Abstract
Circulating microRNAs (miRNAs) are emerging as promising biomarkers for colorectal cancer (CRC). In this study, we sought to examine the diagnostic and prognostic role of serum miR-206 in CRC. A total of 73 CRC cases and 45 healthy control subjects were enrolled in this study. Quantitative reverse transcription PCR (qRT-PCR) was used to measure the relative serum miR-206 levels. Serum miR-206 levels were significantly decreased in CRC patients than normal controls. In addition, serum miR-206 demonstrated good diagnostic performance for discriminating CRC patients from healthy controls. miR-206 levels were increased in the blood samples of CRC patients who received surgery treatment. Moreover, serum miR-206 expression was closely associated with TNM stage and lymph node metastasis. CRC patients with low miR-206 level suffered more unfavorable overall survival and disease free survival compared to those with high miR-206 level. Furthermore, serum miR-206 was confirmed to be an independent prognostic indicator for CRC. In summary, serum miR-206 might be a promising biomarker for the diagnosis and prognosis of CRC.
Keywords: Serum miR-206, colorectal cancer, diagnosis, prognosis, biomarker
Introduction
Colorectal cancer (CRC) is one of the most frequent malignancy and has become a major global health problem. It is the third leading cause of cancer mortality in the USA and ranks the fourth in China [1,2]. Great progress has been made for the treatment of CRC in the past decades, and the 5-year postoperative survival rate for early CRC patients was currently favorable. However, most of CRC patients were diagnosed at advanced stages, leading to adverse clinical outcome [3,4]. Therefore, identification of novel and reliable biomarkers for early detection as well as prognosis prediction of CRC is urgently required.
MicroRNAs (miRNAs) are a class of small (18-25 nucleotides long) non-coding RNAs that act as post-transcriptional regulators by binding to the 3’-untranslated region (3’-UTR) of target messenger RNA (mRNA), resulting in mRNA degradation and/or translational repression [5,6]. Numerous studies have showed that dysregulation of miRNAs play crucial roles in the initiation and progression of various types of cancers [7,8]. More importantly, miRNAs can be stably detected in blood, and some circulating miRNAs show great potential for the detection and prognosis prediction of CRC [9]. For instance, serum levels of miR-21, miR-29a, and miR-125b were significantly increased in patients with early colorectal neoplasm, and could effectively discriminated CRC patients from healthy controls [10]. Similarly, a four-miRNA signature including miR-23a-3p, miR-27a-3p, miR-142-5p and miR-376c-3p could distinguish CRC patients from healthy subjects with high accuracy. In addition, serum miR-23a-3p and miR-376c-3p levels seem closely correlated with the prognosis of CRC [11].
MiR-206, locates on chromosome 6p12.2, is highly conserved in genomic organization and sequence [12]. Accumulating evidence has showed that miR-206 involved in the initiation and progression of CRC [13-15]. However, the clinical significance of serum or plasma miR-206 in CRC had not yet been explored. In the current study, we aimed to evaluate the diagnostic and prognostic value of serum miR-206 in patients with CRC.
Materials and methods
Ethics statement
All subjects provided written informed consent, and our study was conducted with the approval of the First Affiliated Hospital of Anhui Medical University. All serum samples were handled and made anonymous according to the ethical and legal standards.
Patient features and blood sample collection
In our study, a total of 73 stage II/III CRC patients aged between 43 and 69 years were recruited. Patients were excluded if they had received chemotherapy or radiotherapy prior to surgery. Tumor stages were evaluated according to the criteria of the Union for International Cancer Control (UICC) Staging System. In addition, we enrolled 45 healthy individuals between 38 to 62 years old as controls. Detailed clinicopathological characteristics of CRC patients were presented in Table 1. All patients accepted routine follow-up. Overall survival (OS) was defined as the time from the date of surgery to the date of death from tumor relapse or last follow-up. Disease-free survival (DFS) was defined as the time from the date of surgery to the date of relapse or death from relapse or last follow-up.
Table 1.
