Abstract
Mutations in mitochondrial DNA (mtDNA) were found to be associated with hypertension. We reported here clinical, genetic and molecular characterization of a Han Chinese family with maternally inherited hypertension. Most strikingly, this family exhibited a high penetrance of hypertension. Sequence analysis of the entire mitochondrial genome showed the presence of the well-known T4363C mutation in tRNAGln, as well as the ND1 T3394C mutation, and a set of polymorphisms belonging to human mitochondrial haplogroup M7b. Of these, the T4363C mutation was localized at the highly conserved nucleotide in the anticodon stem of tRNAGln (position 38), may result the failure in tRNA metabolism. Moreover, the homoplasmic ND1 T3394C mutation, which had been reported to be associated with Leber’s hereditary optic neuropathy (LHON), was regarded as a pathogenic mutation associated with mitochondrial diseases. Thus, the combination of ND1 T3394C and tRNAGln T4363C mutations may contribute to the high penetrance and expressivity of hypertension in this Chinese family.
Keywords: Mitochondrial mutation, T3394C, T4363C, tRNA metabolism, hypertension
Introduction
Hypertension is a major public health problem, affecting approximately 1 billion people worldwide [1]. Hypertension can be classified as either essential (primary) or secondary. Essential hypertension indicates that no specific medical cause can be found to explain a patient’s condition. Secondary hypertension means that the high blood pressure is a result of another condition, such as kidney disease or certain tumors (especially of the adrenal gland) [2]. To date, the etiology of hypertension is not well understood, it is now generally believed that this disease can be caused by single gene or multifactorial conditions, resulting from interactions between the environment and inherited risk factors. Of hereditary factors, maternal transmissions of hypertension have been implicated in some pedigrees, suggesting that mutations in mitochondrial DNA (mtDNA) were responsible for this phenotype [3,4]. Mitochondrial dysfunction caused by mtDNA mutations resulted in oxidative stress, uncoupling of the oxidative pathways for ATP synthesis and subsequent failure of cellular energetic processes [5]. In addition, an inefficient metabolism caused by mitochondrial dysfunctions in skeletal and vascular smooth muscles would lead to the elevation of systolic blood pressure and therefore may be involved in the development of hypertension [6,7]. Most recently, several mtDNA mutations have been reported to be associated with hypertension, such as the A4295G in tRNAIle [8], A4435G in tRNAMet [9], and A1555G in 12S rRNA [10].
However, these genetic factors in mitochondrial genome remained poorly identified. To understand the association between mtDNA mutations and hypertension, we recently initiated a systematic and extensive mutational screening for mitochondrial genomes in patients with hypertension. In this study, we reported here the molecular characterization of a three-generation Han Chinese family with maternally inherited hypertension. Sequence analysis of the mitochondrial genome led us to identify the homoplasmic ND1 T3394C and tRNAGln T4363C mutations.
Materials and methods
Subjects
As the part of genetic screening program for hypertension, a Han Chinese family, as shown in Figure 1, was ascertained in the Department of Cardiology, Affiliated Wenling Hospital, Wenzhou Medial University. Informed consent was obtained from members before their participation in the study, in accordance with the Ethics Committee of Affiliated Wenling Hospital, Wenzhou Medial University. In addition, we selected 300 unrelated healthy subjects from the same area with age-matched as controls.
Figure 1.
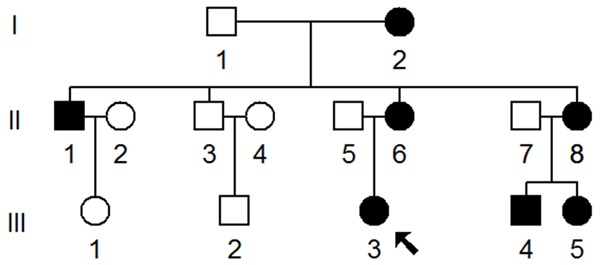
One Han Chinese family with hypertension, hypertension individuals were indicated by filled symbols, arrow denoted the proband.
Clinical examinations
Members of this Chinese family underwent a physical examination, laboratory assessment of cardiovascular disease risk factors and routine electrocardiography. A physician measured the systolic and diastolic blood pressures of subjects using a mercury column sphygmomanometer and a standard protocol. The first and the fifth Korotkoff sounds were taken as indicators of systolic and diastolic blood pressure, respectively. The average of 3 such systolic and diastolic blood pressure readings was taken as the examination blood pressure. Hypertension was defined according to the recommendation of the Joint National Committee on Detection, Evaluation and Treatment of High Blood Pressure (JNCVI) and the World Health Organization-International Society of Hypertension as a systolic blood pressure of ≥140 mmHg and/or a diastolic blood pressure of ≥90 mmHg [11].
