Abstract
Cynanchum auriculatum is a traditional herbal medicine in China and can grow in saline soils. However, little is known in relation to the underlying molecular mechanisms. In the present study, C. auriculatum seedlings were exposed to 3.75‰ and 7.5‰ salinity. Next, transcriptome profiles of leaves were compared. Transcriptome sequencing showed 35,593 and 58,046 differentially expressed genes (DEGs) in treatments with 3.75‰ and 7.5‰, compared with the control, respectively. Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of these DEGs enriched various defense-related biological pathways, including ROS scavenging, ion transportation, lipid metabolism and plant hormone signaling. Further analyses suggested that C. auriculatum up-regulated Na+/H+ exchanger and V-type proton ATPase to avoid accumulation of Na+. The flavonoid and phenylpropanoids biosynthesis pathways were activated, which might increase antioxidant capacity in response to saline stress. The auxin and ethylene signaling pathways were upregulated in response to saline treatments, both of which are important plant hormones. Overall, these results raised new insights to further investigate molecular mechanisms underlying resistance of C. auriculatum to saline stress.
Subject terms: RNA sequencing, Physiology
Introduction
Tuberous root of Cynanchum auriculatum is a traditional herbal medicine in Asian countries. It has been listed in Chinese Pharmacopoeia and Korean Pharmacopoeia, namely “Baishouwu” and “Baekshuoh”, respectively1. Pharmacological researches revealed that application of C. auriculatum could enhance immunity2, display activities of anti-tumor3, antioxidant4 and gastro protection2. Thus, C. auriculatum has high economic values.
In China, C. auriculatum is cultivated in Shandong, Jiangsu, and Anhui provinces. Approximately 95% of C. auriculatum are produced at Binhai County, Yancheng City (China), which is a coastal city. Due to human activities and natural seawater erosion, most agricultural lands there show severe salinization. Cultivation on saline soils and/or using brackish water resources has attracted widespread attentions in recent years. Planting C. auriculatum on saline soils may be an alternative approach to solve the problem of insufficient agricultural lands, thus increasing the total production and decreasing the unit price of C. auriculatum. However, there are no reports investigating responses of C. auriculatum to saline stress.
The genus Cynanchum (Linn.) includes approximately 200 species in the world, which are widely distributed in eastern Africa, Mediterranean region, tropical, subtropical and temperate regions of Eurasia5. There are 53 species and 12 varieties of this genus in China, which are mainly distributed in southwest provinces6. The complete chloroplast genome sequence has been sequenced for C. auriculatum and C. wilfordii, and phylogenetic analysis revealed that these two species are evolutionarily close7,8. According to previous studies, plants in Cynanchum showed strong tolerance to salinity. It was found that the seed germination and radicle length of swallowwort (Cynanchum acutum L.) decreased with increased saline stress, which is more tolerant to salinity than common milkweed, hairy beggarticks, and scotch thistle9. As a halophyte speices, Cynanchum chinense increases the content of osmotic regulators such as soluble sugar, betaine and organic acids in saline-alkali environments, and its osmotic regulators content is higher than plants in Poaceae, Leguminosae, Cyperaceae and Boraginaceae families10. C. auriculatum is closely related to C. acutum and C. chinense11, and may also have similar tolerance to saline stress.
Besides antioxidant regulation, plants have also evolved other physiological and molecular mechanisms to resist saline stress12. For example, plant hormones, such as abscisic acid (ABA), auxin and ethylene, could function as essential endogenous signal molecules and regulate plant development and tolerance to salinity13,14. The WRKY and MYB transcription factors are important to the regulation of transcriptional reprogramming involved in abiotic stress responses15,16, which have been demonstrated to regulate the contents of chlorophyll, proline, soluble sugar as well as activities of superoxide dismutase (SOD), and catalase (CAT) under saline stress17,18. In response to saline stress, metabolism of unsaturated fatty acid was activated to produce various compounds, which then function as signal molecules or participate in plant defense system as antimicrobial substances19–21. Whether these mechanisms also function in C. auriculatum resistance to salinity should be investigated.
Transcriptome sequencing is an ideal method to investigate molecular changes in response to saline stress in plants22–24. In the present study, to investigate molecular changes of C. auriculatum to saline environments, seedlings were exposed to saline treatments. Next, leaves were subjected to transcriptome sequencing, and bioinformatics analyses were conducted to globally predict effects of salinity on physiology of C. auriculatum. These results might contribute theoretical basis to guide C. auriculatum cultivation on saline soils.
Materials and Methods
Ethics statement
No specific permit is required for the present study in P. R. China.
Cultivation of C. auriculatum
Seeds of C. auriculatum were collected from the planting base at Xinyang Agricultural Experiment Station of Yancheng City (Yancheng, P. R. China) on 2017. Full and healthy C. auriculatum seeds were washed with sterile water for 2–3 times, soaked in 2% NaClO solution for 2 min, and then washed with sterile water for 3 times. After germination in sterile water, the seedlings were cultured in 1/2 Hoagland’s solution at 25 °C under a photoperiod of 16 h:8 h (light:dark). The light intensity was 14,400 lux.
Treatments with salinity
Based on the preliminary results, saline treatments included two salinities, 3.75‰ and 7.5‰ (dissolving commercial sea salts in 1/2 Hoagland’s solution). The experiments were carried out in plastic boxes (20 cm * 10 cm * 8 cm). In each box, 1 L of solution was added. For saline treatments, suitable amount of sea salts were added in 1/2 Hoagland’s solution to achieve salinities of 3.75‰ and 7.5‰. 1/2 Hoagland’s solution without extra addition of sea salts was also prepared as the control (CT). Each treatment consisted of five individuals and repeated three times independently. To avoid effects of evaporation on salinity, the culture media were renewed every three days. After 15 days, the top four leaves from five individuals in each treatment were collected and mixed as one sample. The plants were taken out, and quickly dried using sterile filter paper. After freezing with liquid nitrogen, leaves were stored at −80 °C until RNA extraction.
Transcriptome sequencing
The Biozol reagent produced by Bioer Technology (Hangzhou, China) was used to extract total RNA according to the manufacture’s protocol. The integrity of RNA was visualized by 1% agarose gel electrophoresis, and its concentration and purity were detected using a NanoDrop Microvolume Spectrophotometers and Fluorometer (ThermoFisher, Shanghai, China). Oligo(dT) magnetic beads (NEB, USA) were used to enrich mRNA in the samples and then sequencing libraries were prepared using NEBNext mRNA Library Prep Master Mix Set for Illumina (NEB, USA) as described in the manufacture’s protocol. AMPure XP beads (Beckman, Germany) were used to purify the DNA fragments with 250–300 bp length, which were further amplified by PCR using Phusion High-Fidelity DNA polymerase. Qubit 2.0 software was used for preliminary quantitative analysis of the libraries. The library was diluted to 1.5 ng/μL to check the distribution of insert size using an Agilent Bioanalyzer 2100 system. Real-time quantitative PCR (RT-qPCR) was applied to determine the accurate concentration of library (concentration > 2 nmol was the effective concentration of the library). Finally, Illumina HiSeq. 4000 platform was used to sequence the DNA libraries.