Correlation between miR-206 expression in serum with different clinicopathological features in CRC
| Factors | Patients (n=73) | Low miR-206 (n=37) | High miR-206 (n=36) | P value |
|---|---|---|---|---|
| Age | .160 | |||
| <60 | 32 | 13 (35.1%) | 19 (52.8%) | |
| ≥60 | 41 | 24 (64.9%) | 17 (47.2%) | |
| Gender | .341 | |||
| Man | 44 | 20 (54.1%) | 24 (66.7%) | |
| Woman | 29 | 17 (45.9%) | 12 (33.3%) | |
| Tumor type | .640 | |||
| Colon cancer | 33 | 18 (48.6%) | 15 (41.7%) | |
| Rectal cancer | 40 | 19 (51.4%) | 21 (58.3%) | |
| TNM stage | .001 | |||
| II | 47 | 17 (45.9%) | 30 (83.3%) | |
| III | 26 | 20 (54.1%) | 6 (16.7%) | |
| Tumor size (cm) | .102 | |||
| <4 | 39 | 16 (43.2%) | 23 (63.9%) | |
| ≥4 | 34 | 21 (56.8%) | 13 (36.1%) | |
| Histological grade | .121 | |||
| Well or moderate | 52 | 23 (62.2%) | 29 (80.6%) | |
| Poor | 21 | 14 (37.8%) | 7 (19.4%) | |
| Lymph node metastasis | .010 | |||
| Yes | 35 | 12 (32.4%) | 23 (63.9%) | |
| No | 38 | 25 (67.6%) | 13 (36.1%) |
Paired blood samples were obtained from 73 CRC patients before and after their cancer resection surgeries. Moreover, venous blood was withdrawn from each healthy volunteer. All blood samples were centrifuged at 3000 r/min for 10 min within 30 min after collection. Supernatants were stored at -80°C until further use.
Total RNA isolation and real-time RT-PCR
Total RNA was isolated from serum samples using a miRcute miRNA isolation kit (Tiangen Biotech, Beijing, China). Reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Real-time PCR was conducted using Maxima SYBR Green qPCR Kit (Thermo Scientific, CA, USA), and carried out on an ABI PRISM 7500 Sequence Detection System (Applied Biosystems, Foster City, USA). Each reaction was independently repeated in triplicate. Relative expression levels of miR-206 were normalized to the reference gene RNU6 and calculated using the 2-ΔΔCt method.
Statistical analysis
All statistical analyses were conducted using MedCalc 15.6.1 (MedCalc, Mariakerke, Belgium). P value less than 0.05 was considered significant. The Mann-Whitney U test was conducted to compare the difference in serum miR-206 levels between two groups. The Fisher’s exact test was used to analyze the correlation between clinical variables and serum miR-206 expression levels. Receiver-operating-characteristic (ROC) curve and the area under the curve (AUC) were used to determine the diagnostic accuracy of serum miR-206. Overall and disease free survival curves were constructed using the Kaplan-Meier method, and differences were compared by log-rank tests. Multivariate Cox proportional hazards model was used to estimate risk ratios for relapse and prognosis.
Results
Serum miR-206 was significantly decreased in CRC patients and its diagnostic value
We detected miR-206 levels in 73 CRC patients and 45 healthy subjects using qRT-PCR. The expression of serum miR-206 was greatly down-regulated in CRC patients compared to the healthy controls (P<0.05, Figure 1). In addition, serum miR-206 levels in patients with stage II patients (n=47) were higher than those in patients with stage III (n=26). Moreover, a significant downregulation in serum miR-206 expression was observed in patients with lymph node metastasis (n=38) when compared to patients without metastasis (n=35) (both P<0.05, Figure 2). Interestingly, the levels of serum miR-206 in post-operative blood samples of CRC patients were dramatically elevated compared to those in paired pre-operative samples (P<0.01, Figure 3).
Figure 1.
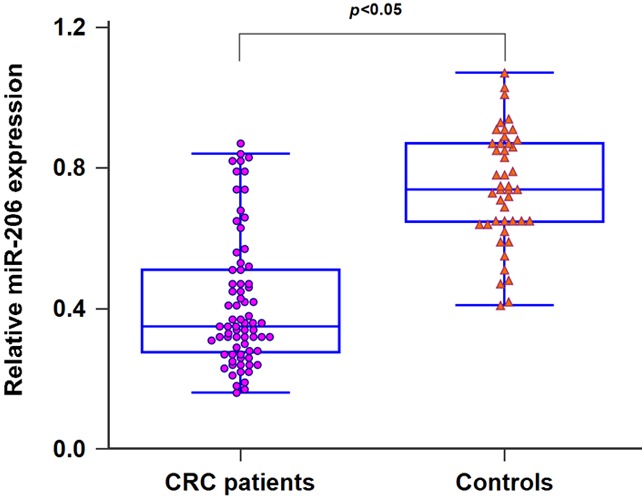
MiR-206 expression was significantly decreased in CRC patients in comparison with healthy controls.
