Abstract
Long non-coding RNAs (LncRNAs) are thought to be involved in several biological processes in carcinomas. The aim of this study is to evaluate the roles of lncRNA-XIST in the tumorigenicity of renal cell carcinoma (RCC) cells via the miR-302c/SDC1 axis. In this study, the expression levels of miR-302c and XIST in RCC tissues and cells were analyzed by qRT-PCR. Cell proliferation was measured using MTT and colony formation assays, and cell apoptosis was detected using flow cytometry. The interaction between XIST and miR-302c was analyzed using a luciferase reporter gene assay. RCC tissues and cells exhibited decreased miR-302c expression and increased lncRNA-XIST expression. Furthermore, XIST negatively regulated miR-302c by directly binding regulatory sites in RCC cells. In addition, XIST silencing with siRNAs significantly inhibited the proliferation and promoted the apoptosis of 786-O and Caki-1 cells. Knockdown of Syndecan-1 (SDC1), a miR-302c target gene, yielded similar results as XIST silencing. In summary, XIST regulated the development and progression of RCC by inhibiting the miR302c/SDC1 axis.
Keywords: LncRNA XIST, miR-302c, SDC1, renal cell carcinoma
Introduction
Renal cell carcinoma (RCC) is the most common type of kidney cancer [1,2] and accounts for approximately 2 percent of all cancers [3]. The incidence of RCC has drastically increased over the past few decades. In addition, thousands of people with RCC die every year, because patents with RCC respond poorly to conventional treatments such as radiation and chemotherapy [4,5]. Therefore, studies elucidating the mechanisms underlying the pathogenesis of RCC with the aim of identifying more effective therapies are extremely urgently needed.
Long non-coding RNAs (lncRNAs) represent the major form of noncoding RNAs and are thought to play very important roles in various human cancers [6-8]. According to previous studies, lncRNAs have significant biological functions in proliferation, development [9], apoptosis [10] and differentiation [11]. The lncRNA XIST (X inactivated-specific transcript) is encoded by the XIST gene, which is the master regulator of X inactivation, and has been reported to be involved in breast cancer, hepatocellular carcinoma, and glioblastoma [12,13]. However, researchers have not clearly determined whether XIST plays an important role in RCC. In this study, we aimed to explore the role of XIST in RCC and postulate that it may represent a new effective treatment for RCC.
MicroRNAs (miRNAs), a class of small noncoding RNAs of approximately 20 nucleotides in length, are thought to regulate the expression of numerous mammalian genes by affecting mRNA translation [14,15]. As shown in previous studies, miRNAs are associated with various human diseases, including cancers such as breast cancer, lung cancer and glioblastoma [16-20]. Among these miRNAs, the miR-302 family (miR-302s), which includes four homologous miRNAs, have been confirmed to be involved in the proliferation, apoptosis and differentiation of human cancer cells [21]. However, the role of miR-302c is less clear, but it may also play a vital role in the development and progression of cancers, similar to other miR-302 family members. In our study, we examined the role of miR-302c and the XIST/miR-302c regulatory network in the development and progression of RCC.
Materials and methods
Patients and tissue samples
Twenty-six pairs of renal cancer tissue samples and adjacent normal renal tissue samples were collected from the First Affiliated Hospital of Soochow University between 2015/05 and 2016/07. The histological diagnosis of renal cancer was confirmed according to the guidelines of the World Health Organization (WHO). Informed consent was obtained from every patient and ethical approval was granted by the Ethics Committee of The First Affiliated Hospital of Soochow University. All tissue samples were stored at -80°C before use.
Cell culture
Human renal cell carcinoma cell lines (786-O, Caki-1, ACHN and 769-P), a human renal proximal tubule epithelial cell line (HK-2), and HEK293T cells were all obtained from ATCC (Rockville, MD, USA). HEK293T, 786-O, and Caki-1 cell lines were routinely cultured in Dulbecco’s modified Eagle medium (DMEM) medium (Gibco, Gaithersburg, MD), whereas HK-2 cells were cultured in F-12K medium (Gibco, Gaithersburg, MD), supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 mg/ml streptomycin in a humidified atmosphere containing 5% CO2 at 37°C.
