Abstract
Genetic association analysis and functional analysis have suggested that telomerase reverse transcriptase (TERT) gene affects the predisposition to various tumors. In this study, we wanted to explore the association between TERT variants and hepatocellular carcinoma (HCC) risk in a Han Chinese population via a case-control study of 473 HCC patients and 564 controls. Sequenom Mass-ARRAY platform was applied to determine the genotype of TERT polymorphisms in these subjects. Odds ratios and 95% confidence intervals that calculated by logistic regression analysis were used to assess the association under the genotype, dominant, recessive, and additive models. The “AA” genotype frequency of TERT rs2242652 in cases was significantly lower than in controls (1.69% versus 3.72%). We found two SNPs were associated with decreased risk of HCC with or without the adjustment for age and gender: rs10069690 under an additive model (adjusted OR = 0.77, 95% CI: 0.60-0.98, P = 0.038); rs2242652 under a dominant model (adjusted OR = 0.72, 95% CI: 0.54-0.95, P = 0.022) and an additive model (adjusted OR = 0.72, 95% CI: 0.56-0.92, P = 0.009). To our knowledge, the present study is the first to show the significant association between TERT polymorphisms and HCC susceptibility in a Han Chinese population from China, which may act as a potential prognostic biomarker in HCC patients.
Keywords: Hepatocellular carcinoma, TERT, single nucleotide polymorphisms (SNPs), association analysis
Introduction
The incidence of HCC, the most common histological subtype of primary hepatic carcinoma, is increasing around the world in recent years, especially in China. HCC is affected by multi-factor, including both environmental and genetic factors. Many candidate genes correlation analysis for this disease have been studied, such as HLA-DP gene polymorphisms have significant association with HCC in the Asian population [1]; HMGB1 variants in the HCC susceptibility and progression in Chinese population [2]; FasL gene polymorphisms confer HCC risk in Egyptian individuals [3]; And SNPs in TERT and CLPTM1L and HCC predisposition in Chinese males [4].
Human telomerase is composed of telomerase reverse transcriptase (TERT), the catalytic subunit that synthesizes the repeat sequence TTAGGG of telomere, and telomerase RNA component (TERC), serves as the reverse transcription template [5]. Telomerase activation is restricted to the early stages of embryonic development and stem cells compartments in adult, however, it also occurs in some human cancers with a high level, such as in glioma, skin cancer, and lung carcinoma [4,6]. Telomere locates at the end of chromosome which is critical for chromosome end protection and genomic stability. Telomere shortening occurs early in the initiation of epithelial carcinogenesis [7]. And telomere dysfunction promotes the chromosomal instability which plays an vital role in the initiation of carcinogenesis, while telomerase activation partially restores telomere length and genomic stability [8].
The TERT gene has been mapped to chromosome 5p15.33 and consisted of 16 exons and 15 introns spanning 35 kb of genomic DNA. It encodes the catalytic subunit of the telomerase reverse transcriptase, adds nucleotide repeats to chromosome ends. It is reported that this gene can influence the risk of various cancers, such as lung adenocarcinoma, upper tract urothelial carcinomas, glioma, and melanoma [9-12]. To date, many studies have suggested that the TERT promoter rs2853669 increases mortality and recurrence risks of HCC in Korean population [13]. There are prominent correlations with TERT polymorphisms and increased risk of HCC in a Han Chinese population from Northeast of China [4]. However, the associations between TERT variants and HCC risk in a Chinese Han population have not been investigated. And whether there are other SNPs in TERT that are correlated with HCC predisposition is still unknown in light of only one variant rs2736098 was reported [4]. Therefore, we performed a case-control study that was composed of 473 HCC patients and 564 controls from China was designed to research the potential association.
Materials and methods
Subjects
This gene association study was approved by the Ethics Committee of Haikou People’s Hospital. The 473 HCC patients were newly diagnosed through clinical and histopathologic examinations in Haikou People’s Hospital from March of 2013 to December of 2015. As controls we selected 564 non-cancer individuals from the Physical Examination Center of the same hospital. All subjects were unrelated Han Chinese population from China. Blood samples were collected from them with informed consent.
DNA isolation and SNPs genotyping
Genomic DNA was extracted from peripheral blood leukocytes using the GoldMag-Mini Purification Kit (GoldMag Co. Ltd. Xi’an city, China). Then, we measured DNA concentration with the NanoDrop 2000 (Thermo Scientific, Waltham, Massachusetts, USA) and quantified and diluted DNA with QIAgility to a final concentration of 20 ng/μl. We selected four TERT SNPs (rs10069690, rs2242652, rs2853677, rs2853676) that have been researched in different types of tumor, such as thyroid cancer, breast cancer, lung carcinoma, pancreatic cancer, and glioma [14-18], with minor allele frequencies more than 5% in Chinese Han Beijing population (International HapMapProject, version 28; http://www.hapmap.org) to preliminarily explore the potential correlation. Primers that were used for the identification of the four TERT SNPs were listed in Table 1. Sequenom Mass-ARRAY RS1000 (Sequenom, SanDiego, CA) was applied to SNP genotyping. And data were analyzed and managed using Sequenom Typer 4.0 Software (Sequenom Co. Ltd) [19].
