Abstract
Occludin is transmembrane protein and a key constituent of tight junction, and might participate in barrier function and fence function of epithelia and endothelia. It has been shown to be aberrantly expressed in malignant tumors and plays a role in carcinogenesis and tumor progression. The prognostic significance of Occludin expression has been implicated in various human cancers. However, the prognostic significance of Occludin expression in esophageal squamous cell carcinoma (ESCC) has not been established. In this study, we screened the tight junction genes aberrantly expressed based on two published gene microarray datasets (GSE20347 and GSE23400), and examined 95 esophageal cancer cases to assess immunohistochemical expression patterns of Occludin based on tissue array. Down-regulation of Occludin expression was shown in ESCC as compared with adjacent non-neoplastic specimens (P = 0.003). Decreased expression of Occludin was correlated with high histological grade (P = 0.017). Decreased expression of Occludin was also correlated with short overall survival (P = 0.014). The results indicated Loss of Occludin expression was associated with poor prognosis in ESCC, and Occludin expression was potentially a good predictor of prognosis in ESCC.
Keywords: Occludin, prognosis, ESCC
Introduction
Esophageal cancer is the eighth most common cancer and the sixth leading cause of cancer-related death globally [1,2]. Squamous cell carcinoma (SCC) is the main histological type of esophageal cancer. The so-called Asian-belt encompassing China has a high incidence of esophageal SCC with more than 100/100,000 each year. The prognosis for esophageal SCC is much poorer because of diagnosis at an advanced stage and a high incidence of metastasis and recurrence post-treatment [3]. The overall 5-year survival ranges only around 10% [4]. The TNM staging is the predominant standard for predicting recurrence and metastasis of esophageal SCC, however, lacks sensitivity and accuracy.
Tight junctions are important components contributing to a constitutive barrier of epithelial and endothelial cells [5]. They have two classical functions-barrier function which regulates the passage of various ions and molecules between cells, and fence function which maintains cellular polarity [6]. However, tight junction proteins are also recognized as having functions beyond “barrier and fence”. They also involve in signal transduction mechanisms and regulation of innate immunity in association with carcinogenesis [6]. A diversity of cancerous tissues showed a differential expression of tight junction proteins in human beings. For example, Cldn1 protein was up-regulated in human cervical adenocarcinoma [7] and colorectal cancer tissues [8]. Moreover, altered expression and distribution of tight junction proteins have also been reported in other human malignancies, including pancreas, lung, ovarian, thyroid, prostate, esophagus and breast [9,10]. In turn, tight junction proteins modulated by signaling pathways, cytokines and growth factors, could influence biological behaviors of malignancies [11]. The loss of cell junctional sealing could involve in proliferation, transformation, infiltration of cancer cells and epithelial-mesenchymal transition in association with metastasis [12,13]. Breast cancer with bone metastasis had significantly lower occludin expression in comparison with those without bone metastasis [14].
TNM staging system is a classical evaluation system for recurrent status and prognosis of malignancies. However, it also can’t evaluate recurrence and prognosis of some cancers accurately. The correlation of tight junction protein with patients’ clinicopathological characteristics could be used to evaluate prognosis of malignancies. Occludin, as a constituent of tight junction, is aberrantly expressed in certain types of cancers. Some studies reported Occludin expression was associated with degree of differentiation, TNM staging, metastasis and patents’ survival in gallbladder adenocarcinoma, gastric and breast cancers [15-17]. However, correlation of Occludin expression with recurrence and prognosis of ESCC has not been cleared. In this study, tissue microarray was performed to analyze Occludin expression in 95 ESCC in order to elucidate whether Occludin expression is correlated with clinicopathological characteristics and clinical outcomes.
Materials and methods
Gene datasets and analysis
Two published studies were selected involving gene expression profiling in human ESCC [18,19]. The raw data were accessible through the GEO datasets available on PubMed (GSE20347 and GSE23400). These two datasets were generated using cDNA array. Various numbers of tissue samples were used in these two studies:
1. The study by Hu N, et al utilized Affymetrix HG-U133A 2.0 gene expression arrays to analyze tissue samples of 17 ESCC patients from a high risk region of China. Normal adjacent and ESCC tissues were obtained from each patient with confirmation of histology [18].
2. The study by Su H, et al utilized Affymetrix HG-U133B 2.0 gene expression arrays to analyze 53 matched samples of ESCC and normal adjacent tissues. Histological diagnosis was obtained from experienced pathologists [19].
In order to determine individual gene differentially expressed between ESCC and normal adjacent tissues, Significance analysis of microarrays (SAM) was utilized to analyze the raw, normalized datasets. Delta values were adjusted until the FDR was less than 1%. An arbitrary one and a half-fold cut-off threshold were then used to the list of significant genes from SAM analysis. Tight junction genes differentially expressed were selected from each of the datasets. These genes were designated overlap gene list. Differential expression of specific gene was confirmed by tissue array.