Lentiviral vector construction
The human lncRNA-XIST DNA was amplified from 786-O cells using RT-PCR. PCR products were cloned into a lentiviral vector. Enhanced green fluorescent protein (GFP) was used as the control. Lentiviral vectors expressing EGFP and targeting lncRNA-XIST were generated. The lentiviral vectors were packaged in HEK293T cells by co-transfecting the cells with packaging vectors (pCMV-VSVG, pMDLg/pRRE and pRSV-REV). Lentiviruses were ultracentrifuged and concentrated, as described in a previous study [22]. The 786-O and Caki-1 cells (1×104 cells/well) were seeded in 24-well plates and transfected with XIST and control vectors using 8 μg/ml polybrene (Sigma). Cells stably expressing the vectors were then screened with 800 μg/ml G418 (Sigma).
siRNAs, oligonucleotides, and transfections
The siRNAs targeting human XIST (si-XIST) and human SDC1 (si-SDC1), and the negative control siRNA (si-Control) were designed by and purchased from GenePharma (Shanghai, China). Oligonucleotides for the miR-302 mimics (5’-TAAGTGCTTCCATGTTTCAGTGG-3’) were ordered from RiboBio (China). The 786-O and Caki-1 cells were seeded into 6-well plates, incubated overnight, and then transfected with 50 nM oligonucleotides using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
Reverse transcription and real-time PCR
Total RNAs were extracted from RCC tissues, relatively normal renal tissues, and the treated 786-O and Caki-1 cells using TRIzol reagent (Invitrogen, CA, USA), according to the manufacturer’s instructions, and reverse transcribed into cDNAs using the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher, no. K1622), according to the manufacturer’s protocol. Real-time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) was performed to determine the expression levels of miR-302c and SDC1 using SYBR Premix Ex Taq (Takara, Japan). The primers for housekeeping genes, GAPDH and U6 (internal loading control), were designed and synthesized by Sangon in China. The sequences of the GAPDH primers are: 5’-CCT CGT CTC ATA GAC AAG ATG GT-3’ (the forward primer) and 5’-GGG TAG AGT CAT ACT GGA ACA TG-3’ (the reverse primer). The sequences of the XIST primers are: 5’-CTC TCC ATT GGG TTC AC-3’ (the forward primer) and 5’-GCG GCA GGT CTT AAG AGA TGA G-3’ (the reverse primer) [23]. The sequences of the SDC1 primers are: 5’-ATG AGA CGC GCG GCG CTC TG-3’ (the forward primer) and 5’-CTG ATT GGC AGT TCC ATC CT-3’ (the reverse primer) [24]. The sequences of the U6 primers are: 5’-CTC GCT TCG GCA GCA CA-3’ (the forward primer) and 5’-AAC GCT TCA CGA ATT TGC GT-3’ (the reverse primer). The sequences of the hsa-miR-302c primers are: 5’-CGG GAC TAG TCG GGA GGG GAG GTC AGA ATA A-3’ (the forward primer) and 5’-GGT CGA CGC GTC AGG CAG CTA CAT CTA CTG CTA AAA-3’ (the reverse primer). The expression levels of the XIST and SDC1 mRNAs were normalized to the GAPDH mRNA levels. The expression level of the hsa-miR-302c mRNA was normalized to the U6 mRNA level. All reactions were performed in triplicate and the 2-ΔΔCt method was employed to evaluate the differences in XIST, SDC1, and hsa-miR-302c expression.
MTT assay
The transfected 786-O and Caki-1 cells were seeded in 96-well plates at a density of 8,000 cells per well and were routinely cultured completed medium for 0, 24 or 48 hrs. Then, cells were stained with 0.5 mg/ml sterile MTT (Sigma, MO, USA) for 4 hrs at 37°C. The original culture medium was discarded and 100 μl of dimethyl sulfoxide (DMSO, Sigma, MO, USA) were added. The absorbance was measured at 450 nm after transfection.
Colony formation assay
The treated cells (500 cells/well) were seeded on dishes and cultured for 2 weeks at 37°C with 5% CO2 (the medium was changed every three days). Then, the colonies were stained with 0.1% crystal violet and the number of colonies was counted.
Flow cytometry analysis
The rate of apoptotic cell death was analyzed by flow cytometry and double staining using Annexin V-APC/7-AAD (BD Pharmingen, San Diego, CA), according to the manufacturer’s instructions. Forty-eight hours after transfection, the cells were harvested, washed twice with cold PBS, and re-suspended in 1× binding buffer at a concentration of 1×106 cells/mL. Annexin V-APC/7-AAD were added to the cells suspended in buffer solution (1×105 cells), incubated for 15 min at RT in the dark, and then 400 μL of 1× binding buffer were added to each sample tube. Cell apoptosis was analyzed using a BD FACSCalibur cytometer (BD Biosciences). The results were analyzed using FlowJo software version 8.8.6 (TreeStar Inc., Ashland, Oregon, USA).