Table 1.
Primers used for the identification of TERT polymorphisms
| SNP | First PCRP (5’→3’) | Second PCRP (5’→3’) | UEP SEQ (5’→3’) |
|---|---|---|---|
| rs10069690 | ACGTTGGATGCCTGTGGCTGCGGTGGCTG | ACGTTGGATGATGTGTGTTGCACACGGGAT | GGGATCCTCATGCCA |
| rs2242652 | ACGTTGGATGACAGCAGGACACGGATCCAG | ACGTTGGATGAGGCTCTGAGGACCACAAGA | gtcgGAGGACCACAAGAAGCAGC |
| rs2853677 | ACGTTGGATGATCCAGTCTGACAGTCGTTG | ACGTTGGATGGCAAGTGGAGAATCAGAGTG | gggtAATCAGAGTGCACCAG |
| rs2853676 | ACGTTGGATGTGTCTCCTGCTCTGAGACC | ACGTTGGATGCAAAACTAAGACCCAAGAGG | agatGGAAGTCTGACGAAGGC |
SNPs: single-nucleotide polymorphisms; PCRP: PCR primer; UEP: Un-extended mini-sequencing primer.
Statistical analysis
We used SPSS 16.0 (SPSS, Chicago, IL, USA) and Microsoft Excel to conduct statistical analysis. Age and gender were compared between the cases and controls using Welch’s t test and Pearson’s χ2 test, respectively. Genotype frequencies of the four TERT SNPs were determined for deviation from Hardy-Weinberg Equilibrium (HWE) using Pearson’s χ2 test. The associations of these polymorphisms genotypes with HCC risk were evaluated by odds ratios and 95% confidence intervals from multivariate logistic regression analysis with or without adjustment for age and gender. And the relationships were also assessed under dominant, recessive, and additive genetic models using PLINK software (http://pngu.mgh.harvard.edu/purcell/plink/). Finally, the SHEsis software platform (http://www.nhgg.org/analysis) and Haploview software package (version 4.2) were used to haplotype construction and analysis [20]. The correlations between TERT haplotypes and HCC risk were also calculated by logistic regression analysis. Statistical significance was set at a two-sided P < 0.05.
Results
A total of 473 HCC cases and 564 controls from China were genotyped for TERT polymorphisms. We listed age and gender distributions in Table 2 and found significant difference in them between cases and controls (P < 0.05). To eliminate the possible confounding effects caused by the difference, unconditional logistic regression analysis with adjustment for age and gender was applied to calculate Odds ratios.
Table 2.
Age and gender characteristics of HCC cases and controls
| Variable | Cases | Controls | P value |
|---|---|---|---|
|
| |||
| (n = 473) | (n = 564) | ||
| Gender | < 0.05 | ||
| Male | 390 (82.5%) | 339 (60.1%) | |
| Female | 83 (17.5%) | 225 (39.9%) | |
| Age, yr | 55.83 | 53.92 | < 0.05 |
Table 3 shows the minor allele frequency distributions of TERT variants and their relationships with HCC susceptibility. The four SNPs were all in line with Hardy-Weinberg equilibrium in controls (P > 0.05). Significant differences were observed in allele frequencies of rs10069690T and rs2242652A between cases and controls (13.5% versus 17.1%; 13.3% versus 17.9%, respectively). And rs10069690T and rs2242652A were significantly correlated with decreased risk of HCC (OR = 0.75, 95% CI: 0.59-0.96, P = 0.021; OR = 0.70, 95% CI: 0.55-0.90, P = 0.004, respectively). In Table 4 significant correlation with a reduced HCC susceptibility was also found in “AA” genotype of rs2242652 when it compared with the wild “GG” genotype with or without adjustment by age and gender (adjusted OR = 0.41, 95% CI: 0.17-0.95, P = 0.037).
Table 3.