Mutational analysis of mitochondrial genome
Genomic DNA was isolated form the whole blood of participants using PAXgene Blood DNA Isolation Kits (QIAGEN, Valencia, CA, USA). First, subject’s DNA fragments spanning the entire mitochondrial ND1 gene were amplified by PCR using oligodeoxynucleotides corresponding to positions 3149-3169 and 3942-3961 [12]. PCR fragments were purified and subsequently analyzed by direct sequencing analysis. In addition, the entire mitochondrial genomes of the proband and the matrilineal relatives carrying the T3394C mutation were PCR amplified in 24 overlapping fragments by using sets of the light-strand and the heavy-strand oligonucleotide primers, as described elsewhere [12]. Each fragment was purified and subsequently analyzed by direct sequencing in an ABI 3700 automated DNA sequencer using the Big Dye Terminator Cycle sequencing reaction kit. The resultant sequence data were compared with the updated consensus Cambridge sequence (GenBank accession number: NC_012920) [13].
Statistical analysis
Statistical analyses were performed using SPSS 17.0 (SPSS Inc., Chicago, IL, USA). Differences in categorical variables were assessed with Fisher’s exact test. We considered P<0.05 as statistically significant.
Results
Clinical features of the Chinese family with hypertension
The proband (III-3) was a 40-year-old woman who came from the Wenling area of Zhejiang province. She began to suffer from hypertension when she was 37 years old, and her blood pressure was 160/90 mmHg. Physical examination, laboratory assessment of cardiovascular disease risk factors and routine electrocardiography showed that she did not have other clinical abnormalities, including diabetes, vision and hearing impairments, renal and neurological disorders. Therefore, she exhibited a typical essential hypertension. As shown in Figure 1 and Table 1, matrilineal relatives in this family had a wide range of severity in hypertension, notably, the age at onset of hypertension in the maternal kindred varied from 35 to 75 years, with an average of 50 years.
Table 1.
Summary of clinical data of several members in this family
| Subjects | Gender | Age of onset (year) | Age at test (year) | Systolic pressure (mmHg) | Diastolic pressure (mmHg) |
|---|---|---|---|---|---|
| I-2 | Female | 75 | 82 | 145 | 95 |
| II-1 | Male | 55 | 60 | 150 | 90 |
| II-6 | Female | 53 | 62 | 180 | 100 |
| II-8 | Female | 58 | 65 | 155 | 85 |
| III-3 | Female | 37 | 40 | 160 | 90 |
| III-4 | Male | 35 | 42 | 150 | 80 |
| III-5 | Female | 38 | 41 | 145 | 75 |
| III-1 | Female | / | 38 | 135 | 70 |
Analysis of mutations in mitochondrial genome
The maternal transmission of hypertension in this family suggested mitochondrial involvement and led us to analyze the mitochondrial genomes of matrilineal relatives (I-2, II-1, II-3, II-6, II-8, III-3, III-4, III-5). For this purpose, the DNA fragments spanning the entire mtDNA were PCR amplified using 24-overlapping primers, after amplification, each fragment was purified and subsequently analyzed by direct sequence [12]. As shown in Table 2, there were 38 sequence variants in mitochondrial genome and belonging to human mitochondrial haplogroup M7b [14]. Of these, there were 9 variants in D-loop, 2 variants in 12S rRNA, 2 variants in 16S rRNA and 1 mutation in tRNA gene, while other variants were mainly localized at protein-coding genes, in addition, there were 8 missense mutations in mitochondrial genome, these missense mutations included T3394C (Y30H) in ND1 gene, G8584A (A20T), A8701G (T59A) and A8860G (T112A) in A6 gene, A10398G (T114A) in ND3 gene, A12361G (T9A) in ND5 gene, C14766T (T7I) and A15326G (T194A) in CytB gene. These variants in RNAs and polypeptides were further evaluated by phylogenetic analysis and sequences from other organisms including mouse [15], bovine [16] and Xenopus laevis [17]. However, all these variants showed no evolutionary conservation excepted for the T3394C and T4363C mutations (Figure 2), suggesting that the T3394C and T4363C mutations may have significantly functional consequence. Moreover, both of these mutations were absent in 300 healthy controls. Fisher’s exact frequency difference test showed that the T3394C and T4363C mutations were of statistical significance with the P<0.05.
Table 2.
