after 48 h of transfection. To construct the stable cell line, MM1.R and RPMI-8226 MM cells were transfected with lentiviruses, and then puromycin was used for a week to select the stable cell line. Finally, quantitative real-time PCR (qRT-PCR) was utilized to confirm the expression of H19.
Cell transient transfection.
MM cells were seeded at a density of 1 × 106 cells per well in six-well plates. Scramble negative control (NC), miR-152-3p mimics, miR-152-3p inhibitor, and pcDNA3.1-H19 were chemically synthesized by GeneChem Company. pcDNA3.1-H19, miR-152-3p mimics, and miR-152-3p inhibitor, as well as the corresponding NC, were transiently transfected into MM cells. The transfection experiments were performed using Lipofectamine 2000 (Invitrogen).
Vector construction and luciferase reporter assay.
The fragment from the 3′ untranslated region (3′-UTR) of BRD4 or H19 containing the predicted miR-152-3p binding site was cloned into pmirGLO vector (Promega, Madison, WI, USA) to form the reporter vector BRD4-wild type (WT-BRD4-3′UTR) or WT-H19. To mutate the putative binding site of miR-152-3p in WT-BRD4-3′UTR or WT-H19, site mutations were performed to generate BRD4-mutated-type (MUT-BRD4-3′UTR) and MUT-H19. The vectors (WT-BRD4-3′UTR or MUT-BRD4-3′UTR or WT-H19 or MUT-H19) and miR-152-3p mimics or miR-152-3p inhibitor then were cotransfected into 293T cells, and the Dual-Luciferase reporter assay system (Promega, USA) was used for testing luciferase activity.
Soft-agar colony formation assay.
MM cells were plated in 0.4% agarose on top of a 1% agarose base supplemented with complete medium and then were incubated under conditions of 5% CO2 and 37°C. After 4 weeks, the colonies that appeared were observed under a microscope. Pictures were taken, and the number of colonies was counted.
Annexin V-PI staining and cell cycle analysis.
For annexin V-propidium iodide (PI) staining, MM cells were harvested and resuspended in 100 μl of binding buffer containing 5 μl of fluorescein isothiocyanate-annexin V (Vazyme) and 1 μl of PI (Beyotime Biotechnology) working solution for 15 min at room temperature. After dilution with 400 μl of binding buffer, stained cells were immediately analyzed by flow cytometry (Beckman, USA). For cell cycle analysis, cells were fixed in 70% ethanol at 4°C overnight and stained with 5 μl PI for 45 min in the dark at room temperature. The stained cells then were immediately analyzed by flow cytometry (Beckman, USA).
RNA extraction and qRT-PCR.
Total RNA from MM patient samples, MM cells, or xenograft tumor tissues was isolated using a TRIzol reagent (Invitrogen). Concentration and integrity of total RNA were detected, and the qRT-PCR was conducted on a sequence detection system (ABI PRISM 7500) by using SYBR green quantitative PCR SuperMix (Invitrogen). For the measurement of miR-152-3p expression, a TaqMan microRNA reverse transcription kit and TaqMan universal master mix II were used for reverse transcription and quantitative PCR. The PCR conditions were 95°C for 2 min, followed by 40 cycles at 95°C for 5 s and at 60°C for 34 s. The primers used for this assay are listed here: H19 forward, 5′-GAGCACCTTGGACATCTGGAG-3′; reverse, 5′-GCCCTCGATCCCCTAAACCT-3′; miR-152-3p forward, 5′-GCGCTCAGTGC ATGACAGA-3′; reverse, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCAAGT-3′; BRD4 forward, 5′-GTGGTGCACATCATCCAGTC-3′; reverse, 5′-CCGACTCTGAGGACGAGAAG-3′; U6 forward, 5′-C TCGCTTCGGCAGCACA-3′; reverse, 5′-AACGCTTCACGAATTTGCGT-3′; GAPDH forward, 5′-TGTGGGCATCAATGGATTTGG-3′; reverse, 5′-ACACCATGTATTCCGGGTCAAT-3′. The 2−ΔΔCT method was used to assess the relative level of mRNA or miRNAs.
Western blotting.