Allele distributions of TERT variants and their relationships with HCC susceptibility
| SNPs | Chromosome | Position | Allele | Minor allele frequency | HWE P value | OR (95% CI) | P | |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Case | Control | |||||||
| rs10069690 | 5p15.33 | 1279790 | T/C | 0.135 | 0.171 | 0.6551 | 0.75 (0.59-0.96) | 0.021* |
| rs2242652 | 5p15.33 | 1280028 | A/G | 0.133 | 0.179 | 0.3914 | 0.70 (0.55-0.90) | 0.004* |
| rs2853677 | 5p15.33 | 1287194 | G/A | 0.370 | 0.369 | 0.7174 | 1.00 (0.84-1.20) | 0.966 |
| rs2853676 | 5p15.33 | 1288547 | T/C | 0.132 | 0.159 | 0.8744 | 0.81 (0.63-1.04) | 0.093 |
SNPs: single-nucleotide polymorphisms; HWE: Hardy-Weinberg equilibrium; OR: odds ratio; 95% CI: 95% confidence interval. P values were calculated from Chi-square test/Fisher’s exact test.
P ≤ 0.05 indicates statistical significance.
Table 4.
Genotype frequencies of rs10069690 and rs2242652 and their associations with HCC risk
| SNPs | Models | Genotype | Cases | Controls | Without adjustment | With adjustment | ||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| OR (95% CI) | P values | OR (95% CI) | P values | |||||
| rs10069690 (C>T) | Genotype | CC | 353 | 386 | 1.00 | 1.00 | ||
| TC | 111 | 156 | 0.78 (0.59-1.03) | 0.082 | 0.80 (0.60-1.07) | 0.138 | ||
| TT | 8 | 18 | 0.49 (0.21-1.13) | 0.094 | 0.48 (0.20-1.15) | 0.098 | ||
| Dominant | CC | 353 | 386 | 1.00 | 1.00 | |||
| TC+TT | 119 | 174 | 0.75 (0.57-0.98) | 0.038* | 0.77 (0.58-1.02) | 0.067 | ||
| Recessive | CC+TC | 464 | 542 | 1.00 | 1.00 | |||
| TT | 8 | 18 | 0.52 (0.22-1.21) | 0.127 | 0.51 (0.21-1.21) | 0.126 | ||
| Additive | - | - | - | 0.75 (0.59-0.96) | 0.022* | 0.77 (0.60-0.98) | 0.038* | |
| rs2242652 (G>A) | Genotype | GG | 355 | 383 | 1.00 | 1.00 | ||
| AG | 110 | 160 | 0.74 (0.56-0.98) | 0.038* | 0.76 (0.57-1.02) | 0.066 | ||
| AA | 8 | 21 | 0.41 (0.18-0.94) | 0.035* | 0.41 (0.17-0.95) | 0.037* | ||
| Dominant | GG | 355 | 383 | 1.00 | 1.00 | |||
| AA+AG | 118 | 181 | 0.70 (0.54-0.92) | 0.012* | 0.72 (0.54-0.95) | 0.022* | ||
| Recessive | GG+AG | 465 | 543 | 1.00 | 1.00 | |||
| AA | 8 | 21 | 0.44 (0.20-1.01) | 0.054 | 0.44 (0.19-1.01) | 0.054 | ||
| Additive | - | - | - | 0.71 (0.56-0.90) | 0.005* | 0.72 (0.56-0.92) | 0.009* | |
SNPs: single-nucleotide polymorphisms; OR: odds ratio; 95% CI: 95% confidence interval. P values were calculated from unconditional logistic regression analysis.
P ≤ 0.05 indicates statistical significance.
After evaluating the potential association under dominant, recessive, and additive genetic models, we found two SNPs were associated with decreased risk of HCC with or without the adjustment: rs10069690 under an additive model (adjusted OR = 0.77, 95% CI: 0.60-0.98, P = 0.038); rs2242652 under a dominant model (adjusted OR = 0.72, 95% CI: 0.54-0.95, P = 0.022) and an additive model (adjusted OR = 0.72, 95% CI: 0.56-0.92, P = 0.009) (Table 4).
In addition, the candidate SNPs (rs10069690-rs2242652) in TERT exhibited strong linkage. In Figure 1, the red squares of the TERT linkage disequilibrium block showed statistically significant linkage between the two polymorphisms. We listed the haplotypes with frequencies of more than 0.05 and their associations with HCC risk in Table 5. Haplotype “TA” was associated with a reduced risk of HCC (adjusted OR = 0.77, 95% CI: 0.60-0.99, P = 0.040), on the contrary, “CG” was correlated with an increased risk of HCC (adjusted OR = 1.37, 95% CI: 1.07-1.75, P = 0.013).
Figure 1.