Figure 2.
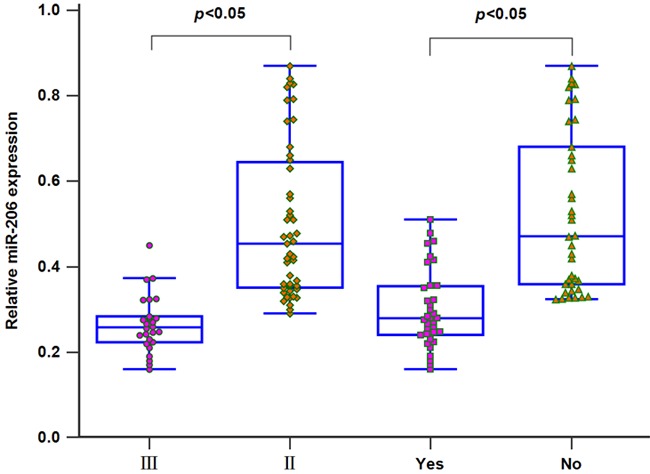
Low miR-206 expression was frequently associated with higher TNM stage and lymph node metastasis.
Figure 3.
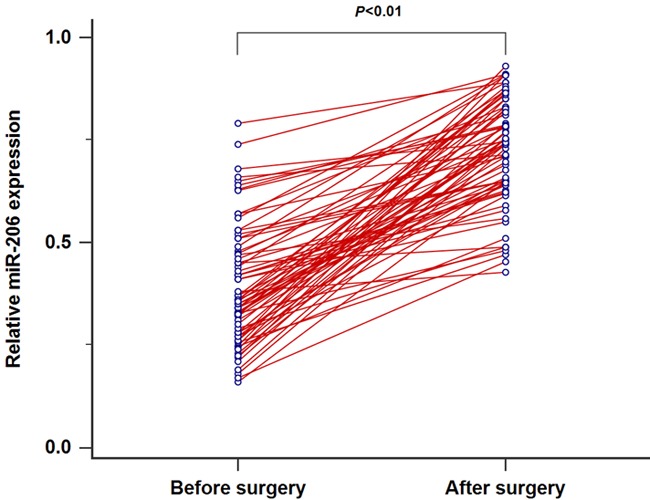
MiR-206 expression was significantly up-regulated in CRC patients after surgery.
Additionally, the results of ROC curve analysis showed that AUC value for serum miR-206 was 0.846, and the specificity and sensitivity were 80.00% and 82.19%, respectively. These findings suggested miR-206 might serve as serum indicator for discriminating patients with CRC from healthy controls (Figure 4).
Figure 4.
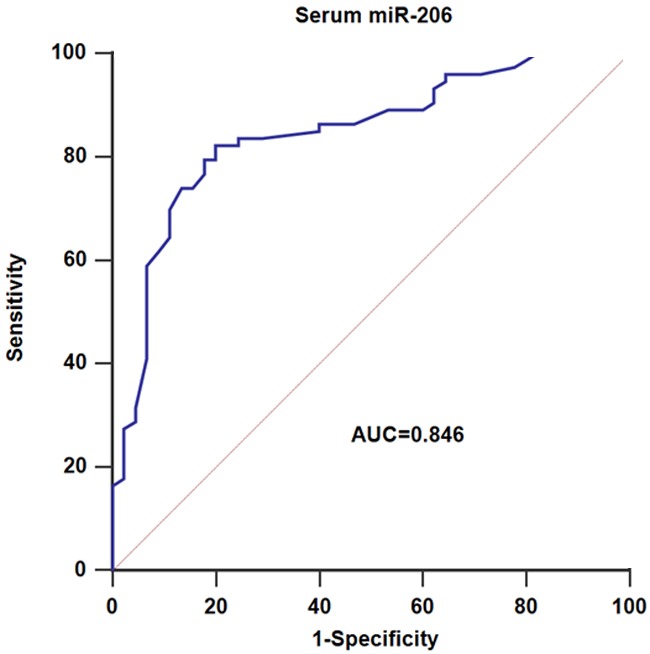
Serum miR-206 yielded AUC values of 0.846 with 82.19% sensitivity and 80.00% specificity in discriminating CRC patients from normal controls.