Bioinformatics analyses
To determine whether sequencing data were suitable for subsequent analyses, quality control (QC) of raw reads was performed first. Reads with low quality (with >20% bases having Phred quality score ≤10), joint contamination and/or high ratio of unknown base (ratio ≥5%) were removed. Clean reads were assembled using the Trinity program25, and the longest transcript of each sequence was selected for the follow-up analysis of unigene. After assembly, the gene expression levels were compared between treatments and the control by calculating FPKM (expected number of fragments per kilobase of transcript sequence per million base pairs sequenced) values of each unigene using RSEM v1.2.826. Genes with fold change ≥ 2 and Q-value (adjusted P-value) ≤ 0.001 were considered as significantly differentially expressed genes (DEGs).
TransDecoder software was used to identify candidate coding sequence (CDS). The longest open reading frame was extracted for each unigene, and then the Pfam protein homologous sequences were searched by blasting against SwissProt and Hmmscan database to predict CDS27. For gene ontology (GO) annotations, the DEGs were blasted against the GO database using Blast2Go program (http://www.blast2go.com/Ver.2.3.5)28. For Kyoto Encyclopedia of Genes and Genomics (KEGG) annotations, all the DEGs were mapped to the KEGG database (https://www.genome.jp/kegg/pathway.html) using BLASTX29. Comparisons with Q-value ≤ 0.05 were considered statistically significant.
Real-time quantitative PCR
To verify the reliability of RNA-seq results, nine unigenes were randomly selected for RT-qPCR. The total RNA samples used for qPCR were the same for RNA-seq. The first strand of cDNA was synthesized by reverse transcription using the BioRT cDNA first strand synthesis kit (Bioer, Hangzhou, China) with oligo(dT) primer (Supplementary Table S1). Beta-actin, which showed similar FPKM values among treatments, was also included as the internal control for qPCR. The qPCR experiments were performed in 20 µl of reaction system using BioEasy master mix (Bioer, Hangzhou, China). The thermal cycles were performed and the signals of SYBR Green were detected using a Gene9600 Plus qPCR machine (Bioer, Hangzhou, China). Melting curves were prepared to reveal the specificity of primers. Expression levels of each gene were compared by calculating their relative fold change using the 2−ΔΔCt method30.
Results and Discussion
Preliminary experiments were conducted to test the tolerance of C. auriculatum to saline stress. Seedlings were treated with salinities ranging from 7.5‰ to 15‰. After treatment for one week, the leaves of C. auriculatum turned yellow, the whole plant was shorter and the bottom leaves withered in the 7.5‰ treatment, compared with the control. In treatments with 10‰, 12‰ and 15‰, the plants died (Fig. S1). These results indicated that C. auriculatum could tolerate 7.5‰ salinity. To explore the underlying molecular mechanisms, treatments with 3.75‰ and 7.5‰ were re-prepared for transcriptome sequencing.
Illumina sequencing
The sequencing data have been deposited in National Center for Biotechnology Information (NCBI) with the bioproject number of PRJNA558052. RNA sequencing resulted in 65 M to 73 M of clean reads for each leaf sample. Q20 and Q30 values were all higher than 96.92% and 88.19%, respectively (Table S2). These results suggested that the quality of sequencing data could satisfy further transcriptome analyses.
De novo assembly
Separate assembly of each sample was tried first. The results showed that the total number of unigenes ranged from 43,123 to 76,090, and the mean length of unigenes ranged from 1,257 bp to 1,521 bp. Alternatively, all sequencing data were assembled together, finally producing 196,199 unigenes with the mean length of 1,673 bp (Table S3). Among them, 15.89% unigenes were shorter than 300 bp and 17.07% were longer than 3,000 bp (Fig. S2). Benchmarking Universal Single-Copy Orthologs (BUSCO)31 analyses revealed that 99% BUSCOs were complete and 1% BUSCOs were missing (Fig. S3). Due to the longer mean length and higher ratio of BUSCOs, the unigene library assembled from nice samples together was used for the subsequent analyses.
Prediction of coding sequences and functional annotation
Prediction of coding sequences detected 122,855 CDSs in total. Their total length and N50 length was 137.21 M bp and 1,431 bp, respectively (Table S4). Among them, 77.44% CDSs were longer than 500 bp, and 42.61% CDSs were longer than 1,000 bp (Fig. S4).
Blasting against the public databases, 73.35%, 64.41%, 57.08%, 59.70%, 60.62%, 57.71%, and 43.61% unigenes could be annotated to the NCBI non-redundant proteins (NR), nucleotide (NT), Swissprot, KEGG, Eukaryotic Orthologous Groups (KOG), Pfam, and GO databases, respectively (Table 1).
Table 1.
BLAST analysis of non-redundant unigenes against public databases.
| Number of unigenes | Percentage (%) | |
|---|---|---|
| Total unigenes | 196,199 | 100 |
| Annotated in NR | 143,913 | 73.35 |
| Annotated in NT | 126,371 | 64.41 |
| Annotated in Swissprot | 111,994 | 57.08 |
| Annotated in KEGG | 117,134 | 59.70 |
| Annotated in KOG | 118,931 | 60.62 |
| Annotated in Pfam | 113,221 | 57.71 |
| Annotated in GO | 85,559 | 43.61 |
| Intersection | 48,166 | 24.55 |
| Overall | 159,285 | 81.19 |
Differentially expressed genes and qPCR validation
Pairwise comparisons revealed that 16,849 and 19,404 unigenes were significantly up-regulated, 18,744 and 38,642 were down-regulated in treatment with 3.75‰ and 7.5‰, respectively, compared with the control. After removing unigenes with FPKM values lower than 2 in all samples, 4,975 and 6,566 unigenes were significantly up-regulated, 4,820 and 6,484 were significantly down-regulated in treatment with 3.75‰ and 7.5‰, respectively (Fig. 1a). Among them, 2,897 up-regulated DEGs and 2,956 down-regulated DEGs were commonly shared between treatments with 3.75‰ and 7.5‰ (Fig. 1b). The top 30 DEGs are listed in Table S5.
Figure 1.
Differentially expressed genes (DEGs) in C. auriculatum leaves in response to saline stress. (a) Numbers of DEGs between treatment with 3.75‰ and 7.5‰ salinity. (b) Venn diagrams of up-regulated and down-regulated DEGs in treatments with 3.75‰ and 7.5‰. The numbers did not include genes with FPKM values lower than 2 in all samples. 3.75‰ vs CT, 3.75‰ salinity compared to the control; 7.5‰ vs CT, 7.5‰ salinity compared to the control.
To validate the DEGs identified by RNA-Seq, nine randomly selected DEGs were determined by qPCR. The results showed similar changing trends between qPCR validation and FPKM calculation for all selected genes (Fig. S5), indicating that the transcriptome data were reliable.
Enrichment of GO categories for DEGs
In both treatments with 3.75‰ and 7.5‰, the top four GO categories were ‘membrane part’, ‘cell’, ‘catalytic activity’ and ‘cellular process’ (Fig. 2). In these categories, more DEGs were included in treatment with 7.5‰ than 3.75‰, suggesting that higher salinity caused more severe effects on C. auriculatum leaves. Besides, the category of ‘binding’ was only highly expressed in treatment with 3.75‰ but not in treatment with 7.5‰.
Figure 2.