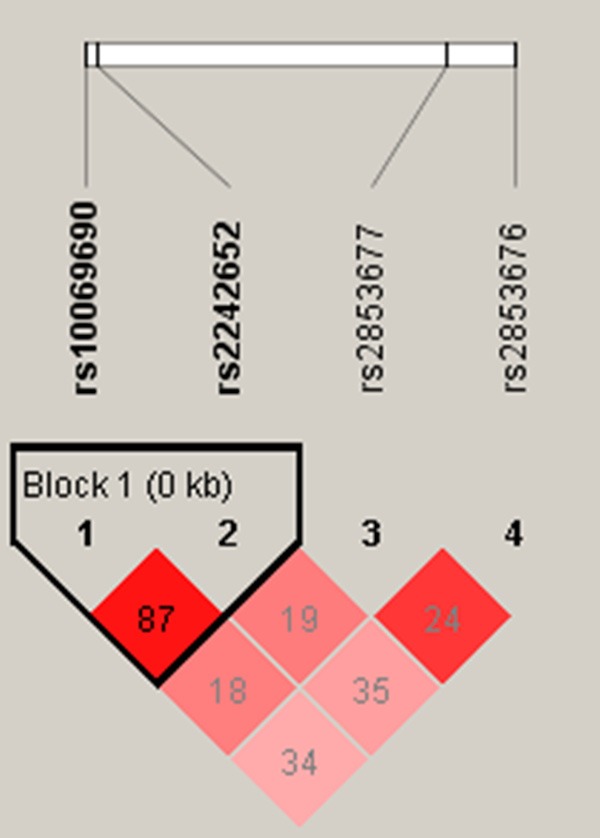
Haplotype block map for SNPs in the TERT gene.
Table 5.
TERT haplotype frequencies and their correlations with the risk of HCC
| Haplotype block | Haplotype frequencies | Without adjustment | With adjustment | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| Case | Control | OR (95% CI) | P | OR (95% CI) | P | |
| TA | 0.13 | 0.17 | 0.75 (0.59-0.96) | 0.018* | 0.77 (0.60-0.99) | 0.040* |
| CG | 0.86 | 0.82 | 1.38 (1.08-1.75) | 0.009* | 1.37 (1.07-1.75) | 0.013* |
OR: odds ratio; 95% CI: 95% confidence interval. P values were calculated from unconditional logistic regression analysis.
P ≤ 0.05 indicates statistical significance.
Discussion
In the present study, we genotyped four TERT polymorphisms (rs10069690, rs2242652, rs2853677, rs2853676) in HCC patients and healthy controls, and our results showed a statistically significant association between TERT variants and HCC susceptibility: the “T” allele of rs10069690 and the “A” allele of rs2242652 were associated with decreased risk of HCC in a Han Chinese population. These findings suggest that some TERT polymorphisms identified in other types of tumor are also correlated with HCC risk.
Telomerase reverse transcriptase, encoded by the TERT gene, is an essential component of telomerase. Telomerase expression is restricted to stem cell and embryonic tissue, but in some human cancers, telomerase activity is higher than in normal tissues in adult. The high expression of telomerase confers a indefinitely replicative potentiality via the restoration of telomere length which may be involved in carcinogenesis [21]. On the other hand, as we know, the p53 is an important molecule involved in regulating cellular response to DNA damage, such as induced by telomere dysfunction and shortening, and repairing or eliminating cells [22]. In the setting of deactivated p53, telomere dysfunction and shortening can result in the chromosomal instability which may promote carcinogenesisin epithelial compartments [5]. Therefore, it is difficult to determine long or short telomere length is correlated with tumor susceptibility.
Studies have demonstrated that TERT rs10069690 could confer an increased risk of thyroid cancer, breast cancer, and ovarian cancer etc or a reduced predisposition to multiple myeloma [14,15,23,24]. As for TERT rs2242652, it was either associated with increased risk of breast cancer or decreased susceptibility to multiple myeloma [24,25]. These differences may be due to the dysregulated TERT expression in most kinds of tumors. A recent study by Bojesen et al. suggested rs10069690 and rs2242652 respectively contain a silencer of the TERT promoter and form a truncated TERT splice variant which further reduce gene expression [26]. Kote-Jarai et al. observed that rs2242652 was related to a decreased TERT expression in prostate cancer [27]. In the present study, we found TERT rs10069690 and rs2242652 served as protective factors for the formation of HCC, which may be due to the reduced expression of TERT protein by these SNPs and synthesis of telomerase with shorter telomere length.
However, this study also had several limitations. First, although age and gender were taken into consideration for the unconditional logistic regression, other risk factors, for instance, infection of hepatitis B virus, drinking and smoking status, and diet were not analyzed in this study. Second, we have not genotyped all the TERT variants which may lead to omission of some significant SNPs. Third, our study is limited by relatively small sample size, and lack of randomization to conditions.
To our knowledge, this study is the first to present the significant correlation between TERT polymorphisms and HCC susceptibility in a Han Chinese population from China, which may provide theoretical foundation for others to further study the potential association and new information for screening of HCC predisposed population in clinical practice.