Serum miR-206 correlated with clinical characteristics in CRC patients
We divided all the 73 CRC patients into two groups based on the median serum miR-206 level. The high expression group had 36 subjects while the low expression group had 37 people. As shown in Table 1, serum miR-206 expression was found to be strongly associated with TNM stage (P=0.001), and lymph node metastasis (P=0.010). However, no correlation was observed between serum miR-206 expression and age (P=0.160), gender (P=0.341), tumor type (P=0.640), tumor size (P=0.102), histological grade (P=0.121).
Association between serum miR-206 expression and survival in CRC patients
The Kaplan-Meier analysis revealed that patients in high serum miR-206 expression group had longer overall survival (P=0.022, Figure 5A) and disease free survival (P=0.013, Figure 5B) than those in low expression group.
Figure 5.
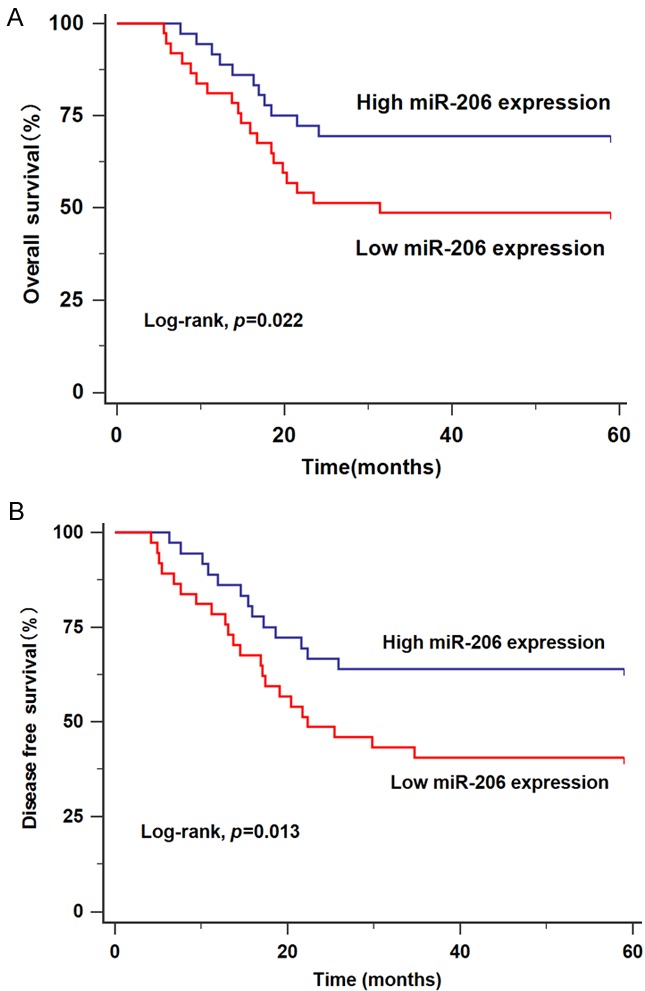
A. CRC patients with low miR-206 expression (n=37) had worse overall survival than those with high miR-206 expression (n=36). B. CRC patients with low miR-206 expression (n=37) had worse disease free survival than those with high miR-206 expression (n=36).
Serum miR-206 was an independent prognostic factor for CRC patients
Multivariate Cox proportional hazard regression model was performed and demonstrated that serum miR-206 expression (OS: RR=1.75, 95% CI=1.14-2.45, P=0.034; DFS: RR=1.94, 95% CI=1.20-2.73, P=0.027), lymph node metastasis (OS: RR=2.54, 95% CI=1.32-3.91, P=0.021; DFS: RR=2.96, 95% CI=1.44-4.78, P=0.014) and TNM stage (OS: RR=3.62, 95% CI=1.63-5.82, P=0.008; DFS: RR=4.15, 95% CI=1.81-6.54, P=0.003) were independent predictors for CRC patients (Table 2).
Table 2.
Multivariate analysis of parameters associated with overall survival and disease free survival of CRC patients
| Parameters | OS | DFS | ||
|---|---|---|---|---|
|
| ||||
| RR (95% CI) | P | RR (95% CI) | P | |
| TNM stage | 3.62 (1.63 to 5.82) | .008 | 4.15 (1.81 to 6.54) | .003 |
| Lymph node metastasis | 2.54 (1.32 to 3.91) | .021 | 2.96 (1.44 to 4.78) | .014 |
| MiR-206 in serum | 1.75 (1.14 to 2.45) | .034 | 1.94 (1.20 to 2.73) | .027 |
Footnote: CI = confidence interval; RR = risk ratio; OS = overall survival; DFS = disease free survival.