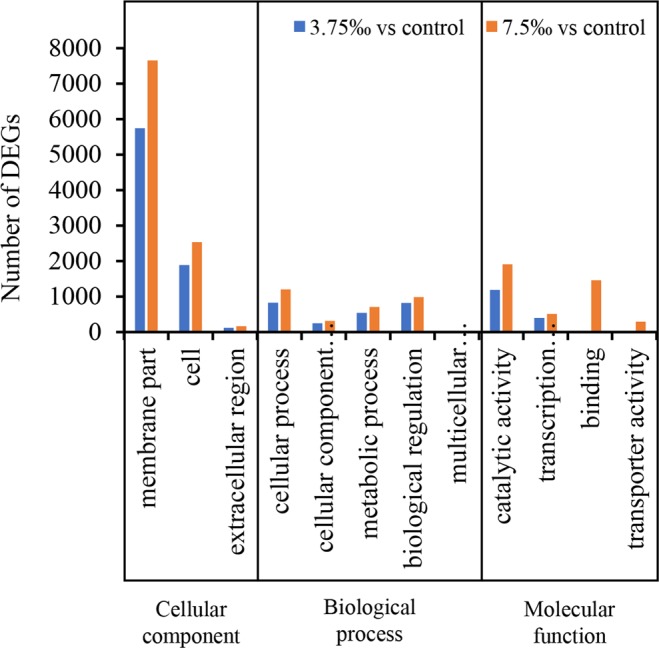
Top 12 GO categories of DEGs in C. auriculatum leaves in response to 3.75‰ and 7.5‰ saline stress.
Enrichment of KEGG pathways for DEGs
KEGG enrichment analysis of down-regulated genes in treatment with 3.75‰ and 7.5‰ revealed 19 and 28 pathways, respectively (Tables 2 and 3). These pathways might represent the inhibitory effects on the molecular processes in C. auriculatum leaves. Analysis of up-regulated genes, which represented mechanisms underlying resistance to saline stress, showed 19 and 23 significantly enriched pathways at 3.75‰ and 7.5‰, respectively, which shared 12 KEGG pathways (ko00500, ko00940, ko04075, ko00563, ko00941, ko00330, ko00511, ko00591, ko04016, ko00270, ko00460 and ko00904; Tables 2 and 3). These enriched pathways mainly involved in oxidative stress-related function, signal transduction, carbohydrate metabolism and lipid metabolism.
Table 2.
Enrichment of KEGG pathway for differentially expressed genes between treatment with 3.75‰ and the control in C. auriculatum.
| Pathway ID | Pathway Name | Enriched genes | P value | Q value |
|---|---|---|---|---|
| Up-regulated genes in treatment with 3.75‰ | ||||
| ko00500 | Starch and sucrose metabolism | 411 | 9.76E-12 | 1.30E-09 |
| ko04075 | Plant hormone signal transduction | 418 | 2.73E-11 | 1.82E-09 |
| ko00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 82 | 8.44E-09 | 3.74E-07 |
| ko00940 | Phenylpropanoid biosynthesis | 293 | 4.17E-06 | 9.24E-05 |
| ko04016 | MAPK signaling pathway - plant | 334 | 4.66E-05 | 0.000886 |
| ko00330 | Arginine and proline metabolism | 105 | 0.000198 | 0.00329 |
| ko00511 | Other glycan degradation | 107 | 0.001072 | 0.012138 |
| ko00591 | Linoleic acid metabolism | 81 | 0.001174 | 0.012138 |
| ko00905 | Brassinosteroid biosynthesis | 37 | 0.001186 | 0.012138 |
| ko00920 | Sulfur metabolism | 71 | 0.001029 | 0.012138 |
| ko00941 | Flavonoid biosynthesis | 75 | 0.001069 | 0.012138 |
| ko00904 | Diterpenoid biosynthesis | 55 | 0.001539 | 0.014622 |
| ko03410 | Base excision repair | 83 | 0.00193 | 0.017116 |
| ko02010 | ABC transporters | 156 | 0.002627 | 0.021835 |
| ko00906 | Carotenoid biosynthesis | 84 | 0.003458 | 0.027052 |
| ko00460 | Cyanoamino acid metabolism | 139 | 0.00424 | 0.031326 |
| ko00270 | Cysteine and methionine metabolism | 166 | 0.006196 | 0.043373 |
| ko00261 | Monobactam biosynthesis | 25 | 0.00766 | 0.048978 |
| ko03430 | Mismatch repair | 75 | 0.007733 | 0.048978 |
| Down-regulated genes in treatment with 3.75‰ | ||||
| ko00460 | Cyanoamino acid metabolism | 181 | 3.78E-09 | 2.51E-07 |
| ko00500 | Starch and sucrose metabolism | 402 | 2.13E-07 | 7.07E-06 |
| ko00592 | alpha-Linolenic acid metabolism | 111 | 3.34E-06 | 8.87E-05 |
| ko00906 | Carotenoid biosynthesis | 100 | 1.95E-05 | 0.00041 |
| ko02010 | ABC transporters | 179 | 2.16E-05 | 0.00041 |
| ko00770 | Pantothenate and CoA biosynthesis | 75 | 3.03E-05 | 0.000503 |
| ko00790 | Folate biosynthesis | 67 | 5.80E-05 | 0.000858 |
| ko00650 | Butanoate metabolism | 71 | 7.42E-05 | 0.000986 |
| ko00591 | Linoleic acid metabolism | 89 | 0.000193 | 0.00233 |
| ko00196 | Photosynthesis - antenna proteins | 23 | 0.000262 | 0.002871 |
| ko04626 | Plant-pathogen interaction | 414 | 0.000281 | 0.002871 |
| ko00564 | Glycerophospholipid metabolism | 159 | 0.001366 | 0.012981 |
| ko00410 | beta-Alanine metabolism | 104 | 0.002037 | 0.018058 |
| ko00051 | Fructose and mannose metabolism | 113 | 0.003898 | 0.027285 |
| ko01210 | 2-Oxocarboxylic acid metabolism | 115 | 0.003775 | 0.027285 |
| ko04075 | Plant hormone signal transduction | 371 | 0.003377 | 0.027285 |
| ko04712 | Circadian rhythm - plant | 135 | 0.003584 | 0.027285 |
| ko00220 | Arginine biosynthesis | 86 | 0.005834 | 0.038797 |
| ko00561 | Glycerolipid metabolism | 142 | 0.007597 | 0.048112 |
Table 3.
Enrichment of KEGG pathway for differentially expressed genes between treatment with 7.5‰ and the control in C. auriculatum.