Cells or xenograft tumor tissues were collected and lysed with radioimmunoprecipitation assay buffer (Cell Signaling Technology, USA), and the protein contents were quantified by a bicinchoninic acid kit (Beyotime Biotechnology, China) by following the manufacturer’s instructions. Proteins then were separated by SDS-PAGE and transferred to a 10% polyvinylidene difluoride (PVDF) membrane (Millipore, Boston, MA). After blocking with 5% bovine serum albumin in Tris-buffered saline with 0.1% Tween 20 (TBST), the PVDF membrane was rinsed and incubated with the primary anti-BRD4 antibody, primary anti-c-Myc, primary antisurvivin antibody, primary anti-cyclin D1 antibody, primary anti-CDK4 antibody, primary anti-Bax antibody, primary anti-Bcl2 antibody, and primary anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (all purchased form Cell Signaling Technology) overnight at 4°C. Membranes then were incubated with peroxidase-conjugated secondary antibody (Santa Cruz) for 1 h at room temperature. Protein expression then was detected by enhanced chemiluminescence (Millipore) after extensive washing, and the band intensities were quantified using ImageJ software (W. Rasband, NIH, USA).
Nude mice xenograft experiments.
Briefly, 1 × 106 MM1.R or RPMI-8226 cells stably transfected with scramble vector (shNC) or sh-H19 vector in 100 μl of phosphate-buffered saline (PBS) were injected subcutaneously into 6-week-old female BALB/c nude mice. The volume of tumors was measured every 5 days until the mice were sacrificed at day 30. The tumor volume was calculated using the formula (length)(width2)/2. All animal experiments were performed under an approved animal study protocol.
IHC staining.
Tumor tissues were fixed with 4% paraformaldehyde and embedded in paraffin. The samples were then sectioned into slices with a thickness of 5 μm. These sections were attached to slides, washed, and incubated with 3% H2O2 in methanol for 10 min to block endogenous peroxidase activity. Afterwards, these sections were blocked and then incubated with primary anti-BRD4 antibody and anti-Ki-67 antibody (Abcam, USA) at 4°C overnight. After washing with PBS, sections were incubated with secondary antibody for 1 h. A DAB immunohistochemistry color development kit (Sangon Biotech, China) then was used to detect the expression of Ki-67 and BRD4 by following the instructions. Hematoxylin was used as a counterstain for visualization. The sections were sealed with coverslips and subjected to fluorescence microscopy (IX-51; Olympus) for imaging.
TUNEL assay.
Apoptotic cells in xenograft tumor sections were detected by using the TUNEL kit (Invitrogen, USA). Briefly, tissue sections were treated with proteinase K (20 pg/ml) in 10 mM Tris-HCI (pH 8.0) for 15 min at room temperature and inactivated by endogenous peroxidases for 5 min in 3% H2O2 at room temperature. The samples then were preincubated for 10 min in TdT buffer, followed by 1 h at 37°C with 25 to 50 μl of TdT buffer supplied with 0.5 U of TdT per μl and 40 μM biotinylated 16-dUTP in a moist chamber. The reaction was stopped by transferring the sections to 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer for 15 min at room temperature. The sections then were incubated in Vectastain ABC peroxidase standard solution (Vector Laboratories, USA) and stained using aminoethyl carbazole. Finally, the reaction was stopped with water. Sections then were coverslipped in Aqua-Poly/Mount (Polysciences, USA) and observed under a microscope.
Statistical analysis.
The quantitative data were presented as means ± standard deviations (SD) and were analyzed with Prism 5.0 (GraphPad Software, USA). Spearman correlation analysis was performed to analyze the correlation among H19, miR-152-3p, and BRD4 in MM tissues. Student's t test (two-tailed) and one-way analysis of variance (ANOVA) followed by Dunnett’s test were applied for two or multiple comparisons, respectively. A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This work was supported by the Jiangxi Provincial Health and Family Planning Commission Science and Technology Plan (no. 20195209) and the National Natural Science Foundation of China (no. 81560030).
We have no competing interests to declare.