Bioinformatics analysis and fluorescent reporter assay
The microRNA targets were predicted using starBase v2.0 and TargetScan v6.2. Four putative XIST target sites were separately cloned into the luciferase expression vector pGL3-Bacsic. Cells (5000 cells/well) were cultured in 24-well plates and co-transfected with the wild type (XIST-WT) or mutant (XIST-Mut) plasmids and miR-302c using Lipofectamine 3000 (Invitrogen). Luciferase activities were detected using the Dual-Luciferase reporter assay according to the manufacturer’s instructions (Promega, Madison, WI).
Western blotting
Cultured cells were lysed in RIPA buffer (0.1% SDS, 1% Triton X-100, 1 mM MgCl2, and 10 mM Tris-HCl (pH 7.4) containing a protease inhibitor cocktail (P8340; Sigma-Aldrich, St. Louis, MO, USA)) for 30 mins at 4°C. Total protein concentrations were measured using a BCA Protein Assay kit (Thermo Fisher Scientific, Rockford, IL, USA). The proteins (50 μg) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes (Millipore, Billerica, MA, USA). Nonspecific binding sites on the membranes were saturated with 5% skim milk in TBST (100 mmol/L Tris-Cl, pH 7.5, 150 mmol/L NaCl, and 0.1% Tween 20) and then incubated with primary antibodies overnight, followed by horseradish peroxidase-conjugated secondary antibodies. The following antibodies were used: anti-SDC1 (1:200, Abcam, Cambridge, UK) and anti-actin (1:2000, Cell Signaling Technology Inc., Danvers, Massachusetts, USA).
Statistical analysis
The statistical significance of the differences was analyzed using GraphPad Prism Software (La Jolla, CA, USA) and SPSS 21.0 software (analysis of variance and Student’s t-test or one-way analysis of variance (ANOVA)). Data are expressed as means ± standard deviations (SD), and P < 0.05 is considered a statistically significant difference.
Results
miR-302c expression was down-regulated and XIST expression was up-regulated in human renal cell carcinoma tissues and cells
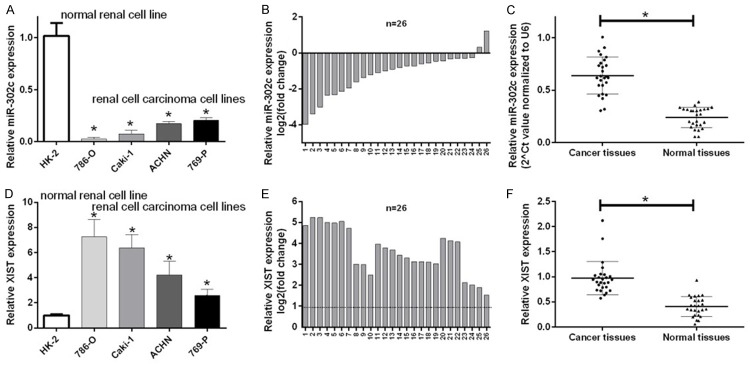
We examined the expression levels of XIST and miR-302c in the normal renal cell line (HK-2), and renal cell carcinoma cell lines (86-O, Caki-1, ACHN, and 769-P) to determine whether XIST and miR-302c were involved in the tumorigenesis of RCC. Compared with normal renal cells (HK-2), miR-302c expression was significantly down-regulated in renal cell carcinoma cells (786-O, Caki-1, ACHN, 769-P) (P < 0.05, Figure 1A). We then examined miR-302c expression in 26 pairs of RCC tissues and their corresponding adjacent normal tissues and showed that miR-302c expression was significantly down-regulated in the 24 of 26 RCC tissues compared with adjacent normal tissues (P < 0.05, Figure 1B and 1C). Similarly, we further investigated the expression of the XIST mRNA in RCC tissues and cells. The expression of the XIST mRNA was drastically up-regulated in RCC cell lines compared to HK-2 cells (P < 0.05, Figure 1D). Simultaneously, the expression of the XIST mRNA was drastically up-regulated in all 26 RCC tissues compared with the normal counterparts (P < 0.05, Figure 1E and 1F). Therefore, XIST and miR302c may play critical roles in RCC progression.
Figure 1.