| Pathway ID | Pathway Name | Enriched genes | P value | Q value |
|---|---|---|---|---|
| Up-regulated genes in treatment with 7.5‰ | ||||
| ko00500 | Starch and sucrose metabolism | 467 | 1.25E-13 | 1.66E-11 |
| ko00940 | Phenylpropanoid biosynthesis | 361 | 1.61E-11 | 1.07E-09 |
| ko04075 | Plant hormone signal transduction | 453 | 1.25E-09 | 5.52E-08 |
| ko00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 91 | 2.54E-09 | 7.49E-08 |
| ko00941 | Flavonoid biosynthesis | 94 | 4.42E-06 | 8.41E-05 |
| ko03030 | DNA replication | 101 | 6.71E-05 | 0.001116 |
| ko00330 | Arginine and proline metabolism | 117 | 0.000148 | 0.002188 |
| ko00511 | Other glycan degradation | 122 | 0.000337 | 0.004481 |
| ko00591 | Linoleic acid metabolism | 92 | 0.000455 | 0.005505 |
| ko04016 | MAPK signaling pathway - plant | 362 | 0.000562 | 0.006225 |
| ko00945 | Stilbenoid, diarylheptanoid and gingerol biosynthesis | 49 | 0.000699 | 0.007147 |
| ko00600 | Sphingolipid metabolism | 115 | 0.001326 | 0.012598 |
| ko00270 | Cysteine and methionine metabolism | 190 | 0.002058 | 0.017484 |
| ko00410 | beta-Alanine metabolism | 110 | 0.002103 | 0.017484 |
| ko00460 | Cyanoamino acid metabolism | 157 | 0.002499 | 0.019553 |
| ko00531 | Glycosaminoglycan degradation | 79 | 0.002989 | 0.022085 |
| ko00592 | alpha-Linolenic acid metabolism | 101 | 0.003212 | 0.022486 |
| ko00904 | Diterpenoid biosynthesis | 59 | 0.003443 | 0.022898 |
| ko03430 | Mismatch repair | 85 | 0.004344 | 0.027509 |
| ko00360 | Phenylalanine metabolism | 72 | 0.006695 | 0.037103 |
| ko00520 | Amino sugar and nucleotide sugar metabolism | 234 | 0.006483 | 0.037103 |
| ko00604 | Glycosphingolipid biosynthesis - ganglio series | 65 | 0.006317 | 0.037103 |
| ko00260 | Glycine, serine and threonine metabolism | 117 | 0.009135 | 0.048599 |
| Down-regulated genes in treatment with 7.5‰ | ||||
| ko00052 | Galactose metabolism | 251 | 1.05E-06 | 4.74E-05 |
| ko04712 | Circadian rhythm - plant | 218 | 1.57E-06 | 5.29E-05 |
| ko00195 | Photosynthesis | 88 | 2.94E-06 | 7.94E-05 |
| ko00906 | Carotenoid biosynthesis | 139 | 1.90E-05 | 0.000427 |
| ko00630 | Glyoxylate and dicarboxylate metabolism | 286 | 6.28E-05 | 0.001212 |
| ko02010 | ABC transporters | 249 | 9.48E-05 | 0.0016 |
| ko00909 | Sesquiterpenoid and triterpenoid biosynthesis | 65 | 0.000108 | 0.001622 |
| ko00790 | Folate biosynthesis | 90 | 0.000162 | 0.002186 |
| ko00945 | Stilbenoid, diarylheptanoid and gingerol biosynthesis | 66 | 0.000226 | 0.002715 |
| ko00966 | Glucosinolate biosynthesis | 18 | 0.000241 | 0.002715 |
| ko00196 | Photosynthesis - antenna proteins | 30 | 0.000296 | 0.002854 |
| ko00650 | Butanoate metabolism | 95 | 0.000295 | 0.002854 |
| ko00460 | Cyanoamino acid metabolism | 220 | 0.00037 | 0.003121 |
| ko00900 | Terpenoid backbone biosynthesis | 134 | 0.000348 | 0.003121 |
| ko00500 | Starch and sucrose metabolism | 542 | 0.000405 | 0.003216 |
| ko00591 | Linoleic acid metabolism | 122 | 0.000504 | 0.003783 |
| ko00604 | Glycosphingolipid biosynthesis - ganglio series | 93 | 0.000542 | 0.00385 |
| ko00531 | Glycosaminoglycan degradation | 109 | 0.000842 | 0.005411 |
| ko00940 | Phenylpropanoid biosynthesis | 418 | 0.000808 | 0.005411 |
| ko00061 | Fatty acid biosynthesis | 103 | 0.001988 | 0.012199 |
| ko00770 | Pantothenate and CoA biosynthesis | 93 | 0.002773 | 0.016275 |
| ko04146 | Peroxisome | 292 | 0.00293 | 0.016479 |
| ko03450 | Non-homologous end-joining | 34 | 0.003761 | 0.020307 |
| ko00603 | Glycosphingolipid biosynthesis - globo and isoglobo series | 37 | 0.004752 | 0.024675 |
| ko00071 | Fatty acid degradation | 139 | 0.006563 | 0.032814 |
| ko00230 | Purine metabolism | 466 | 0.007961 | 0.038381 |
| ko00240 | Pyrimidine metabolism | 380 | 0.009931 | 0.046232 |
| ko00780 | Biotin metabolism | 48 | 0.010923 | 0.049154 |
Ion transportation
Under saline conditions, the first response of plants is ion toxicity, and generally Na+ displays more severe influence than Cl−, since Na+ would reach toxic concentrations earlier than Cl−. Thus, most studies focused on Na+ uptake, transportation and exclusion in plants28. In the present study, compared with the control, Na+/H+ exchanger was significantly up-regulated in treatment with 3.75‰ (P < 0.05), but K+ efflux antiporter, cation/H+ antiporter and sodium/proton antiporter were significantly down-regulated at 7.5‰ salinity (P < 0.05). V-type proton ATPase (V-ATPase) was significantly up-regulated, while cyclic nucleotide-gated ion channel (CNGC) were significantly down-regulated at 3.75‰ salinity (P < 0.05) (Table 4). The up-regulation of Na+/H+ exchanger might alleviate cytosolic salt accumulation in the vacuole and other cellular compartments32,33. Similar results were observed in Arabidopsis34, which assisted Na+ extrusion from plant cells. Activation of V-ATPase in plants might reduce Na+ accumulation by interacting with H+ pumps to resist saline stress35. CNGCs are non-selective ion channels involved in mediation of Na+ transport. The expression level of CNGC significantly decreased at 3.75‰ salinity compared with the control, which was in agreement with the findings reported in Arabidopsis that CNGC10 expression was inhibited when exposed to high salinity or low salinity for long time36. K+ efflux antiporter was significantly down-regulated at 7.5‰ salinity, similar to the transcriptome analysis of grapevine (Vitis vinifera L.) under saline conditions37, implying the replacement of K+ by Na+.
Table 4.
FPKM values of selected genes in saline treatment and the control in C. auriculatum.