Discussion
Previous reports have revealed that miR-206 played a tumor suppressive role in CRC. For instance, Wang et al showed that over-expression of miR-206 repressed CRC cell proliferation, migration and induced apoptosis [13]. Moreover, Ren et al found that forced miR-206 expression greatly inhibited CRC cell invasion and promoted apoptosis by directly targeting FMNL2 [14]. In addition, Sun and colleagues demonstrated that miR-206 level was markedly lower in CRC tissues in comparison with adjacent normal controls. Down-regulation of miR-206 was closely associated with more unfavorable disease characteristics and shorter overall survival [15]. In the present study, we demonstrated that serum miR-206 levels were remarkably lower in CRC patients and showed good diagnostic performance for discriminating CRC patients from controls. Also, serum miR-206 levels were elevated in those patients who received surgery. Subsequently, a positive association was found between decreased serum miR-206 expression and worse clinical outcome as well as unfavorable OS/DFS. Furthermore, multivariate analysis confirmed that serum miR-206 was an independent prognostic predictor for CRC. Therefore, these data provided strong evidence that miR-206 might potentially function as a tumor suppressor in CRC.
Besides CRC, miR-206 was also found to exhibit tumor suppressive properties in some other tumor types. For instance, down-regulation of miR-206 in breast cancer tissues was correlated with larger tumor size and advanced clinical stage. Additionally, enforced miR-206 expression suppressed cancer cell growth, proliferation and colony formation in vitro by targeting cyclinD2 [16]. In gastric cancer, enhanced miR-206 expression restrained cell growth and metastasis. Moreover, gastric cancer patients with lower miR-206 expression suffered poorer prognosis [17,18]. Zhang et al showed that miR-206 expression was significantly reduced in both osteosarcoma tissues and serum. In addition, down-regulation of miR-206 was positively associated with worse clinical outcome and poorer survival [19]. In hepatocellular carcinoma, miR-206 expression was greatly underexpressed in cancer tissues compared with paracancerous normal tissues. Moreover, low miR-206 expression was associated with poor tumor differentiation, multiple tumor nodes, lymph node metastasis, and advanced TNM stage [20]. In non-small cell lung cancer, Zhang and colleagues demonstrated that miR-206 was decreased in cancer tissues compared with adjacent normal tissues. Furthermore, increased miR-206 expression significantly suppressed cancer cell proliferation, migration and invasion via targeting SOX9 [21]. In glioblastoma, enforced miR-206 level remarkably restrained cancer cell proliferation in vitro and inhibited tumorigenicity in vivo through regulating BCL-2 [22]. All these results consisted with our findings, and suggested that miR-206 acted as a tumor suppressor. It should be noted that the present work lacked a larger cohort of CRC cases, and our results needs further validation in future analysis.
In conclusion, the current study provided compelling evidence that serum miR-206 had great potential to be used as diagnostic and prognostic biomarker for CRC.