Acknowledgements
This research was supported by (a) National Natural Science Foundation of China grants (81460450); (b) Natural Science Foundation of Hainan Province grants (309115 and 817379 and 20168312); (c) Hainan Province Natural Science Foundation of Innovation Research Team Project (2017CXTD010).
Disclosure of conflict of interest
None.
References
- 1.Zhang X, Zheng C, Zhou ZH, Li M, Gao YT, Jin SG, Sun XH, Gao YQ. Relationship between HLA-DP gene polymorphisms and the risk of hepatocellular carcinoma: a meta-analysis. Genet Mol Res. 2015;14:15553–15563. doi: 10.4238/2015.December.1.6. [DOI] [PubMed] [Google Scholar]
- 2.Wang B, Yeh CB, Lein MY, Su CM, Yang SF, Liu YF, Tang CH. Effects of HMGB1 polymorphisms on the susceptibility and progression of hepatocellular carcinoma. Int J Med Sci. 2016;13:304–309. doi: 10.7150/ijms.14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khalifa RH, Bahgat DM, Darwish HA, Shahin RM. Significant association between FasL gene -844T/C polymorphism and risk to hepatocellular carcinoma in Egyptian patients. Immunol Lett. 2016;172:84–88. doi: 10.1016/j.imlet.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Su LY, Li XL, Shen L, Zhang Y, Zhao MM, Yin ZH, Su HY, Zhou BS. Polymorphisms of TERT and CLPTM1L and the risk of hepatocellular carcinoma in Chinese males. Asian Pac J Cancer Prev. 2014;15:8197–8201. doi: 10.7314/apjcp.2014.15.19.8197. [DOI] [PubMed] [Google Scholar]
- 5.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet. 2005;6:611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 7.Meeker AK, Hicks JL, Iacobuzio-Donahue CA, Montgomery EA, Westra WH, Chan TY, Ronnett BM, De Marzo AM. Telomere length abnormalities occur early in the initiation of epithelial carcinogenesis. Clin Cancer Res. 2004;10:3317–3326. doi: 10.1158/1078-0432.CCR-0984-03. [DOI] [PubMed] [Google Scholar]
- 8.Rudolph KL, Millard M, Bosenberg MW, DePinho RA. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat Genet. 2001;28:155–159. doi: 10.1038/88871. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Xu X, Fang J, Wang L, Mu Y, Zhang P, Yao Z, Ma Z, Liu Z. Rs2853677 modulates Snail1 binding to the TERT enhancer and afGifects lung adenocarcinoma susceptibility. Oncotarget. 2016;7:37825–37838. doi: 10.18632/oncotarget.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan X, Meng Y, Li P, Ge N, Kong F, Yang L, Bjorkholm M, Zhao S, Xu D. The association between the TERT rs2736100 AC genotype and reduced risk of upper tract urothelial carcinomas in a Han Chinese population. Oncotarget. 2016;7:31972–9. doi: 10.18632/oncotarget.7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh KM, Codd V, Smirnov IV, Rice T, Decker PA, Hansen HM, Kollmeyer T, Kosel ML, Molinaro AM, McCoy LS, Bracci PM, Cabriga BS, Pekmezci M, Zheng S, Wiemels JL, Pico AR, Tihan T, Berger MS, Chang SM, Prados MD, Lachance DH, O’Neill BP, Sicotte H, Eckel-Passow JE, van der Harst P, Wiencke JK, Samani NJ, Jenkins RB, Wrensch MR. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46:731–735. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llorca-Cardenosa MJ, Pena-Chilet M, Mayor M, Gomez-Fernandez C, Casado B, Martin-Gonzalez M, Carretero G, Lluch A, Martinez-Cadenas C, Ibarrola-Villava M, Ribas G. Long telomere length and a TERT-CLPTM1 locus polymorphism association with melanoma risk. Eur J Cancer. 2014;50:3168–3177. doi: 10.1016/j.ejca.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 13.Ko E, Seo HW, Jung ES, Kim BH, Jung G. The TERT promoter SNP rs2853669 decreases E2F1 transcription factor binding and increases mortality and recurrence risks in liver cancer. Oncotarget. 2016;7:684–699. doi: 10.18632/oncotarget.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong L, Xu Y, Hu YQ, Ding QJ, Yi CH, Huang W, Zhou M. hTERT gene polymorphism correlates with the risk and the prognosis of thyroid cancer. Cancer Biomark. 2016;17:195–204. doi: 10.3233/CBM-160631. [DOI] [PubMed] [Google Scholar]