REFERENCE
- 1.Harding T, Baughn L, Kumar S, Van Ness B. 2019. The future of myeloma precision medicine: integrating the compendium of known drug resistance mechanisms with emerging tumor profiling technologies. Leukemia 33:863–883. doi: 10.1038/s41375-018-0362-z. [DOI] [PubMed] [Google Scholar]
- 2.Vélez R, Turesson I, Landgren O, Kristinsson SY, Cuzick J. 2016. Incidence of multiple myeloma in Great Britain, Sweden, and Malmö, Sweden: the impact of differences in case ascertainment on observed incidence trends. BMJ Open 6:e009584. doi: 10.1136/bmjopen-2015-009584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leng S, Chen Y, Tsai W-Y, Bhutani D, Hillyer GC, Lim E, Accordino MK, Wright JD, Hershman DL, Lentzsch S, Neugut AI. 2019. Use of bisphosphonates in elderly patients with newly diagnosed multiple myeloma. J Natl Compr Canc Netw 17:22–28. doi: 10.6004/jnccn.2018.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brioli A, Klaus M, Sayer H, Scholl S, Ernst T, Hilgendorf I, Scherag A, Yomade O, Schilling K, Hochhaus A, Mügge L-O, von Lilienfeld-Toal M. 2019. The risk of infections in multiple myeloma before and after the advent of novel agents: a 12-year survey. Ann Hematol 98:713–722. doi: 10.1007/s00277-019-03621-1. [DOI] [PubMed] [Google Scholar]
- 5.Chen D, Zhou D, Xu J, Zhou R, Ouyang J, Chen B. 2019. Prognostic value of 1q21 gain in multiple myeloma. Clin Lymphoma Myeloma Leuk 19:e159–e164. doi: 10.1016/j.clml.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Senín A, García-Pallarols F, Azaiz RB, Martínez-Serra L, Montesdeoca S, Román D, Ferraro M, Párraga I, Besses C, Abella E. 2019. Study of the frequency and reasons for discontinuation of different lines of treatment in patients with multiple myeloma. Ann Hematol 98:705–711. doi: 10.1007/s00277-019-03601-5. [DOI] [PubMed] [Google Scholar]
- 7.Lambert J-P, Picaud S, Fujisawa T, Hou H, Savitsky P, Uusküla-Reimand L, Gupta GD, Abdouni H, Lin Z-Y, Tucholska M, Knight JDR, Gonzalez-Badillo B, St-Denis N, Newman JA, Stucki M, Pelletier L, Bandeira N, Wilson MD, Filippakopoulos P, Gingras A-C. 2019. Interactome rewiring following pharmacological targeting of BET bromodomains. Mol Cell 73:621–638. doi: 10.1016/j.molcel.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren C, Zeng L, Zhou MM. 2016. Chapter fourteen–preparation, biochemical analysis, and structure determination of the bromodomain, an acetyl-lysine binding domain. Methods Enzymol 573:321–343. doi: 10.1016/bs.mie.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 9.Ren W, Wang C, Wang Q, Zhao D, Zhao K, Sun D, Liu X, Han C, Hou J, Li X, Zhang Q, Cao X, Li N. 2017. Bromodomain protein Brd3 promotes Ifnb1 transcription via enhancing IRF3/p300 complex formation and recruitment to Ifnb1 promoter in macrophages. Sci Rep 7:39986. doi: 10.1038/srep39986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hensel T, Giorgi C, Schmidt O, Calzada-Wack J, Neff F, Buch T, Niggli FK, Schäfer BW, Burdach S, Richter G. 2016. Targeting the EWS-ETS transcriptional program by BET bromodomain inhibition in Ewing sarcoma. Oncotarget 7:1451–1463. doi: 10.18632/oncotarget.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stubbs MC, Burn TC, Sparks R, Maduskuie T, Diamond S, Rupar M, Wen X, Volgina A, Zolotarjova N, Waeltz P, Favata M, Jalluri R, Liu H, Liu XM, Li J, Collins R, Falahatpisheh N, Polam P, DiMatteo D, Feldman P, Dostalik V, Thekkat P, Gardiner C, He X, Li Y, Covington M, Wynn R, Ruggeri B, Yeleswaram S, Xue C-B, Yao W, Combs AP, Huber R, Hollis G, Scherle P, Liu PCC. 2019. The novel bromodomain and extraterminal domain inhibitor INCB054329 induces vulnerabilities in myeloma cells that inform rational combination strategies. Clin Cancer Res 25:300–311. doi: 10.1158/1078-0432.CCR-18-0098. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt J, Braggio E, Kortuem KM, Egan JB, Zhu YX, Xin CS, Tiedemann RE, Palmer SE, Garbitt VM, McCauley D, Kauffman M, Shacham S, Chesi M, Bergsagel PL, Stewart AK. 2013. Genome-wide studies in multiple myeloma identify XPO1/CRM1 as a critical target validated using the selective nuclear export inhibitor KPT-276. Leukemia 27:2357–2365. doi: 10.1038/leu.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu YJ, Zhou R, Zong JF, Lin WS, Tong S, Guo QJ, Lin C, Lin SJ, Chen YX, Chen MR. 2019. Epstein-Barr virus-coded miR-BART13 promotes nasopharyngeal carcinoma cell growth and metastasis via targeting of the NKIRAS2/NF-κB pathway. Cancer Lett 447:33–40. doi: 10.1016/j.canlet.2019.01.022. [DOI] [PubMed] [Google Scholar]
- 14.Bogedale K, Jagannathan V, Gerber V, Unger L. 2019. Differentially expressed miRNAs, including a large microRNA cluster on chromosome 24, are associated with equine sarcoid and squamous cell carcinoma. Vet Comp Oncol 17:155. doi: 10.1111/vco.12458. [DOI] [PubMed] [Google Scholar]
- 15.Jiang J, Chang W, Fu Y, Gao Y, Zhang S. 2019. SAV1, regulated by microRNA-21, suppresses tumor growth in colorectal cancer. Biochem Cell Biol 97:91–99. doi: 10.1139/bcb-2018-0034. [DOI] [PubMed] [Google Scholar]
- 16.Ramchandani D, Lee SK, Yomtoubian S, Han MS, Tung CH, Mittal V. 2019. Nanoparticle delivery of miR-708 mimetic impairs breast cancer metastasis. Mol Cancer Ther 18:579–591. doi: 10.1158/1535-7163.MCT-18-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Ha T, Liu L, Hu Y, Kao R, Kalbfleisch J, Williams D, Li C. 2018. TLR3 mediates repair and regeneration of damaged neonatal heart through glycolysis dependent YAP1 regulated miR-152 expression. Cell Death Differ 25:966–982. doi: 10.1038/s41418-017-0036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Z-W, Du M-Y, Qian L-X, Zhang N, Gu J-J, Ding K, Wu J, Zhu H-M, He X, Yin L. 2018. MiR-152 functioning as a tumor suppressor that interacts with DNMT1 in nasopharyngeal carcinoma. Onco Targets Ther 11:1733. doi: 10.2147/OTT.S154464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Wang YE, Zhang Y, Leleu X, Reagan M, Zhang Y, Mishima Y, Glavey S, Manier S, Sacco A, Jiang B, Roccaro AM, Ghobrial IM. 2014. Global epigenetic regulation of microRNAs in multiple myeloma. PLoS One 9:e110973. doi: 10.1371/journal.pone.0110973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang M, Zhou L, Yu F, Zhang Y, Li P, Wang K. 2019. The functional roles of exosomal long non-coding RNAs in cancer. Cell Mol Life Sci 76:2059–2076. doi: 10.1007/s00018-019-03018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Gao L, Ma S, Ma J, Wang Y, Li S, Ding Z. 2019. MALAT1-KTN1-EGFR regulatory axis promotes the development of cutaneous squamous cell carcinoma. Cell Death Differ 26:2061–2073. doi: 10.1038/s41418-019-0288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J, Xu J, Zhang L, Liu S, Ma Y, Wen X, Hao J, Li Z, Ni Y, Li X, Zhou F, Li Q, Wang F, Wang X, Si Y, Zhang P, Liu C, Bartolomei M, Tang F, Liu B, Yu J, Lan Y. 2019. Combined single-cell profiling of lncRNAs and functional screening reveals that H19 is pivotal for embryonic hematopoietic stem cell development. Cell Stem Cell 24:285–298. doi: 10.1016/j.stem.2018.11.023. [DOI] [PubMed] [Google Scholar]
- 23.Pan Y, Chen H, Shen X, Wang X, Ju S, Lu M, Cong H. 2018. Serum level of long noncoding RNA H19 as a diagnostic biomarker of multiple myeloma. Clin Chim Acta 480:199–205. doi: 10.1016/j.cca.2018.02.019. [DOI] [PubMed] [Google Scholar]
- 24.Mercurio A, Adriani G, Catalano A, Carocci A, Rao L, Lentini G, Cavalluzzi MM, Franchini C, Vacca A, Corbo F. 2017. A mini-review on thalidomide: chemistry, mechanisms of action, therapeutic potential and anti-angiogenic properties in multiple myeloma. Curr Med Chem 24:2736. doi: 10.2174/0929867324666170601074646. [DOI] [PubMed] [Google Scholar]