Patients and follow-up
In this research, ninety-five ESCC patients who underwent radical esophagectomy were included between July 2006 and December 2008. The tissue samples were stored at the biobank center in National Engineering Center for Biochip at Shanghai. All the tissue samples were obtained with informed written consent, and ethical approval was obtained from the ethnical committee of biobank center related hospital. Formalin-fixed and paraffin-embedded (FFPE) tissue samples and clinicopathological data were collected. Clinical diagnosis and tumor differentiation assessment were according to WHO grading criteria [20]. Pathological staging was determined according to NCCN Esophageal Cancer Guideline [21].
The patients after surgery were followed up ranging from 5.8 years to 7.8 years (median 6.9 years). Tumor relapse was monitored by enhancement thoracic and abdominal CT every 3 months for the first and second years, and 6 months thereafter. Cerebral CT and radioisotope bone scanner were performed if necessary. Any new masses in the related organs were considered as recurrence. The follow-up interruption and death other than tumor relapse was considered as a censoring event.
Construction of tissue microarray (TMA)
Representative cancerous region and its adjacent non-malignant specimen were selected by two separate pathologists from each tissue block. A cylinder-shaped hole were created by a tissue arraying instrument (Beecher Instruments, Sun Prarie, WI) in a square recipient paraffin block. The tissue cores from the donor blocks of cancerous and adjacent tissue samples were removed by a hollow needle with an inner diameter of 1.5 mm. The cores from the donor blocks were inserted into a recipient paraffin block in a precisely spaced, array pattern. A series of 5-μm sections were prepared with a microtome, and a perfect piece of which was placed on polylysine-coated slides. Sections were stained with H&E to confirm the presence of tumor within each core, and immunostaining of Occludin was performed as described below.
Immunostaining and evaluation
The TMA slides were heated at 60°C for 20 min, deparaffinized and rehydrated in an ethanol gradient, washed with Tris-buffered saline (TBS). Antigen retrieval was performed for 15 min at water boiling point under high pressure in sodium citrate buffer (PH = 6), followed by 3% hydrogen peroxide in phosphate buffer saline (PBS), followed by PBS for 5 min three times. Upon quenching endogenous peroxidase activity and blocking non-specific binding, anti-occludin polyclonal antibody (PA5-30230, Invitrogen, Camarrilo, CA) was added at a dilution of 1:800. The slides were incubated with primary antibody overnight in a humid chamber at 4°C. The corresponding secondary biotinylated antibody was used for 30 min at room temperature. After further washing with TBS, sections were incubated with StrepABC complex/horseradish peroxidase (1:100, DAKO) for 30 min at room temperature. 3,3-diaminobenzidine terahydrochloride at a concentration of 0.05% was used for chromogenic immunolocalization. Sections were counterstained with hematoxylin before dehydration and mounting. The cores containing breast carcinoma served as positive control.
Positive immunostaining of occludin was defined mainly in the location of cytoplasm. It was graded by the intensity and percentage of cells with positive staining. The immunoreactivity was assessed by two pathologists simultaneously, and a consensus was reached for each core. The staining intensity of occludin was scored from 0 to 3 (0 = negative; 1 = weak; 2 = moderate; 3 = strong). The percentage of positive cells was also graded into 6 categories: 0 (negative), 1 (1%-20%), 2 (21%-40%), 3 (41%-60%), 4 (61%-80%), 5 (81%-100%). In the cases with a discrepancy between duplicated cores, the average score from the duplicated cores was taken as the final score. The whole level of occludin expression in carcinoma was evaluated by immunoreactive score (IRS), which was calculated by multiplying the scores of staining intensity and the percentage of positive cells. Based on IRS, staining pattern was defined as low expression (IRS≤6) and high expression (IRS>6).
Statistical analysis
The differential expression of occludin between cancerous and adjacent tissues was determined by Wilcoxon signed rank test. Correlation analysis between clinicopathological and molecular parameters was analyzed by Spearman’s rank analysis. Survival curves were performed with the Kaplan-Meier analysis and compared by the log-rank test. A multivariate analysis was applied by the Cox regression model to evaluate independent prognostic factors for ESCC. A two-tailed P value of less than 0.05 was considered as statistical significance. All the statistical analysis was performed with SPSS 19.0 version (SPSS Inc, Chicago, IL).
Results
Differential expression of tight junction genes in ESCC
GSE23400 data showed 5 tight junction genes were over-expressed, and the expression of 2 tight junction genes was down-regulated. GSE20347 data included 7 tight junction genes down-regulated (Table 1). It was observed only the expression of Occludin was down-regulated in both datasets.
Table 1.
Differential expression of tight junction genes in ESCC
The expression of occludin was down regulated in ESCC specimens comparing with the corresponding adjacent specimens
The expression of Occludin was down-regulated in ESCC in comparison with that in adjacent tissues based on 2 datasets (Table 1). Furthermore, TMA was used to investigate Occludin protein expression in ESCC and adjacent tissues. The specific staining was semi-quantitatively scored by IRS. In this study, the specific staining for Occludin within ESCC and adjacent tissues was primarily located in the cytoplasm. The expression of Occludin protein in ESCC was down-regulated as comparison with that in adjacent tissues (Table 2). The representative images of immunostaining were shown in Figure 1.