- 15.Li ZY, Dong YL, Feng Y, Zhang Z, Cao XZ. Polymorphisms in the telomerase reverse transcriptase promoter are associated with risk of breast cancer: a meta-analysis. J Cancer Res Ther. 2016;12:1040–1044. doi: 10.4103/0973-1482.164701. [DOI] [PubMed] [Google Scholar]
- 16.Gao L, Thakur A, Liang Y, Zhang S, Wang T, Chen T, Meng J, Wang L, Wu F, Jin T, Li X, Liu JJ, Chen C, Chen M. Polymorphisms in the TERT gene are associated with lung cancer risk in the Chinese Han population. Eur J Cancer Prev. 2014;23:497–501. doi: 10.1097/CEJ.0000000000000086. [DOI] [PubMed] [Google Scholar]
- 17.Campa D, Rizzato C, Stolzenberg-Solomon R, Pacetti P, Vodicka P, Cleary SP, Capurso G, Bueno-de-Mesquita HB, Werner J, Gazouli M, Butterbach K, Ivanauskas A, Giese N, Petersen GM, Fogar P, Wang Z, Bassi C, Ryska M, Theodoropoulos GE, Kooperberg C, Li D, Greenhalf W, Pasquali C, Hackert T, Fuchs CS, Mohelnikova-Duchonova B, Sperti C, Funel N, Dieffenbach AK, Wareham NJ, Buring J, Holcatova I, Costello E, Zambon CF, Kupcinskas J, Risch HA, Kraft P, Bracci PM, Pezzilli R, Olson SH, Sesso HD, Hartge P, Strobel O, Malecka-Panas E, Visvanathan K, Arslan AA, Pedrazzoli S, Soucek P, Gioffreda D, Key TJ, Talar-Wojnarowska R, Scarpa A, Mambrini A, Jacobs EJ, Jamroziak K, Klein A, Tavano F, Bambi F, Landi S, Austin MA, Vodickova L, Brenner H, Chanock SJ, Delle Fave G, Piepoli A, Cantore M, Zheng W, Wolpin BM, Amundadottir LT, Canzian F. TERT gene harbors multiple variants associated with pancreatic cancer susceptibility. Int J Cancer. 2015;137:2175–2183. doi: 10.1002/ijc.29590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G, Jin TB, Wei XB, He SM, Liang HJ, Yang HX, Cui Y, Chen C, Cai LB, Gao GD. Selected polymorphisms of GSTP1 and TERT were associated with glioma risk in Han Chinese. Cancer Epidemiol. 2012;36:525–527. doi: 10.1016/j.canep.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009;Chapter 2:Unit 2.12. doi: 10.1002/0471142905.hg0212s60. [DOI] [PubMed] [Google Scholar]
- 20.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez-Marcelo T, Sanchez-Pernaute A, Pascua I, De Juan C, Head J, Torres-Garcia AJ, Iniesta P. Clinical relevance of telomere status and telomerase activity in colorectal cancer. PLoS One. 2016;11:e0149626. doi: 10.1371/journal.pone.0149626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 23.Lee AW, Bomkamp A, Bandera EV, Jensen A, Ramus SJ, Goodman MT, Rossing MA, Modugno F, Moysich KB, Chang-Claude J, Rudolph A, Gentry-Maharaj A, Terry KL, Gayther SA, Cramer DW, Doherty JA, Schildkraut JM, Kjaer SK, Ness RB, Menon U, Berchuck A, Mukherjee B, Roman L, Pharoah PD, Chenevix-Trench G, Olson S, Hogdall E, Wu AH, Pike MC, Stram DO, Pearce CL Ovarian Cancer Association Consortium. A splicing variant of TERT identified by GWAS interacts with menopausal estrogen therapy in risk of ovarian cancer. Int J Cancer. 2016;139:2646–2654. doi: 10.1002/ijc.30274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campa D, Martino A, Varkonyi J, Lesueur F, Jamroziak K, Landi S, Jurczyszyn A, Marques H, Andersen V, Jurado M, Brenner H, Petrini M, Vogel U, Garcia-Sanz R, Buda G, Gemignani F, Rios R, Vangsted AJ, Dumontet C, Martinez-Lopez J, Moreno MJ, Stepien A, Watek M, Moreno V, Dieffenbach AK, Rossi AM, Butterbach K, Jacobsen SE, Goldschmidt H, Sainz J, Hillengass J, Orciuolo E, Dudzinski M, Weinhold N, Reis RM, Canzian F. Risk of multiple myeloma is associated with polymorphisms within telomerase genes and telomere length. Int J Cancer. 2015;136:E351–358. doi: 10.1002/ijc.29101. [DOI] [PubMed] [Google Scholar]