| Gene name | Control | 3.75‰ | 7.5‰ |
|---|---|---|---|
| Antioxidant enzymes | |||
| Superoxide dismutase [Cu-Zn] | 235.51 ± 24.41 | 264.66 ± 10.57 | 289.7 ± 28.82 |
| Superoxide dismutase [Mn] | 54.59 ± 10.99 | 72.45 ± 12.22 | 117.67 ± 31.78* |
| Superoxide dismutase [Fe] | 28.52 ± 2.81 | 54.61 ± 12.01* | 45.86 ± 18.52 |
| Copper chaperone for superoxide dismutase | 7.34 ± 1.45 | 7.54 ± 1.28 | 6.95 ± 2.10 |
| Flavonoid | |||
| Chalcone synthase (CHS) | 68.77 ± 9.01 | 89.72 ± 21.40 | 93.91 ± 64.69 |
| Chalcone isomerase (CHI) | 19.59 ± 3.36 | 27.24 ± 6.99 | 32.4 ± 11.73* |
| Flavonol synthase (FLS) | 13.31 ± 2.75 | 14.32 ± 1.78 | 15.66 ± 6.44 |
| Trans-cinnamate 4-monooxygenase (C4H) | 13.68 ± 2.29 | 25.12 ± 3.85* | 34.80 ± 17.19* |
| Bifunctional dihydroflavonol 4-reductase (DFR) | 42.88 ± 1.63 | 44.43 ± 12.11 | 38.85 ± 2.82 |
| Phenylpropanoid | |||
| Phenylalanine ammonia-lyase (PAL) | 16.98 ± 1.38 | 24.99 ± 6.69* | 36.10 ± 17.76* |
| 4-coumarate–CoA ligase (4CL) | 26.64 ± 1.96 | 25.73 ± 6.86 | 39.11 ± 17.74 |
| Carotenoid | |||
| Phytoene synthase (PSY) | 38.58 ± 6.00 | 27.06 ± 5.75 | 25.08 ± 7.01 |
| Phytoene desaturase (PDS) | 2.70 ± 0.29 | 2.79 ± 1.05 | 0.75 ± 0.27* |
| Zeta-carotene desaturase (Zds) | 41.30 ± 3.65 | 45.52 ± 13.66 | 40.05 ± 13.97 |
| Nine-cis-epoxycarotenoid dioxygenase (NCED) | 10.02 ± 2.77 | 3.89 ± 0.86* | 9.11 ± 2.93 |
| Beta-carotene 3-hydroxylase (CrtR-b) | 3.31 ± 1.02 | 3.32 ± 0.92 | 3.19 ± 1.15 |
| Ion transprotation | |||
| Sodium/hydrogen exchanger | 83.49 ± 2.97 | 93.32 ± 5.81* | 95.84 ± 16.92 |
| K+ efflux antiporter | 147.81 ± 8.44 | 124.32 ± 22.92 | 110.41 ± 19.92* |
| Cation/H+ antiporter | 26.04 ± 3.85 | 22.68 ± 1.18 | 19.55 ± 4.05* |
| Sodium/proton antiporter | 4.61 ± 1.34 | 4.52 ± 0.57 | 2.83 ± 0.51* |
| V-type proton ATPase (V-ATPase) | 1022.52 ± 68.32 | 1200.48 ± 109.60* | 1315.03 ± 167.55 |
| Cyclic nucleotide-gated ion channel (CNGCs) | 182.29 ± 14.62 | 124.27 ± 7.17* | 162.24 ± 16.93 |
| Potassium channel | 143.87 ± 28.09 | 127.9 ± 2.31 | 150.15 ± 10.90 |
| Two-pore potassium channel | 24.78 ± 2.70 | 22.58 ± 0.62 | 26.38 ± 1.61 |
| Chloride channel protein (CLC) | 167.38 ± 29.56 | 152.79 ± 8.84 | 138.88 ± 8.4 |
| Choline monooxygenase (CMO) | 24.10 ± 5.08 | 21.21 ± 7.30 | 21.44 ± 7.01 |
| Lipid metabolisms | |||
| Lipoxygenase | 40.09 ± 6.61 | 19.49 ± 5.35* | 26.36 ± 11.97 |
| Hydroperoxide dehydratase | 82.68 ± 49.35 | 19.90 ± 0.62* | 40.49 ± 9.74 |
| Allene oxide cyclase | 40.38 ± 5.28 | 41.88 ± 5.00 | 62.92 ± 15.02 |
| 12-oxophytodienoic acid reductase | 79.17 ± 11.09 | 93.19 ± 15.21 | 87.5 ± 4.66 |
Data represent mean ± standard deviation of FPKM values (n = 3). *Significantly different from the control (P < 0.05, Student’s t-test).
Saline treatments induced antioxidant mechanisms in C. auriculatum
Oxidative stress is a general negative effect on plants caused by saline stress. To resist saline stress, antioxidant mechanisms were activated38. Among them, SOD is an important antioxidant enzyme39. SODs have three isozymes including copper-zinc SOD (Cu/Zn-SOD), manganese SOD (Mn-SOD) and iron SOD (Fe-SOD). In the present study, three isozymes were all detected. In comparison to the control, Mn-SOD was significantly up-regulated for 2.15 times in 7.5‰ treatment, while Fe-SOD increased significantly for 1.91 times at 3.75‰ salinity (P < 0.05) (Table 4). These changes might enhance tolerance of C. auriculatum to saline stress, since it has been reported that overexpression of Mn-SOD enhanced salt-tolerance in Arabidopsis40. In pea, the activity of Mn-SOD in salt-tolerant plants is higher than that in salt-sensitive plants under NaCl treatment. Further analysis found that NaCl-tolerant plants had protective mechanisms against salt-induced O2− production by increasing the mitochondrial Mn-SOD activity41. According to these results, C. auriculatum might be salt-tolerant, but the specific functions of SODs need further study.
Flavonoid biosynthesis pathway plays a pivotal role and protects the plant cells from oxidative damage by scavenging free radicals42,43. KEGG enrichment analyses of up-regulated DEGs significantly enriched flavonoid biosynthesis pathway. Within this pathway, trans-cinnamate 4-monooxygenase (C4H) and chalcone isomerase (CHI) are key enzymes, directly related to the synthesis of flavonoids44. In the present study, compared with the control, CHI and C4H were significantly up-regulated in treatments with 3.75‰ and/or 7.5‰ (Table 4). These results implied that more flavonoids were synthesized, which might contribute to the total antioxidant capacity in response to saline stress in C. auriculatum. Similarly, Walia et al.45 reported that a large number of genes were up-regulated in the flavonoid biosynthesis pathway under salinity stress, which played an important protective role against saline stress.
Phenylpropanoids are involved in antioxidant activity in cell walls and lignin biosynthesis in secondary metabolites, and play an important role in plant response to abiotic stress, which connects to flavonoid biosynthesis46. Phenylalanine ammonia-lyase (PAL) is the first enzyme that converts phenylalanine into cinnamic acid and is transcriptionally regulated in response to many environmental cues47. PAL was a marker during molecular breeding to improve resistance of plants to injury and saline stress48. In the present study, PAL was significantly up-regulated for 1.47 and 2.13 times at 3.75‰ and 7.5‰ salinity (P < 0.05), respectively, compared with the control (Table 4). Similar findings were observed in salinity-stressed Olea europaea49.
Carotenoids reveal antioxidant activity in plants. In the carotenoid biosynthesis pathways, phytoene synthase (PSY) is a rate-limiting enzyme, related to abiotic stress and mainly involved in plant stress response50. In the present study, compared with the control, the expression level of phytoene desaturase (PDS) was significantly lower in treatment with 7.5‰ and that of nine-cis-epoxycarotenoid dioxygenase (NCED) was significantly lower in treatment with 3.75‰. Besides, the expression level of phytoene synthase (PSY) was lower in saline treatments than that in the control, although no statistical significance was detected (P > 0.05; Table 4). Similarly, PSY1 gene was reduced in response to enhanced NaCl level51, and NCED gene was clearly repressed under drought and saline stress in cotton52. The decreased expression of key enzymes in carotenoid biosynthesis pathway revealed that carotenoid biosynthesis might not be the antioxidant mechanism in salinity-stressed C. auriculatum.
Saline treatments regulated lipid metabolism in C. auriculatum
Six lipid metabolism-related pathways were significantly enriched when up-regulated or down-regulated genes were subjected to KEGG enrichment analyses, including alpha-linolenic acid (LnA) metabolism, linoleic acid (LA) metabolism, glycerophospholipid metabolism, fatty acid biosynthesis, fatty acid degradation, and sphingolipid metabolism. In response to adverse factors, alpha-linolenic acid and linoleic acid were released and became the initial step of unsaturated fatty acid metabolism53,54. LnA was catalyzed by different functional lipoxygenase (LOX) to produce hydroperxy linolenic acid (HPOT), and then metabolized into a series of compounds with certain biological activities, such as jasmonic acid (JA), 3-hexenol and traumatic acid, which related to plant defense responses to abiotic stresses54–56. Previous studies have shown that JA biosynthesis genes were up-regulated in Arabidopsis and sunflower roots in saline treatments57,58. In the present study, compared with the control, LOX and hydroperoxide dehydratase were significantly down-regulated at 3.75‰ (P < 0.05) (Table 4), revealing that unsaturated fatty acid metabolism might be inhibited by saline stress in C. auriculatum.