The expression of miR-302c was down-regulated and lncRNA XIST expression was upregulated in renal cell carcinoma cells and tissues. A. The expression of miR-302c was detected by qRT-PCR in a human renal proximal tubule epithelial cell line (HK-2) and four RCC cell lines (786-O, Caki-1, ACHN and 769-P) (means ± SD, *P < 0.05). B and C. The relative miR-302c expression levels in 26 pairs of RCC tissues and their corresponding adjacent normal tissues were analyzed using qRT-PCR (means ± SD, *P < 0.05). D. XIST expression in normal HK-2 cells and RCC cell lines (786-O, Caki-1, ACHN and 769-P) was measured using qRT-PCR (means ± SD, *P < 0.05). E and F. The relative XIST expression levels were detected in 26 pairs of RCC tissues and their corresponding adjacent normal tissues (means ± SD, *P < 0.05).
XIST is negatively regulated by miR-302c in RCC cells
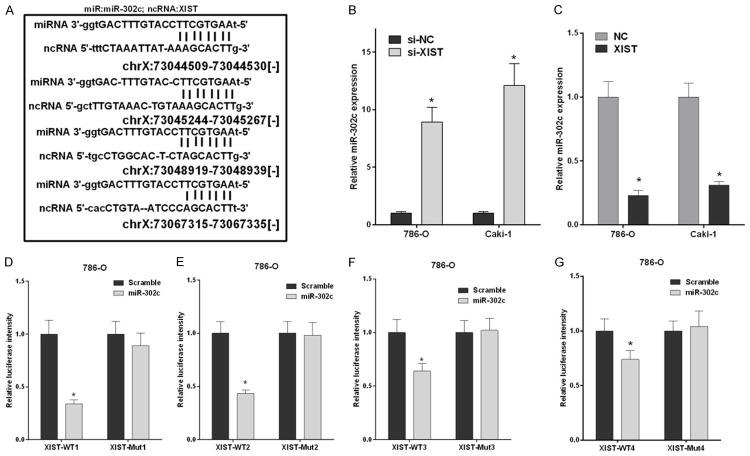
Based on accumulating evidence, numerous lncRNA transcripts function as competing endogenous RNAs (CeRNA) by competitively binding miRNAs. Thus, we predicted the interaction between miR-302c and potential lncRNAs using starBase v2.0 and identified four miR-302c binding sites in the XIST sequence (Table 1; Figure 2A). The 786-O and Caki-1 cells were transfected with si-XIST and si-NC, as well as XIST and NC and miR-302c expression was detected by qRT-PCR to further examine the relationship between XIST and miR-302c. 786-O and Caki-1 cells that had been transfected with si-XIST exhibited a significant increase in miR-302 expression compared to cells transfected with si-NC (P < 0.05, Figure 2B). Meanwhile, miR-302c expression was substantially decreased in 786-O and Caki-1 cells that had been transfected with XIST compared to cells transfected with NC (P < 0.05, Figure 2C). We further cloned the four miR-302c binding sites in XIST into pGL3/luciferase vectors (XIST-WT1, XIST-WT2, XIST-WT3, and XIST-WT4) and constructed their corresponding mutant pGL3/luciferase vectors (XIST-Mut1, XIST-Mut2, XIST-Mut3, and XIST-Mut4). Then, 786-O cells were co-transfected with XIST-WT1, WT2, WT3, or WT4 and miR-302c, or were simultaneously co-transfected with XIST-Mut1, Mut 2, Mut 3, or Mut 4 and miR-302c. Luciferase activities were obviously decreased in cells expressing wild type XISTs (XIST-WT1, XIST-WT2, XIST-WT3, and XIST-WT4) compared with the scrambled groups. However, after co-transfection with XIST-Mut1, Mut2, Mut3, or Mut4 and miR-302c, no changes in the luciferase activities of mutant type XISTs (XIST-Mut1, XIST-Mut2, XIST-Mut3, and XIST-Mut4) were observed compared with cells transfected with the scrambled sequence (P < 0.05, Figure 2D-G). Therefore, XIST was negatively regulated by miR-302c in RCC cells.
Table 1.