Mitochondrial DNA sequence variants in this family with hypertension
| Gene | Position | Replacement | Conservation (H/B/M/X)a | CRSb | Previously reportedc |
|---|---|---|---|---|---|
| D-loop | 73 | A to G | A | Yes | |
| 146 | T to C | T | Yes | ||
| 152 | T to C | T | Yes | ||
| 249 | Del A | A | Yes | ||
| 310 | T to TC | T | Yes | ||
| 489 | T to C | T | Yes | ||
| 16192 | C to T | C | Yes | ||
| 16223 | C to T | C | Yes | ||
| 16298 | T to C | T | Yes | ||
| 12S rRNA | 750 | A to G | A/A/A/- | A | Yes |
| 1438 | A to G | A/A/A/G | A | Yes | |
| 16S rRNA | 2706 | A to G | A/G/A/A | A | Yes |
| 3107 | Del N | C | Yes | ||
| ND1 | 3394 | T to C (Tyr to His) | Y/Y/Y/Y | T | Yes |
| tRNAGln | 4363 | T to C | T/T/T/T | T | Yes |
| ND2 | 4769 | A to G | A | Yes | |
| CO1 | 6752 | A to G | A | Yes | |
| 7028 | C to T | C | Yes | ||
| 7196 | C to A | C | Yes | ||
| CO2 | 8227 | T to C | T | Yes | |
| A6 | 8584 | G to A (Ala to Thr) | A/V/V/I | G | Yes |
| 8701 | A to G (Thr to Ala) | T/S/L/Q | A | Yes | |
| 8860 | A to G (Thr to Ala) | T/A/A/T | A | Yes | |
| 9090 | T to C | T | Yes | ||
| CO3 | 9540 | T to C | T | Yes | |
| ND3 | 10398 | A to G (Thr to Ala) | T/T/T/A | A | Yes |
| 10400 | C to T | C | Yes | ||
| ND4 | 10873 | T to C | T | Yes | |
| 11719 | G to A | G | Yes | ||
| ND5 | 12361 | T to A (Thr to Ala) | T/S/I/N | C | Yes |
| 12705 | C to T | C | Yes | ||
| 12996 | A to G | A | Yes | ||
| CytB | 14766 | C to T (Thr to Ile) | T/S/T/S | C | Yes |
| 14783 | T to C | T | Yes | ||
| 15043 | G to A | G | Yes | ||
| 15301 | G to A | G | Yes | ||
| 15326 | A to G (Thr to Ala) | T/M/I/I | A | Yes | |
| 15487 | A to T | A | Yes |
Conservation of amimo acid for polypeptides of nucleotide for rRNAs in human (H), bovine (B), mouse (M), and Xenopus laevis (X);
CRS: Cambridge reference sequence;
See the online mitochondrial genome database www.mitomap.org.
Figure 2.
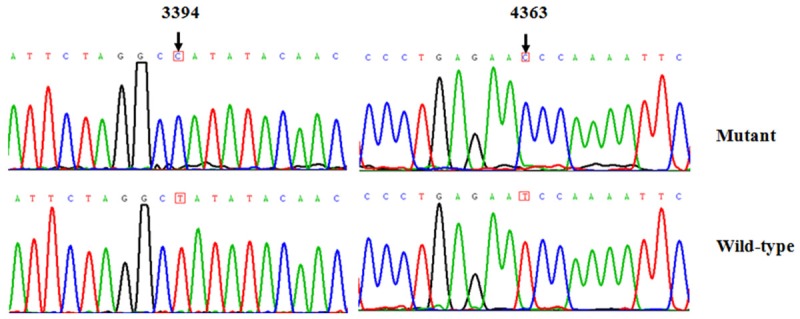
Identification of T3394C and T4363C mutations in mitochondrial genome. Partial sequence chromatograms of mitochondrial genome from the affected proband and control. Arrows indicated the locations of the base changes at position 3394 and 4363.
Discussion
In this study, we have performed clinical, genetic and molecular characterization of a Han Chinese family with high penetrance and expressivity of hypertension. Hypertension as the sole clinical phenotype was only presented in the maternal lineage of this pedigree. The inherited pattern provided a clear indication for the mtDNA mutations being responsible for the phenotype. Sequence analysis of the complete mitochondrial genomes in this pedigree showed the distinct sets of mtDNA polymorphisms, in addition to the identical T3394C (Y30H) mutation in ND1 gene and the T4363C mutation in tRNAGln gene. Indeed, the T3394C mutation was present in homoplasy only in the maternal lineage of this pedigree. The tyrosine at amino acid position 30 was extremely conserved in ND1 polypeptide among different organisms [18]. This mutation had been associated with other clinical abnormalities including LHON [19], deafness [20] and metabolic disorders [21]. In fact, the occurrence of the T3394C mutation in these matrilineal relatives affected by hypertension strongly indicated that this mutation was involved in the pathogenesis of hypertension.