Changes of plant hormone signaling transduction pathways under saline stress
Plants respond to abiotic stress through a complex coordination of various phytohormone signaling pathways, mainly including auxin (AUX), cytokinine (CTK), gibberellin (GA), abscisic acid (ABA), ethylene (ETH), brassinosteroid (BR), jasmonic acid (JA) and salicylic acid (SA)59. In the present study, the functional analysis of DEGs by KEGG enrichment analysis showed that a large number of genes were involved in plant hormone signaling transduction and hormone responses under saline stress, highlighting the key roles of plant hormones in regulating C. auriculatum response to saline stress. In the JA signal pathway, the expression level of jasmonate ZIM domain-containing protein (JAZ) was significantly down-regulated at 3.75‰ salinity (Table S6), indicating that JA signaling transduction was inhibited by saline stress. This result was consistent with the inhibition of unsaturated fatty acid metabolism.
As previously reported, the BR signaling pathway was activated under saline stress in Arabidopsis60, and the exogenous application of BR effectively alleviated the adverse effects of abiotic stress or increased plant resistance to stresses61,62. In the present study, compared with the control, brassinosteroid insensitive 1 (BRI1), BR-signaling kinase (BSK), protein brassinosteroid insensitive 2 (BIN2), brassinosteroid resistant 1/2 (BZR1/2), xyloglucan: xyloglucosyl transferase (TCH4) and cyclin-D3 (CYCD3) were significantly up-regulated in treatments with 3.75‰ and/or 7.5‰ (Table S6), suggesting that activation of BR signaling pathway might participate in resistance to saline stress.
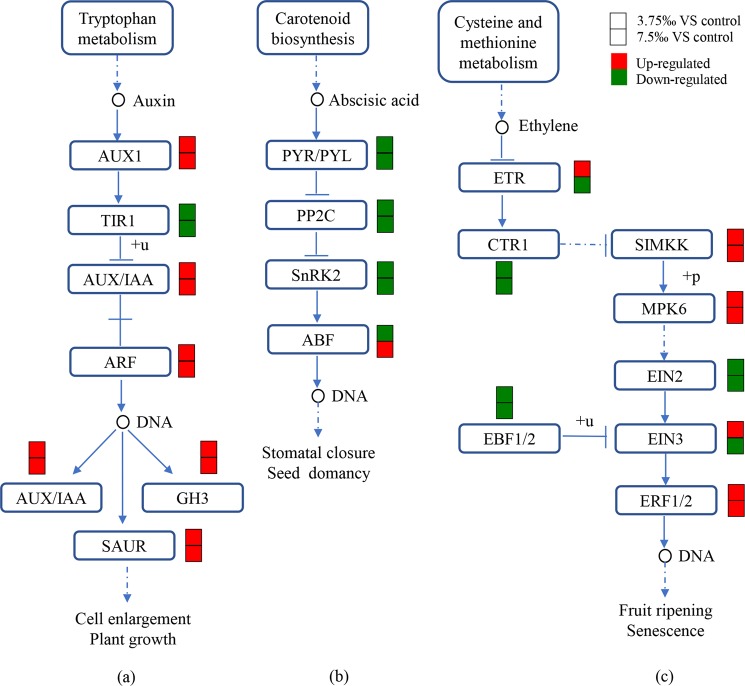
Auxin can induce expression of auxin-response factor gene (ARF), auxin early response gene (Aux/IAA), gretchen hagen3 (GH3) and small auxin-up RNAs (SAUR) rapidly and instantaneously63. In the present study, compared with the control, GH3 and SAUR were significantly up-regulated in response to saline treatments. However, the expression levels of protein transport inhibitor response (TIR1) was significantly down-regulated under saline stress (P < 0.05; Fig. 3). Since GH3 and SAUR were downstream genes of auxin signaling pathway, the present results suggested that auxin signaling pathway was activated in C. auriculatum in response to saline stress.
Figure 3.
DEGs mapped to the plant hormone signaling transduction pathways in leaves. (a) Auxin signaling pathway, (b) ABA signaling pathway and (c) ethylene signaling pathway. Upper and lower color box indicate difference between 3.75‰ and the control, between 7.5‰ and the control, respectively. Red: up-regulated compared with the control. Green: down-regulated compared with the control.
The abscisic (ABA) signaling pathway is well known to be associated with responses to saline stress in plants and the increase of ABA content could help plants adapt and survive under saline condition by reducing the accumulation of Na+ and improving osmotic adjustment15. Besides, carotenoid biosynthesis pathway provides precursors for plant hormone abscisic acid (ABA) synthesis64. In the present study, ABA receptors (PYR/PYL) were significantly down-regulated in treatment with 7.5‰ (Fig. 3), which might be resulted from the inhibition of carotenoid biosynthesis. Similar results were also detected in Corchorus spp. and Vitis vinifera L37,65. Besides, downstream of PYR/PYL, serine/threonine-protein kinase 2 (SNRK2) was also significantly down-regulated for 2.03 and 1.97 times in treatment with 3.75‰ and 7.5‰, respectively (P < 0.05; Fig. 3). Overall, these results suggested that ABA signaling pathway was inhibited by saline treatments in C. auriculatum.
Ethylene is a stress hormone regulating myriad stress responses66. Ethylene receptors (ETR) serve as negative regulators of the ethylene signaling transduction pathway, while ethylene-insensitive protein 2 (EIN2) and ethylene-insensitive protein 3 (EIN3) are positive regulators. Thus, a decrease in receptor levels is predicted to sensitize the plant such that it responds to lower levels of ethylene than usual67. In the present study, compared with the control, mitogen-activated protein kinase kinase (SIMKK) and ethylene-responsive transcription factor 1/2 (ERF1/2) were significantly up-regulated in treatment with 7.5‰, while serine/threonine-protein kinase CTR1 (CTR1) and EIN3-binding F-box protein 1/2 (EBF1/2) were significantly down-regulated in treatment with 3.75‰ and 7.5‰, respectively (P < 0.05; Fig. 3, Table S6). No significant differences in expression levels of ETR, EIN2 and EIN3 were detected between saline treatments and the control. Considering ERF1/2 is a downstream gene of CTR1 and EBF1/2 (Fig. 3), the present results implied that ethylene signaling pathway might be activated in C. auriculatum under saline conditions.
In summary, the changes of gene expression profile in C. auriculatum leaves under saline stress were explored using the transcriptome sequencing to investigate regulatory mechanisms in response to saline stress. Compared with the control, C. auriculatum showed suppressed carotenoid biosynthesis, lipid metabolisms, abscisic acid and jasmonic acid signaling transduction, which might damage plant cells. Expression levels of Na+/H+ exchanger and V-type proton ATPase, the flavonoid and phenylpropanoids biosynthesis pathways, the auxin and ethylene signaling pathways were upregulated to improve the antioxidant capacity of C. auriculatum in response to saline treatments. These changes might facilitate resistance of C. auriculatum to saline stress. Flavonoid and phenylpropanoids appear to play roles in resistance to salt tolerance in C. auriculatum, possibly due to its pharmacodynamics.
Supplementary information
Acknowledgements
This work was supported by the Natural Science Foundation of Jiangsu Province (No. BK20141255) and the Jiangsu Agriculture Science and Technology Innovation Fund [No: CX(17)2014].