The binding sites between hsa-miR-302c-3p and potential lncRNAs were predicted by starBase v2.0
| Name | mirAccession | GeneName | TargetSites |
|---|---|---|---|
| hsa-miR-302c-3p | MIMAT0000717 | RP11-197N18.2 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | LINC00657 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-392M18.5 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-355O1.11 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-68L18.1 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | HNRNPU-AS1 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | AL589743.1 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | SNHG16 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | CT49 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | XIST | 4 |
| hsa-miR-302c-3p | MIMAT0000717 | MIR4720 | 2 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-145M9.4 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | HOXA-AS2 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-139H15.1 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-379K17.11 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-197P3.5 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | AC108142.1 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-31E23.1 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-498C9.15 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | MAP3K14 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | COX10-AS1 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-701H24.4 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-315C6.3 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | CASP8AP2 | 3 |
| hsa-miR-302c-3p | MIMAT0000717 | C11orf95 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-156E6.1 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-615I2.7 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-305N23.1 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | CTD-2162K18.5 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | LINC00338 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | CTC-444N24.11 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP5-1024G6.5 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-80H5.7 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | CTB-92J24.2 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | hsa-mir-6080 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-701H24.2 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | AC005083.1 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | GS1-358P8.4 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-220I1.1 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP1-39G22.7 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | SCAMP1 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | LINC00472 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-73M18.8 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | CTD-2037K23.2 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-140H17.1 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP5-1024G6.8 | 2 |
| hsa-miR-302c-3p | MIMAT0000717 | CTA-204B4.6 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | HCG18 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-429D19.1 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-399O19.8 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP4-773N10.5 | 2 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-509J21.3 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | AL163636.6 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | RP11-96D1.6 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | MAGI1-IT1 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | KB-1460A1.5 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | PPP1R9B | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | ZNF518A | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | FGD5-AS1 | 2 |
| hsa-miR-302c-3p | MIMAT0000717 | MIR17HG | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | BX322557.10 | 1 |
| hsa-miR-302c-3p | MIMAT0000717 | BAIAP2-AS1 | 1 |
Figure 2.
XIST targeted miR-302c in RCC cells. A. Four possible miR-302c binding sites in XIST are shown and named XIST-WT1, XIST-WT2, XIST-WT3, and XIST-WT4. B. The relative miR-302c expression levels in 786-O and Caki-1 cells transfected with XIST siRNAs (si-XIST) or the negative control (si-NC) were examined by qRT-PCR (*P < 0.05). C. The relative miR-302c expression levels in 786-O and Caki-1 cells transfected with XIST and the negative control (NC) were examined by qRT-PCR (*P < 0.05). D-G. 786-O cells were co-transfected with miR-302c and luciferase reporter gene vectors harboring four putative target sites (wild type XIST) or four mutated sites (mutant type XIST), and fluorescence was detected using the luciferase reporter gene assay. The relative luciferase activity was plotted as the means ± SD of three independent experiments (*P < 0.05).
XIST knockdown inhibited cell proliferation and promoted cell apoptosis in vitro
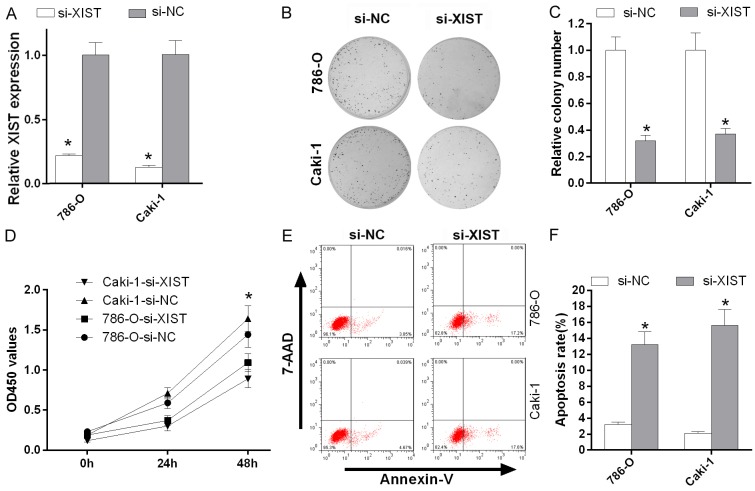
We inhibited XIST expression using XIST siRNAs in two RCC cell lines (786-O and Caki-1) to explore the roles of XIST in RCC cells. XIST expression was significantly decreased in 786-O and Caki-1 cells transfected si-XIST compared to cells transfected with si-NC (P < 0.05, Figure 3A). Based on the results of the colony formation assay and MTT assay, XIST knockdown significantly suppressed the proliferation of 786-O and Caki-1 cells (P < 0.05, Figure 3B-D). Furthermore, according to the results of the annexin V-APC/7-AAD staining assay, XIST silencing with siRNAs promoted the apoptosis of 786-O and Caki-1 cells (P < 0.05, Figure 3E and 3F).
Figure 3.