- 25.Pellatt AJ, Wolff RK, Torres-Mejia G, John EM, Herrick JS, Lundgreen A, Baumgartner KB, Giuliano AR, Hines LM, Fejerman L, Cawthon R, Slattery ML. Telomere length, telomererelated genes, and breast cancer risk: the breast cancer health disparities study. Genes Chromosomes Cancer. 2013;52:595–609. doi: 10.1002/gcc.22056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, Edwards SL, Pickett HA, Shen HC, Smart CE, Hillman KM, Mai PL, Lawrenson K, Stutz MD, Lu Y, Karevan R, Woods N, Johnston RL, French JD, Chen X, Weischer M, Nielsen SF, Maranian MJ, Ghoussaini M, Ahmed S, Baynes C, Bolla MK, Wang Q, Dennis J, McGuffog L, Barrowdale D, Lee A, Healey S, Lush M, Tessier DC, Vincent D, Bacot F, Vergote I, Lambrechts S, Despierre E, Risch HA, Gonzalez-Neira A, Rossing MA, Pita G, Doherty JA, Alvarez N, Larson MC, Fridley BL, Schoof N, Chang-Claude J, Cicek MS, Peto J, Kalli KR, Broeks A, Armasu SM, Schmidt MK, Braaf LM, Winterhoff B, Nevanlinna H, Konecny GE, Lambrechts D, Rogmann L, Guenel P, Teoman A, Milne RL, Garcia JJ, Cox A, Shridhar V, Burwinkel B, Marme F, Hein R, Sawyer EJ, Haiman CA, Wang-Gohrke S, Andrulis IL, Moysich KB, Hopper JL, Odunsi K, Lindblom A, Giles GG, Brenner H, Simard J, Lurie G, Fasching PA, Carney ME, Radice P, Wilkens LR, Swerdlow A, Goodman MT, Brauch H, Garcia-Closas M, Hillemanns P, Winqvist R, Durst M, Devilee P, Runnebaum I, Jakubowska A, Lubinski J, Mannermaa A, Butzow R, Bogdanova NV, Dork T, Pelttari LM, Zheng W, Leminen A, Anton-Culver H, Bunker CH, Kristensen V, Ness RB, Muir K, Edwards R, Meindl A, Heitz F, Matsuo K, du Bois A, Wu AH, Harter P, Teo SH, Schwaab I, Shu XO, Blot W, Hosono S, Kang D, Nakanishi T, Hartman M, Yatabe Y, Hamann U, Karlan BY, Sangrajrang S, Kjaer SK, Gaborieau V, Jensen A, Eccles D, Hogdall E, Shen CY, Brown J, Woo YL, Shah M, Azmi MA, Luben R, Omar SZ, Czene K, Vierkant RA, Nordestgaard BG, Flyger H, Vachon C, Olson JE, Wang X, Levine DA, Rudolph A, Weber RP, Flesch-Janys D, Iversen E, Nickels S, Schildkraut JM, Silva Idos S, Cramer DW, Gibson L, Terry KL, Fletcher O, Vitonis AF, van der Schoot CE, Poole EM, Hogervorst FB, Tworoger SS, Liu J, Bandera EV, Li J, Olson SH, Humphreys K, Orlow I, Blomqvist C, Rodriguez-Rodriguez L, Aittomaki K, Salvesen HB, Muranen TA, Wik E, Brouwers B, Krakstad C, Wauters E, Halle MK, Wildiers H, Kiemeney LA, Mulot C, Aben KK, Laurent-Puig P, Altena AM, Truong T, Massuger LF, Benitez J, Pejovic T, Perez JI, Hoatlin M, Zamora MP, Cook LS, Balasubramanian SP, Kelemen LE, Schneeweiss A, Le ND, Sohn C, Brooks-Wilson A, Tomlinson I, Kerin MJ, Miller N, Cybulski C, Henderson BE, Menkiszak J, Schumacher F, Wentzensen N, Le Marchand L, Yang HP, Mulligan AM, Glendon G, Engelholm SA, Knight JA, Hogdall CK, Apicella C, Gore M, Tsimiklis H, Song H, Southey MC, Jager A, den Ouweland AM, Brown R, Martens JW, Flanagan JM, Kriege M, Paul J, Margolin S, Siddiqui N, Severi G, Whittemore AS, Baglietto L, McGuire V, Stegmaier C, Sieh W, Muller H, Arndt V, Labreche F, Gao YT, Goldberg MS, Yang G, Dumont M, McLaughlin JR, Hartmann A, Ekici AB, Beckmann MW, Phelan CM, Lux MP, Permuth-Wey J, Peissel B, Sellers TA, Ficarazzi F, Barile M, Ziogas A, Ashworth