Acknowledgements
This study was partly supported by the Department of Gastrointestinal Surgery, the First Affiliated Hospital of Anhui Medical University.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Zheng ZX, Zheng RS, Zhang SW, Chen WQ. Colorectal cancer incidence and mortality in China, 2010. Asian Pac J Cancer Prev. 2014;15:8455–8460. doi: 10.7314/apjcp.2014.15.19.8455. [DOI] [PubMed] [Google Scholar]
- 3.Akkoca AN, Yanik S, Ozdemir ZT, Cihan FG, Sayar S, Cincin TG, Cam A, Ozer C. TNM and modified dukes staging along with the demographic characteristics of patients with colorectal carcinoma. Int J Clin Exp Med. 2014;7:2828–2835. [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh JM, Terdiman JP. Colorectal cancer screening: scientific review. JAMA. 2003;289:1288–1296. doi: 10.1001/jama.289.10.1288. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 7.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brainspecific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 8.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng G, Du L, Yang X, Zhang X, Wang L, Yang Y, Li J, Wang C. Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br J Cancer. 2014;111:1985–1992. doi: 10.1038/bjc.2014.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada A, Horimatsu T, Okugawa Y, Nishida N, Honjo H, Ida H, Kou T, Kusaka T, Sasaki Y, Yagi M, Higurashi T, Yukawa N, Amanuma Y, Kikuchi O, Muto M, Ueno Y, Nakajima A, Chiba T, Boland CR, Goel A. Serum miR-21, miR-29a, and miR-125b are promising biomarkers for the early detection of colorectal neoplasia. Clin Cancer Res. 2015;21:4234–4242. doi: 10.1158/1078-0432.CCR-14-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vychytilova-Faltejskova P, Radova L, Sachlova M, Kosarova Z, Slaba K, Fabian P, Grolich T, Prochazka V, Kala Z, Svoboda M, Kiss I, Vyzula R, Slaby O. Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer. Carcinogenesis. 2016;37:941–950. doi: 10.1093/carcin/bgw078. [DOI] [PubMed] [Google Scholar]
- 12.Townley-Tilson WH, Callis TE, Wang D. MicroRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int J Biochem Cell Biol. 2010;42:1252–1255. doi: 10.1016/j.biocel.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang XW, Xi XQ, Wu J, Wan YY, Hui HX, Cao XF. MicroRNA-206 attenuates tumor proliferation and migration involving the downregulation of NOTCH3 in colorectal cancer. Oncol Rep. 2015;33:1402–1410. doi: 10.3892/or.2015.3731. [DOI] [PubMed] [Google Scholar]
- 14.Ren XL, He GY, Li XM, Men H, Yi LZ, Lu GF, Xin SN, Wu PX, Li YL, Liao WT, Ding YQ, Liang L. MicroRNA-206 functions as a tumor suppressor in colorectal cancer by targeting FMNL2. J Cancer Res Clin Oncol. 2016;142:581–592. doi: 10.1007/s00432-015-2053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun P, Sun D, Wang X, Liu T, Ma Z, Duan L. miR-206 is an independent prognostic factor and inhibits tumor invasion and migration in colorectal cancer. Cancer Biomark. 2015;15:391–396. doi: 10.3233/CBM-150489. [DOI] [PubMed] [Google Scholar]
- 16.Zhou J, Tian Y, Li J, Lu B, Sun M, Zou Y, Kong R, Luo Y, Shi Y, Wang K, Ji G. miR-206 is downregulated in breast cancer and inhibits cell proliferation through the up-regulation of cyclinD2. Biochem Biophys Res Commun. 2013;433:207–212. doi: 10.1016/j.bbrc.2013.02.084. [DOI] [PubMed] [Google Scholar]
- 17.Zheng Z, Yan D, Chen X, Huang H, Chen K, Li G, Zhou L, Zheng D, Tu L, Dong XD. MicroRNA-206: effective inhibition of gastric cancer progression through the c-Met pathway. PLoS One. 2015;10:e0128751. doi: 10.1371/journal.pone.0128751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Q, Zhang C, Huang B, Li H, Zhang R, Huang Y, Wang J. Downregulation of microRNA-206 is a potent prognostic marker for patients with gastric cancer. Eur J Gastroenterol Hepatol. 2013;25:953–957. doi: 10.1097/MEG.0b013e32835ed691. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C, Yao C, Li H, Wang G, He X. Serum levels of microRNA-133b and microRNA-206 expression predict prognosis in patients with osteosarcoma. Int J Clin Exp Pathol. 2014;7:4194–4203. [PMC free article] [PubMed] [Google Scholar]
- 20.Yunqiao L, Vanke H, Jun X, Tangmeng G. MicroRNA-206, down-regulated in hepatocellular carcinoma, suppresses cell proliferation and promotes apoptosis. Hepatogastroenterology. 2014;61:1302–1307. [PubMed] [Google Scholar]
- 21.Zhang YJ, Xu F, Zhang YJ, Li HB, Han JC, Li L. miR-206 inhibits non small cell lung cancer cell proliferation and invasion by targeting SOX9. Int J Clin Exp Med. 2015;8:9107–9113. [PMC free article] [PubMed] [Google Scholar]
- 22.Hao W, Luo W, Bai M, Li J, Bai X, Guo J, Wu J, Wang M. microRNA-206 inhibited the progression of glioblastoma through BCL-2. J Mol Neurosci. 2016;60:531–538. doi: 10.1007/s12031-016-0824-6. [DOI] [PubMed] [Google Scholar]