XIST knockdown inhibited cell proliferation and promoted cell apoptosis in vitro. A. The relative XIST expression levels in 786-O and Caki-1 cells transfected with si-XIST and si-NC were detected using qRT-PCR. B and C. The proliferation of 786-O and Caki-1 cells transfected with si-XIST and si-NC was detected using the colony forming assay (*P < 0.05). D. Silencing of XIST expression with siRNAs significantly inhibited the proliferation of 786-O and Caki-1 cells. Proliferation was detected using the MTT assay (*P < 0.05). E and F. The apoptosis of 786-O and Caki-1 cells transfected with si-XIST and si-NC was measured using Annexin V-APC/7-AAD staining (*P < 0.05).
miR-302c inhibited cell proliferation and promoted apoptosis by down-regulating SDC1 expression
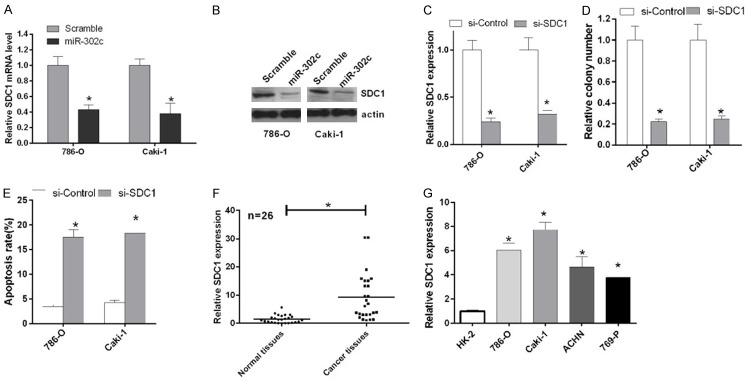
We used TargetScan v6.2 to predict potential miR-302c target genes and identified miR-302c complementary binding sites in the 3’UTR of the SDC1 mRNA. Based on the qRT-PCR results, the A expression level of the SDC1 mRNA was significantly reduced in 786-O and Caki-1 cells transfected miR-302c compared with groups transfected with the scrambled miRNA (P < 0.05, Figure 4A). According to the western blot results, miR-302c also inhibited the expression of the SDC1 protein (Figure 4B). We silenced SDC1 by transfecting 786-O and Caki-1 cells with si-SDC1 to examine the role of SDC1 in RCC cell proliferation. SDC1 expression was significantly decreased in 786-O and Caki-1 cells transfected with si-SDC1 compared with cells transfected with si-Control (P < 0.05, Figure 4C). Based on the results of the colony formation assay, SDC1 knockdown suppressed the proliferation of 786-O and Caki-1 cells (P < 0.05, Figure 4D), and SDC1 knockdown promoted cell apoptosis in the flow cytometry analysis (P < 0.05, Figure 4E). We also examined SDC1 expression in RCC cell lines and tissues and found that SDC1 was expressed at high levels in the RCC cell lines and tissues compared with the normal cells and tissues (P < 0.05, Figure 4F and 4G).
Figure 4.
miR-302c inhibited cell proliferation and promoted cell apoptosis by down-regulating SDC1 expression. A. The relative expression of the SDC1 mRNA in 786-O and Caki-1 cells transfected with miR-302c and the scrambled sequence was detected by qRT-PCR (*P < 0.05). B. Western blotting was used to examine the expression of the protein SDC1 in 786-O and Caki-1 cells transfected with miR-302c and the scrambled sequence. Actin was used as a protein loading control. C. The relative SDC1 levels in 786-O and Caki-1 cells transfected with si-control and si-SDC1 were detected by qRT-PCR (*P < 0.05). D. The proliferation of 786-O and Caki-1 cells transfected with si-control and si-SDC1 was detected using the colony forming assay (*P < 0.05). E. The apoptosis of 786-O and Caki-1 cells transfected with si-SDC1 and si-control was measured using flow cytometry (*P < 0.05). F. The relative expression of the SDC1 mRNA in 26 pairs of RCC tissues and their corresponding adjacent normal tissues was analyzed by qRT-PCR (*P < 0.05). G. The expression of miR-302c in HK-2 cells and RCC cell lines (786-O, Caki-1, ACHN and 769-P) was detected by qRT-PCR (*P < 0.05).