- 25.De BN, Menu E, Bertrand MJ, Favreau M, De BE, Maes K, De VK, Radwanska M, Samali A, Magez S. 2017. Experimental African trypanosome infection suppresses the development of multiple myeloma in mice by inducing intrinsic apoptosis of malignant plasma cells. Oncotarget 8:52016–52025. doi: 10.18632/oncotarget.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalia BL, Lau SK, Boutros PC, Fereshteh K, Igor J, Andrulis IL, Tsao MS, Penn LZ. 2006. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res 66:5330. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 27.Pan Y, Zhang Y, Liu W, Huang Y, Shen X, Jing R, Pu J, Wang X, Ju S, Cong H. 2019. LncRNA H19 overexpression induces bortezomib resistance in multiple myeloma by targeting MCL-1 via miR-29b-3p. Cell Death Dis 10:106. doi: 10.1038/s41419-018-1219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y, Pan J, Zhang N, Wei W, Yu S, Ai L. 2017. Knockdown of long non-coding RNA H19 inhibits multiple myeloma cell growth via NF-κB pathway. Sci Rep 7:18079. doi: 10.1038/s41598-017-18056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C, Chen Z, Fang J, Xu A, Zhang W, Wang Z. 2016. AB144. H19-derived miR-675 contributes to bladder cancer cell proliferation by regulating p53 activation. Tumour Biol 37:263–270. doi: 10.21037/tau.2016.s144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu G, Xiang T, Wu QF, Wang WX. 2016. Long noncoding RNA H19-derived miR-675 enhances proliferation and invasion via RUNX1 in gastric cancer cells. Oncol Res 23:99–107. doi: 10.3727/096504015X14496932933575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Liu CY, Wan Y, Peng L, Li WF, Qiu JX. 2016. Long non-coding RNA H19 promotes the proliferation of fibroblasts in keloid scarring. Oncol Lett 12:2835. doi: 10.3892/ol.2016.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y, Chen B, George SK, Liu B. 2015. Downregulation of microRNA-152 contributes to high expression of DKK1 in multiple myeloma. RNA Biol 12:1314–1322. doi: 10.1080/15476286.2015.1094600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun J, Tian X, Zhang J, Huang Y, Lin X, Chen L, Zhang S. 2017. Regulation of human glioma cell apoptosis and invasion by miR-152-3p through targeting DNMT1 and regulating NF2. J Exp Clin Cancer Res 36:100. doi: 10.1186/s13046-017-0567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He ZY, Wei TH, Zhang PH, Zhou J, Huang XY. 2019. Long noncoding RNA-antisense noncoding RNA in the INK4 locus accelerates wound healing in diabetes by promoting lymphangiogenesis via regulating miR-181a/Prox1 axis. J Cell Physiol 234:4627–4640. doi: 10.1002/jcp.27260. [DOI] [PubMed] [Google Scholar]
- 35.Chen DL, Lu YX, Zhang JX, Wei XL, Wang F, Zeng ZL, Pan ZZ, Yuan YF, Wang FH, Pelicano H, Chiao PJ, Huang P, Xie D, Li YH, Ju HQ, Xu RH. 2017. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics 7:4836–4849. doi: 10.7150/thno.20942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding J, Yeh CR, Sun Y, Lin C, Chou J, Ou Z, Chang C, Qi J, Yeh S. 2018. Estrogen receptor beta promotes renal cell carcinoma progression via regulating LncRNA HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene 37:5037–5053. doi: 10.1038/s41388-018-0175-6. [DOI] [PubMed] [Google Scholar]
- 37.Cazalla D, Yario T, Steitz JA, Steitz J. 2010. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White ME, Fenger JM, Carson WE. 2019. Emerging roles of and therapeutic strategies targeting BRD4 in cancer. Cell Immunol 337:48. doi: 10.1016/j.cellimm.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo NH, Zheng JF, Zi FM, Cheng J. 2019. I-BET151 suppresses osteoclast formation and inflammatory cytokines secretion by targeting BRD4 in multiple myeloma. Biosci Rep 39:BSR20181245. doi: 10.1042/BSR20181245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiang T, Bai JY, She C, Yu DJ, Zhou XZ, Zhao TL. 2018. Bromodomain protein BRD4 promotes cell proliferation in skin squamous cell carcinoma. Cell Signal 42:106–113. doi: 10.1016/j.cellsig.2017.10.010. [DOI] [PubMed] [Google Scholar]