A, Gentry-Maharaj A, Jones M, Ramus SJ, Orr N, Menon U, Pearce CL, Bruning T, Pike MC, Ko YD, Lissowska J, Figueroa J, Kupryjanczyk J, Chanock SJ, Dansonka-Mieszkowska A, Jukkola-Vuorinen A, Rzepecka IK, Pylkas K, Bidzinski M, Kauppila S, Hollestelle A, Seynaeve C, Tollenaar RA, Durda K, Jaworska K, Hartikainen JM, Kosma VM, Kataja V, Antonenkova NN, Long J, Shrubsole M, Deming-Halverson S, Lophatananon A, Siriwanarangsan P, Stewart-Brown S, Ditsch N, Lichtner P, Schmutzler RK, Ito H, Iwata H, Tajima K, Tseng CC, Stram DO, van den Berg D, Yip CH, Ikram MK, Teh YC, Cai H, Lu W, Signorello LB, Cai Q, Noh DY, Yoo KY, Miao H, Iau PT, Teo YY, McKay J, Shapiro C, Ademuyiwa F, Fountzilas G, Hsiung CN, Yu JC, Hou MF, Healey CS, Luccarini C, Peock S, Stoppa-Lyonnet D, Peterlongo P, Rebbeck TR, Piedmonte M, Singer CF, Friedman E, Thomassen M, Offit K, Hansen TV, Neuhausen SL, Szabo CI, Blanco I, Garber J, Narod SA, Weitzel JN, Montagna M, Olah E, Godwin AK, Yannoukakos D, Goldgar DE, Caldes T, Imyanitov EN, Tihomirova L, Arun BK, Campbell I, Mensenkamp AR, van Asperen CJ, van Roozendaal KE, Meijers-Heijboer H, Collee JM, Oosterwijk JC, Hooning MJ, Rookus MA, van der Luijt RB, Os TA, Evans DG, Frost D, Fineberg E, Barwell J, Walker L, Kennedy MJ, Platte R, Davidson R, Ellis SD, Cole T, Bressacde Paillerets B, Buecher B, Damiola F, Faivre L, Frenay M, Sinilnikova OM, Caron O, Giraud S, Mazoyer S, Bonadona V, Caux-Moncoutier V, Toloczko-Grabarek A, Gronwald J, Byrski T, Spurdle AB, Bonanni B, Zaffaroni D, Giannini G, Bernard L, Dolcetti R, Manoukian S, Arnold N, Engel C, Deissler H, Rhiem K, Niederacher D, Plendl H, Sutter C, Wappenschmidt B, Borg A, Melin B, Rantala J, Soller M, Nathanson KL, Domchek SM, Rodriguez GC, Salani R, Kaulich DG, Tea MK, Paluch SS, Laitman Y, Skytte AB, Kruse TA, Jensen UB, Robson M, Gerdes AM, Ejlertsen B, Foretova L, Savage SA, Lester J, Soucy P, Kuchenbaecker KB, Olswold C, Cunningham JM, Slager S, Pankratz VS, Dicks E, Lakhani SR, Couch FJ, Hall P, Monteiro AN, Gayther SA, Pharoah PD, Reddel RR, Goode EL, Greene MH, Easton DF, Berchuck A, Antoniou AC, Chenevix-Trench G, Dunning AM. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013;45:371–384. 384e1–2. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kote-Jarai Z, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Dadaev T, Jugurnauth-Little S, Ross-Adams H, Al Olama AA, Benlloch S, Halim S, Russell R, Dunning AM, Luccarini C, Dennis J, Neal DE, Hamdy FC, Donovan JL, Muir K, Giles GG, Severi G, Wiklund F, Gronberg H, Haiman CA, Schumacher F, Henderson BE, Le Marchand L, Lindstrom S, Kraft P, Hunter DJ, Gapstur S, Chanock S, Berndt SI, Albanes D, Andriole G, Schleutker J, Weischer M, Canzian F, Riboli E, Key TJ, Travis RC, Campa D, Ingles SA, John EM, Hayes RB, Pharoah P, Khaw KT, Stanford JL, Ostrander EA, Signorello LB, Thibodeau SN, Schaid D, Maier C, Vogel W, Kibel AS, Cybulski C, Lubinski J, Cannon-Albright L, Brenner H, Park JY, Kaneva R, Batra J, Spurdle A, Clements JA, Teixeira MR, Govindasami K, Guy M, Wilkinson RA, Sawyer EJ, Morgan A, Dicks E, Baynes C, Conroy D, Bojesen SE, Kaaks R, Vincent D, Bacot F, Tessier DC COGSCRUK GWAS-ELLIPSE (Part of GAME-ON) Initiative; UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology; UK ProtecT Study Collaborators; PRACTICAL Consortium. Easton DF, Eeles RA. Fine-mapping identifies multiple prostate cancer risk loci at 5p15, one of which associates with TERT expression. Hum Mol Genet. 2013;22:2520–2528. doi: 10.1093/hmg/ddt086. [DOI] [PMC free article] [PubMed] [Google Scholar]