Discussion
LncRNAs, which are more than 200 nucleotides in length [25], have been shown to play key roles in regulating the expression of some genes by competitively suppressing miRNAs [26,27]. LncRNAs are reported to participate in the progression of human cancers by regulating cell proliferation, apoptosis, and invasion [28,29]. The lncRNA XIST, which belongs to the master regulator of X chromosome [30], has been shown to be involved in several human cancers, including renal cell carcinoma [31,32]. In the present study, we first examined XIST expression in RCC cell lines and found that XIST was expressed at high levels in RCC cells compared with normal HK-2 cells. Then, we further verified this finding in 26 paired RCC tissues and their corresponding normal tissues and found that XIST expression was drastically upregulated in all 26 RCC tissues compared with their normal counterparts. In addition, XIST knockdown with a siRNA inhibited renal cancer cell proliferation and tumor formation, and promoted cell apoptosis in vitro.
According to numerous studies, miR-302c is associated with different diseases [33-35]. In our study, we examined miR-302c expression in RCC cell lines and found that miR302c was significantly downregulated in RCC cells. Moreover, miR-302c expression was significantly down-regulated in RCC tissues, which strongly supports the findings of previous studies. In addition, XIST regulated miR-302c expression. Furthermore, we postulated that a ceRNA (competing endogenous RNAs) mechanism underlies these effects and concluded that XIST, miR-302c and SDC1 participate in the tumorigenesis of RCC.
SDC1, which is predominantly expressed by epithelial cells, is a heparin-sulfate proteoglycan (HSPG) that is involved in the tumorigenesis of human cancers [36]. SDC1 knockdown was previously shown to inhibit the development and progression of colorectal cancer [37]. In our study, the SDC1 gene was a direct target of the XIST/miR-302c axis, and SDC1 silencing also inhibited renal cancer cell proliferation and promoted cell apoptosis. In summary, our results provide solid evidence that XIST is highly expressed in renal cell carcinoma and regulates the tumorigenicity of renal cell carcinoma cells via the miR-302c/SDC1 axis, representing a potential therapy for RCC.
Acknowledgements
This work was support by National Natural Science Foundation of China (No. 81272839). And we thank the fund support from Professor Pu Jinxian, Department of Urology, and The First Affiliated Hospital of Suzhou University.
Disclosure of conflict of interest
None.
References
- 1.Motzer RJ, Hutson TE, Cella D, Reeves J, Hawkins R, Guo J, Nathan P, Staehler M, de Souza P, Merchan JR, Boleti E, Fife K, Jin J, Jones R, Uemura H, De Giorgi U, Harmenberg U, Wang J, Sternberg CN, Deen K, McCann L, Hackshaw MD, Crescenzo R, Pandite LN, Choueiri TK. Pazopanib versus sunitinib in metastatic renal-cell carcinoma. N Engl J Med. 2013;369:722–731. doi: 10.1056/NEJMoa1303989. [DOI] [PubMed] [Google Scholar]
- 2.Diaz JI, Mora LB, Hakam A. The Mainz classification of renal cell tumors. Cancer Control. 1999;6:571–579. doi: 10.1177/107327489900600603. [DOI] [PubMed] [Google Scholar]
- 3.Roberts JA, Kaack MB, Baskin G, Svenson SB. Prevention of renal scarring from pyelonephritis in nonhuman primates by vaccination with a synthetic escherichia coli serotype O8 oligosaccharide-protein conjugate. Infect Immun. 1993;61:5214–5218. doi: 10.1128/iai.61.12.5214-5218.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng J, Xiao W, Yu G, Yao W, Zhou H, Li H, Pan Y, Li A, Ye Z, Wang J, Xu H, Huang Q. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015;6:38005–38015. doi: 10.18632/oncotarget.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linehan WM. Genetic basis of kidney cancer: role of genomics for the development of disease-based therapeutics. Genome Res. 2012;22:2089–2100. doi: 10.1101/gr.131110.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J, Li X, Xu Y, Yang T, Yang Q, Yang C, Jiang Y. Identification of a long non-coding RNA NR_026689 associated with lung carcinogenesis induced by NNK. Oncotarget. 2016;7:14486–14498. doi: 10.18632/oncotarget.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson JM, Edwards S, Shoemaker D, Schadt EE. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Furuno M, Pang KC, Ninomiya N, Fukuda S, Frith MC, Bult C, Kai C, Kawai J, Carninci P, Hayashizaki Y, Mattick JS, Suzuki H. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eissmann M, Gutschner T, Hammerle M, Gunther S, Caudron-Herger M, Gross M, Schirmacher P, Rippe K, Braun T, Zornig M, Diederichs S. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9:1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossi MN, Antonangeli F. LncRNAs: new players in apoptosis control. Int J Cell Biol. 2014;2014:473857. doi: 10.1155/2014/473857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zequn N, Xuemei Z, Wei L, Zongjuan M, Yujie Z, Yanli H, Yuping Z, Xia M, Wei W, Wenjing D, Na F, Shuanying Y. The role and potential mechanisms of LncRNA-TATDN1 on metastasis and invasion of non-small cell lung cancer. Oncotarget. 2016;7:18219–18228. doi: 10.18632/oncotarget.7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun. 2016;478:811–817. doi: 10.1016/j.bbrc.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 13.Weakley SM, Wang H, Yao Q, Chen C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg. 2011;35:1751–1756. doi: 10.1007/s00268-010-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 16.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 17.Matsubara H, Takeuchi T, Nishikawa E, Yanagisawa K, Hayashita Y, Ebi H, Yamada H, Suzuki M, Nagino M, Nimura Y, Osada H, Takahashi T. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26:6099–6105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
- 18.Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 19.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 20.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 21.Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holkers M, Maggio I, Liu J, Janssen JM, Miselli F, Mussolino C, Recchia A, Cathomen T, Gonçalves MA. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013;41:e63. doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tantai JC, Hu DZ, Yang Y, Geng JF. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:7887–95. [PMC free article] [PubMed] [Google Scholar]
- 24.Stepp MA, Daley WP, Bernstein AM, Pal-Ghosh S, Tadvalkar G, Shashurin A, Palsen S, Jurjus RA, Larsen M. Syndecan-1 regulates cell migration and fibronectin fibril assembly. Exp Cell Res. 2010;316:2322–2339. doi: 10.1016/j.yexcr.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Zhang HM, Yang FQ, Chen SJ, Che J, Zheng JH. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36:2947–2955. doi: 10.1007/s13277-014-2925-6. [DOI] [PubMed] [Google Scholar]
- 27.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitehead J, Pandey GK, Kanduri C. Regulation of the mammalian epigenome by long noncoding RNAs. Biochim Biophys Acta. 2009;1790:936–947. doi: 10.1016/j.bbagen.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Feng T, Lian Y, Zhang G, Garen A, Song X. Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci U S A. 2009;106:12956–12961. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 31.Wu ZS, Lee JH, Kwon JA, Kim SH, Han SH, An JS, Lee JH, Lee ES, Park HR, Kim YS. Genetic alterations and chemosensitivity profile in newly established human renal collecting duct carcinoma cell lines. BJU Int. 2009;103:1721–1728. doi: 10.1111/j.1464-410X.2008.08290.x. [DOI] [PubMed] [Google Scholar]
- 32.Nomura S, Baxter T, Yamaguchi H, Leys C, Vartapetian AB, Fox JG, Lee JR, Wang TC, Goldenring JR. Spasmolytic polypeptide expressing metaplasia to preneoplasia in H. felis-infected mice. Gastroenterology. 2004;127:582–594. doi: 10.1053/j.gastro.2004.05.029. [DOI] [PubMed] [Google Scholar]
- 33.Rosa A, Brivanlou AH. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leivonen SK, Makela R, Ostling P, Kohonen P, Haapa-Paananen S, Kleivi K, Enerly E, Aakula A, Hellstrom K, Sahlberg N, Kristensen VN, Borresen-Dale AL, Saviranta P, Perala M, Kallioniemi O. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene. 2009;28:3926–3936. doi: 10.1038/onc.2009.241. [DOI] [PubMed] [Google Scholar]
- 35.Shen R, Liyanarachchi S, Li W, Wakely PE Jr, Saji M, Huang J, Nagy R, Farrell T, Ringel MD, de la Chapelle A, Kloos RT, He H. MicroRNA signature in thyroid fine needle aspiration cytology applied to “atypia of undetermined significance” cases. Thyroid. 2012;22:9–16. doi: 10.1089/thy.2011.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gharbaran R. Insights into the molecular roles of heparan sulfate proteoglycans (HSPGs-syndecans) in autocrine and paracrine growth factor signaling in the pathogenesis of Hodgkin’s lymphoma. Tumour Biol. 2016;37:11573–11588. doi: 10.1007/s13277-016-5118-7. [DOI] [PubMed] [Google Scholar]
- 37.Mitselou A, Galani V, Skoufi U, Arvanitis DL, Lampri E, Ioachim E. Syndecan-1, epithelial-mesenchymal transition markers (E-cadherin/beta-catenin) and neoangiogenesis-related proteins (PCAM-1 and endoglin) in colorectal cancer. Anticancer Res. 2016;36:2271–2280. [PubMed] [Google Scholar]