In addition to the identified T3394C mutation, another mutation: T4363C in tRNAGln was also found during our mutational screening. Notably, the T4363C mutation was localized at the immediate 3 prime end to the anticodon, corresponding to the position 38 of tRNAGln (Figure 3), nucleotide at this position was extremely conserved and often modified, thus, contributed to the high fidelity of codon and anticodon interaction. Therefore, it can be speculated that the T4363C mutation will reduce the steady state level of tRNAGln. Previous studies showed that the T4363C mutation was associated with deafness, development delay and pseudoexfoliation glaucoma [22,23]. Therefore, the T4363C should be regarded as a pathogenic mutation associated with hypertension.
Figure 3.
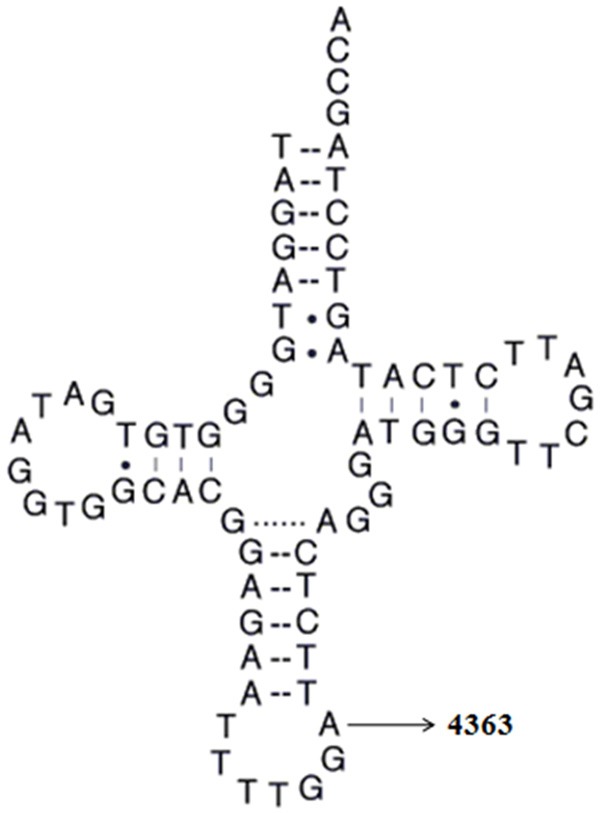
The location of T4363C mutation in tRNAGln gene. Cloverleaf structure of human mt-tRNAGln was derived from Mitomap database (http://www/mitomap.org). Arrow indicated the position of the T4363C mutation.
In conclusion, the clinical and genetic analysis of this family revealed the presence of the homoplasmic T3394C mutation in ND1 gene and the T4363C mutation in tRNAGln. However, matrilineal relatives in this family exhibited different severity of hypertension, moreover, we noticed that one of the matrilineal relatives (II-3) did not manifestate hypertension, suggesting that the T3394C and T4363C mutations were insufficient to produce the clinical phenotype, thus, other modified factors such as nuclear genes, environmental factors and epigenetic modification were necessary for the phenotypic manifestation of the T4363C and T3394C mutations. Taken together, our date indicated that the combination of the T3394C and T4363C mutations in mitochondrial genome may account for the high penetrance and expressivity of hypertension in this family.
Acknowledgements
This work is supported by the grants from Wenling Foundation of Science and Technology (no. 2015C3120551) and Projects of Medical and Health Technology Program in Zhejiang Province (no. 2017KY720).
Disclosure of conflict of interest
None.