Author contributions
M.Z., L.Z.H. and G.Z. designed experiment and did the analysis. M.F.G. and C.D.W. conducted the experiment. C.D.W. wrote the paper. G.Z. supervised the experiment and revised the manuscript. All authors have approved the final article.
Data availability
The original sequencing files have been deposited in the National Center for Biotechnology Information (NCBI) with the bioproject number of PRJNA558052.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ming Zhang, Li-Zhou Hong and Min-Feng Gu.
Contributor Information
Cheng-Dong Wu, Email: syzwcd@126.com.
Gen Zhang, Email: zhanggen1988@163.com.
Supplementary information
is available for this paper at 10.1038/s41598-019-57219-8.
References
- 1.Jiang Y, et al. Chemical constituents of Cynanchum wilfordii and the chemotaxonomy of two species of the family Asclepiadacease, C. wilfordii and C. auriculatum. Arch. Pharm. Res. 2011;34:2021–2017. doi: 10.1007/s12272-011-1203-z. [DOI] [PubMed] [Google Scholar]
- 2.Shan L, et al. Gastroprotective effect of a traditional Chinese herbal drug “Baishouwu” on experimental gastric lesions in rats. J. Ethnopharmacol. 2006;107:389–394. doi: 10.1016/j.jep.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 3.Shan L, et al. Antitumor activity of crude extract and fractions from root tuber of Cynanchum auriculatum Royle ex Wight. Phytother. Res. 2005;19:259–261. doi: 10.1002/ptr.1678. [DOI] [PubMed] [Google Scholar]
- 4.Song JM, Ding XL. Study on the scavenging effect of Baishouwu on superoxide free radicals. Chin. Wild Plant Resour. 1997;17:1–4. [Google Scholar]
- 5.Wu, Z., Ding, L. & Zhao, S. Chemical constituents and pharmacological effects of Cynanchum Linn. World Phytomed. 6, 147–154 [In Chinese] (1991).
- 6.Gu XJ, Hao DC. Recent advances in phytochemistry and pharmacology of C21 steroid constituents from Cynanchum plants. Chin. J. Nat. Med. 2016;14:321–334. doi: 10.3724/SP.J.1009.2016.00321. [DOI] [PubMed] [Google Scholar]
- 7.Jang W, et al. The complete chloroplast genome sequence of Cynanchum auriculatum Royle ex Wight (Apocynaceae) Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27:4549–4550. doi: 10.3109/19401736.2015.1101557. [DOI] [PubMed] [Google Scholar]
- 8.Park HS, et al. The complete chloroplast genome sequence of an important medicinal plant Cynanchum wilfordii (Maxim.) Hemsl. (Apocynaceae) Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27:3747–3748. doi: 10.3109/19401736.2015.1079887. [DOI] [PubMed] [Google Scholar]
- 9.Pahlevani AH, Rashed MH, Ghorbani R. Effects of environmental factors on germination and emergence of swallowwort. Weed Technol. 2008;22:303–308. doi: 10.1614/WT-07-055.1. [DOI] [Google Scholar]
- 10.Liu, Z. X. Comparison of osmotic adjustment and ion balance strategies of eleven alkali-tolerant halophytes during adaptation to salt-alkalinized habitats. Northeast Normal University [In Chinese] (2011).
- 11.Liede S, Täuber A. Circumscription of the Genus Cynanchum (Apocynaceae—Asclepiadoideae) Syst. Bot. 2002;27:789–800. [Google Scholar]
- 12.Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He XJ, et al. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2006;44:903–916. doi: 10.1111/j.1365-313X.2005.02575.x. [DOI] [PubMed] [Google Scholar]
- 14.Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 15.You J, Chan Z. ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 2015;6:1092. doi: 10.3389/fpls.2015.01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong W, Liu X, Li D, Gao T, Song Y. Transcriptional profiling reveals that a MYB transcription factor MsMYB4 contributes to the salinity stress response of alfalfa. Plos One. 2018;13:e0204033. doi: 10.1371/journal.pone.0204033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, et al. Overexpression of the alfalfa WRKY11 gene enhances salt tolerance in soybean. Plos One. 2018;13:e0192382. doi: 10.1371/journal.pone.0192382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Y, et al. Overexpression of a MYB family gene, OsMYB6, increases drought and salinity stress tolerance in transgenic rice. Front. Plant Sci. 2019;10:168. doi: 10.3389/fpls.2019.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber H. Fatty acid-derived signals in plants. Trends Plant Sci. 2002;7:217–224. doi: 10.1016/S1360-1385(02)02250-1. [DOI] [PubMed] [Google Scholar]
- 20.Blee E. Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 2002;7:315–322. doi: 10.1016/S1360-1385(02)02290-2. [DOI] [PubMed] [Google Scholar]
- 21.Turkan I, Bor M, Ozdemir F, Koca H. Differential responses of lipid peroxidation and antioxidants in the leaves of drought-tolerant P. acutifolius Gray and drought-sensitive P. vulgaris L. subjected to polyethylene glycol mediated water stress. Plant Sci. 2005;168:223–231. doi: 10.1016/j.plantsci.2004.07.032. [DOI] [Google Scholar]
- 22.Ben-Abdallah S, et al. Salt stress (NaCl) affects plant growth and branch pathways of carotenoid and flavonoid biosyntheses in Solanum nigrum. Acta Physiol. Plant. 2016;38:72. doi: 10.1007/s11738-016-2096-8. [DOI] [Google Scholar]
- 23.Payyavula Raja S, Navarre Duroy A, Kuhl Joseph C, Alberto P, Pillai Syamkumar S. Differential effects of environment on potato phenylpropanoid and carotenoid expression. BMC Plant Biol. 2012;12:39. doi: 10.1186/1471-2229-12-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez J, Jiménez A, Mullineaux P, Sevilla F. Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defences. Plant Cell Environ. 2000;23:853–862. doi: 10.1046/j.1365-3040.2000.00602.x. [DOI] [Google Scholar]
- 25.Grabherr MG, et al. Nat. Biotechnol. 2011. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data; pp. 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinfor. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HS, et al. Identification of xenobiotic biodegradation and metabolism-related genes in the copepod Tigriopus japonicus whole transcriptome analysis. Mar. Genomics. 2015;3:207–208. doi: 10.1016/j.margen.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 28.Haider MS, et al. Insights into grapevine defense response against drought as revealed by biochemical, physiological and RNA-Seq analysis. Sci. Rep. 2017;7:13134. doi: 10.1038/s41598-017-13464-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Waterhouse R, et al. BUSCO Applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2017;35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munns R, Tester M. Mechanisms salinity tolerance. Annu. Rev. Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 33.Rajendran K, Tester MR, Stuart J. Quantifying the three main components of salinity tolerance in cereals. Plant Cell Environ. 2010;32:237–249. doi: 10.1111/j.1365-3040.2008.01916.x. [DOI] [PubMed] [Google Scholar]
- 34.Shi H, Quintero FJ, Pardo JM, Zhu JK. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu JK, et al. Enrichment of vitronectin- and fibronectin-like proteins in NaCI-adapted plant cells and evidence for their involvement in plasma membrane-cell wall adhesion. Plant J. 2010;3:637–646. doi: 10.1111/j.1365-313X.1993.00637.x. [DOI] [PubMed] [Google Scholar]
- 36.Jin Y, Jing W, Zhang Q, Zhang W. Cyclic nucleotide gated channel 10 negatively regulates salt tolerance by mediating Na+ transport in Arabidopsis. J. Plant Res. 2015;128:211–220. doi: 10.1007/s10265-014-0679-2. [DOI] [PubMed] [Google Scholar]
- 37.Guan L, et al. Transcriptome sequence analysis elaborates a complex defensive mechanism of grapevine (Vitis vinifera L.) in response to salt stress. Int. J. Mol. Sci. 2018;19:4019. doi: 10.3390/ijms19124019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang X, Mu X, Shao H, Wang H, Brestic M. Global plant-responding mechanisms to salt stress: physiological and molecular levels and implications in biotechnology. Crit. Rev. Biotechnol. 2014;35:425–437. doi: 10.3109/07388551.2014.889080. [DOI] [PubMed] [Google Scholar]
- 39.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Ying Y, Chen J, Wang X. Transgenic Arabidopsis overexpressing Mn-SOD enhanced salt-tolerance. Plant Sci. 2004;167:671–677. doi: 10.1016/j.plantsci.2004.03.032. [DOI] [Google Scholar]
- 41.Hernández JA, Corpas FJ, Gómez M, del Río LA, Sevilla F. Salt-induced oxidative stress mediated by activated oxygen species in pea leaf mitochondria. Physiol. Plant. 1993;89:103–110. doi: 10.1111/j.1399-3054.1993.tb01792.x. [DOI] [Google Scholar]
- 42.Bharti P, Mahajan M, Vishwakarma AK, Bhardwaj J, Yadav SK. AtROS1 overexpression provides evidence for epigenetic regulation of genes encoding enzymes of flavonoid biosynthesis and antioxidant pathways during salt stress in transgenic tobacco. J. Exp. Bot. 2015;66:5959–5969. doi: 10.1093/jxb/erv304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma P, Jha A, Shanker Dubey R, Pessarakli M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012;2012:1–26. doi: 10.1155/2012/217037. [DOI] [Google Scholar]
- 44.Wang Y, Li J, Xia RX. Expression of chalcone synthase and chalcone isomerase genes and accumulation of corresponding flavonoids during fruit maturation of Guoqing No. 4 satsuma mandarin (Citrus unshiu Marcow) Sci. Hortic. 2010;125:110–116. doi: 10.1016/j.scienta.2010.02.001. [DOI] [Google Scholar]
- 45.Harkamal W, et al. Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol. 2005;139:822–835. doi: 10.1104/pp.105.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonawitz ND, et al. REF4 and RFR1, subunits of the transcriptional coregulatory complex mediator, are required for phenylpropanoid homeostasis in Arabidopsis. J. Biol. Chem. 2012;287:5434–5445. doi: 10.1074/jbc.M111.312298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hemmati S. Phenylalanine ammonia-lyase through evolution: a bioinformatic approach. Trends Pharmacol. Sci. 2015;1:10–14. [Google Scholar]
- 48.Dehghan S, et al. Differential inductions of phenylalanine ammonia-lyase and chalcone synthase during wounding, salicylic acid treatment, and salinity stress in safflower, Carthamus tinctorius. Biosci. Rep. 2014;34:e00114. doi: 10.1042/BSR20140026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rossi L, et al. Salt stress induces differential regulation of the phenylpropanoid pathway in Olea europaea cultivars Frantoio (salt-tolerant) and Leccino (salt-sensitive) J. Plant Physiol. 2016;204:8–15. doi: 10.1016/j.jplph.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 50.Li F, Vallabhaneni R, Wurtzel ET. PSY3, a new member of the phytoene synthase gene family conserved in the poaceae and regulator of abiotic stress-induced root carotenogenesis. Plant Physiol. 2008;146:1333–1345. doi: 10.1104/pp.107.111120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abdallah SB, et al. Salt stress (NaCl) affects plant growth and branch pathways of carotenoid and flavonoid biosyntheses in Solanum nigrum. Acta Physiol. Plant. 2016;38:1–13. doi: 10.1007/s11738-015-2023-4. [DOI] [Google Scholar]
- 52.Huiru Y, et al. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 2014;55:2060–2076. doi: 10.1093/pcp/pcu133. [DOI] [PubMed] [Google Scholar]
- 53.Porta H, Rocha-Sosa M. Plant Lipoxygenases. physiological and molecular features. Plant Physiol. 2002;130:15. doi: 10.1104/pp.010787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howe GA, Schilmiller AL. Oxylipin metabolism in response to stress. Curr. Opin. Plant Biol. 2002;5:230–236. doi: 10.1016/S1369-5266(02)00250-9. [DOI] [PubMed] [Google Scholar]
- 55.Vancanneyt G, et al. Hydroperoxide lyase depletion in transgenic potato plants leads to an increase in aphid performance. Proc. Nat. Acad. Sci. USA. 2001;98:8139–8144. doi: 10.1073/pnas.141079498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pérez AG, Sanz C, Olías R, Olías JM. Lipoxygenase and hydroperoxide lyase activities in ripening strawberry fruits. J. Agric. Food Chem. 1999;47:249–253. doi: 10.1021/jf9807519. [DOI] [PubMed] [Google Scholar]
- 57.Wang P, et al. Transcriptome analysis reveals sunflower cytochrome P450 CYP93A1 responses to high salinity treatment at the seedling stage. Genes Genom. 2017;39:581–591. doi: 10.1007/s13258-017-0523-x. [DOI] [Google Scholar]
- 58.Yu G, et al. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell. 2013;25:2132–2154. doi: 10.1105/tpc.113.117028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ryu H, Cho YG. Plant hormones in salt stress tolerance. J. Plant Biol. 2015;58:147–155. doi: 10.1007/s12374-015-0103-z. [DOI] [Google Scholar]
- 60.Krishna P. Brassinosteroid-mediated stress responses. J. Plant Growth Regul. 2003;22:289–297. doi: 10.1007/s00344-003-0058-z. [DOI] [PubMed] [Google Scholar]
- 61.Ram Rao SS, Vardhini BV, Edupuganti S, Anuradha S. Brassinosteroids: a new class of phytohormones. Curr. Sci. 2002;82:1239–1245. [Google Scholar]
- 62.Fariduddin Q, Yusuf M, Ahmad A. Comparative roles of brassinosteroids and polyamines in salt stress tolerance. Acta Physiol. Plant. 2013;35:2037–2053. doi: 10.1007/s11738-013-1263-4. [DOI] [Google Scholar]
- 63.Wang SK, et al. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct. Integr. Genomics. 2010;10:533–546. doi: 10.1007/s10142-010-0174-3. [DOI] [PubMed] [Google Scholar]
- 64.Olson JA, Krinsky NI. Introduction: the colorful, fascinating world of the carotenoids: important physiologic modulators. Faseb J. 1995;9:1547–1550. doi: 10.1096/fasebj.9.15.8529833. [DOI] [PubMed] [Google Scholar]
- 65.Yang Z, et al. Salt-stress response mechanisms using de novo transcriptome sequencing of salt-tolerant and sensitive Corchorus spp. genotypes. Genes. 2017;8:226. doi: 10.3390/genes8090226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ryu H, Cho YG. Plant hormones in salt stress tolerance. J. Palnt Biol. 2015;58:147–155. doi: 10.1007/s12374-015-0103-z. [DOI] [Google Scholar]
- 67.Weiner JJ, Peterson FC, Volkman BF, Cutler SR. Structural and functional insights into core ABA signaling. Curr. Opin. Plant Biol. 2010;13:495–502. doi: 10.1016/j.pbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original sequencing files have been deposited in the National Center for Biotechnology Information (NCBI) with the bioproject number of PRJNA558052.