References
- 1.Roth TS, Aboulhosn JA. Pulmonary hypertension and congenital heart disease. Cardiology Clinics. 2016;34:391–400. doi: 10.1016/j.ccl.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Gu D, Reynolds K, Wu X, Chen J, Duan X, Muntner P, Huang G, Reynolds RF, Su S, Whelton PK. Prevalence, awareness, treatment, and control of hypertension in China. Hypertension. 2002;40:920–927. doi: 10.1161/01.hyp.0000040263.94619.d5. [DOI] [PubMed] [Google Scholar]
- 3.Marian A. Mitochondrial genetics and human systemic hypertension. Circ Res. 2011;108:784–786. doi: 10.1161/CIRCRESAHA.111.242768. [DOI] [PubMed] [Google Scholar]
- 4.Ding Y, Xia B, Yu J, Leng J, Huang J. Mitochondrial DNA mutations and essential hypertension (Review) Int J Mol Med. 2013;32:768–774. doi: 10.3892/ijmm.2013.1459. [DOI] [PubMed] [Google Scholar]
- 5.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson FH, Hariri A, Farhi A, Zhao H, Petersen KF, Toka HR, Nelson-Williams C, Raja KM, Kashgarian M, Shulman GI. A cluster of metabolic defects caused by mutation in a mitochondrial tRNA. Science. 2004;306:1190–1194. doi: 10.1126/science.1102521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wisløff U, Najjar SM, Ellingsen Ø, Haram PM, Swoap S, Al-Share Q, Fernström M, Rezaei K, Lee SJ, Koch LG. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science. 2005;307:418–420. doi: 10.1126/science.1108177. [DOI] [PubMed] [Google Scholar]
- 8.Merante F, Myint T, Tein I, Benson L, Robinson B. An additional mitochondrial tRNAIle point mutation (A-to-G at nucleotide 4295) causing hypertrophic cardiomyopathy. Hum Mutat. 1996;8:216–222. doi: 10.1002/(SICI)1098-1004(1996)8:3<216::AID-HUMU4>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Li R, Li Z, Wang XJ, Yang L, Wang S, Guan MX. Mitochondrial transfer RNAMet 4435A> G mutation is associated with maternally inherited hypertension in a Chinese pedigree. Hypertension. 2009;53:1083–1090. doi: 10.1161/HYPERTENSIONAHA.109.128702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santorelli FM, Tanji K, Manta P, Casali C, Krishna S, Hays AP, Mancini DM, DiMauro S, Hirano M. Maternally inherited cardiomyopathy: an atypical presentation of the mtDNA 12S rRNA gene A1555G mutation. Am J Hum Genet. 1999;64:295–300. doi: 10.1086/302188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jones DW, Materson BJ, Oparil S, Wright JT. Seventh report of the joint national committee on prevention, detection, evaluation, and treatment of high blood pressure. Hypertension. 2003;42:1206–1252. doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- 12.Rieder MJ, Taylor SL, Tobe VO, Nickerson DA. Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res. 1998;26:967–973. doi: 10.1093/nar/26.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147–147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 14.Kong QP, Bandelt HJ, Sun C, Yao YG, Salas A, Achilli A, Wang CY, Zhong L, Zhu CL, Wu SF. Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet. 2006;15:2076–2086. doi: 10.1093/hmg/ddl130. [DOI] [PubMed] [Google Scholar]
- 15.Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 16.Gadaleta G, Pepe G, De Candia G, Quagliariello C, Sbisa E, Saccone C. The complete nucleotide sequence of theRattus norvegicus mitochondrial genome: cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol. 1989;28:497–516. doi: 10.1007/BF02602930. [DOI] [PubMed] [Google Scholar]
- 17.Roe BA, Ma DP, Wilson R, Wong J. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985;260:9759–9774. [PubMed] [Google Scholar]
- 18.Fearnley IM, Walker JE. Conservation of sequences of subunits of mitochondrial complex I and their relationships with other proteins. Biochim Biophys Acta. 1992;1140:105–134. doi: 10.1016/0005-2728(92)90001-i. [DOI] [PubMed] [Google Scholar]
- 19.Liang M, Guan M, Zhao F, Zhou X, Yuan M, Tong Y, Yang L, Wei QP, Sun YH, Lu F. Leber’s hereditary optic neuropathy is associated with mitochondrial ND1 T3394C mutation. Biochem Biophys Res Commun. 2009;383:286–292. doi: 10.1016/j.bbrc.2009.03.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Yuan H, Lu J, Liu X, Wang G, Zhu Y, Cheng J, Wang X, Han B, Yang L. Mutations at position 7445 in the precursor of mitochondrial tRNA Ser (UCN) gene in three maternal Chinese pedigrees with sensorineural hearing loss. Mitochondrion. 2008;8:285–292. doi: 10.1016/j.mito.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Saxena R, de Bakker PI, Singer K, Mootha V, Burtt N, Hirschhorn JN, Gaudet D, Isomaa B, Daly MJ, Groop L. Comprehensive association testing of common mitochondrial DNA variation in metabolic disease. Am J Hum Genet. 2006;79:54–61. doi: 10.1086/504926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abu-Amero KK, Bosley TM, Morales J. Analysis of nuclear and mitochondrial genes in patients with pseudoexfoliation glaucoma. Mol Vis. 2008;14:29–36. [PMC free article] [PubMed] [Google Scholar]
- 23.Wong LJ, Liang MH, Kwon H, Park J, Bai RK, Tan DJ. Comprehensive scanning of the entire mitochondrial genome for mutations. Clin Chem. 2002;48:1901–1912. [PubMed] [Google Scholar]