Table 2.
Expression of Occludin protein within ESCC and adjacent normal tissues based on TMA by IRS
| Mean ± Std. Deviation | P Value | Z Value | |
|---|---|---|---|
| ESCC | 3.596 ± 3.039 | 0.003 | -2.944 |
| Adjacent normal | 4.438 ± 2.739 |
Figure 1.
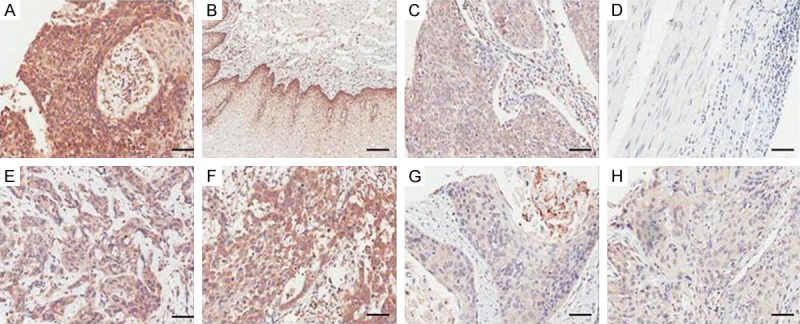
Immunohistochemical expression patterns of Occludin. A. The strong staining intensity of Occludin. B. The moderate staining intensity of Occludin. C. The weak staining intensity of Occludin. D. The negative staining intensity of Occludin. E, F. The high expression of Occludin in ESCC. G, H. The low expression of Occludin in ESCC. The scale bar represented 50 µm.
Correlation analysis between occludin protein expression and clinicopathological characteristics
The clinicopathological characteristics for ESCC patients were presented in Table 3. TMA in combination with immunostaining was used to investigate the correlation of Occludin protein expression with clinicopathological characteristics. The expression of Occludin protein within ESCC was significantly associated with that in adjacent tissues (r = 0.573, P = 0.00) and histological grade (r = -0.248, P = 0.017). There were no statistical differences among Occludin protein expression, gender, age, tumor size, T stage, lymph node metastasis and TNM staging (Table 4).
Table 3.
Clinicopathological characteristics of ESCC patients
| Characteristic | Sample number | Percent |
|---|---|---|
| Gender | ||
| Male | 73 | 76.8 |
| Female | 22 | 23.2 |
| Age (years) | ||
| ≤65 | 49 | 51.6 |
| >65 | 46 | 48.4 |
| Histological grade | ||
| I | 7 | 7.4 |
| II | 64 | 67.4 |
| III | 24 | 25.2 |
| Tumor size (cm) | ||
| ≤5 | 58 | 61.1 |
| >5 | 28 | 29.5 |
| Unknown | 9 | 9.4 |
| T stage | ||
| T1-2 | 17 | 17.9 |
| T3-4 | 74 | 77.9 |
| Unknown | 4 | 10.2 |
| Lymph nodes metastasis | ||
| N0 | 45 | 47.4 |
| N1-3 | 49 | 51.6 |
| Unknown | 1 | 1 |
| TNM stage | ||
| TNM1-2 | 47 | 49.5 |
| TNM3-4 | 44 | 46.3 |
| Unknown | 4 | 4.2 |
Table 4.
Correlation analysis between Occludin expression and clinicopathological characteristics
| Occludin expression in ESCC | |||
|---|---|---|---|
|
|
|||
| Correlation coefficient | Sig. (2-tailed) | Sample number | |
| Occludin expression in adjacent | 0.573 | 0.000 | 76 |
| Gender | 0.033 | 0.755 | 93 |
| Age | -0.006 | 0.951 | 93 |
| Tumor size | 0.034 | 0.759 | 84 |
| Histological grade | -0.248 | 0.017 | 93 |
| T stage | 0.009 | 0.933 | 89 |
| Lymph nodes metastasis | 0.113 | 0.284 | 92 |
| TNM stage | 0.046 | 0.671 | 89 |
Bold black indicated the significance of correlation analysis.
Univariate survival analysis and multivariate cox regression analysis
Univariate analysis was performed by Kaplan-Meier survival curve and log-rank test. It was demonstrated that the overall survival (OS) of ESCC with low expression of Occludin was inferior to those with high expression (P = 0.014), whereas the OS of adjacent tissues with low expression of Occludin was no statistical difference comparing with that with high expression (P = 0.081; Figure 2). Moreover, inferior OS was also observed in patients with male (P = 0.019), larger tumor size (P = 0.012), lymph node metastasis (P = 0.001), advanced TNM stage (P = 0.001) and the advanced depth of tumor invasion (P = 0.025).
Figure 2.
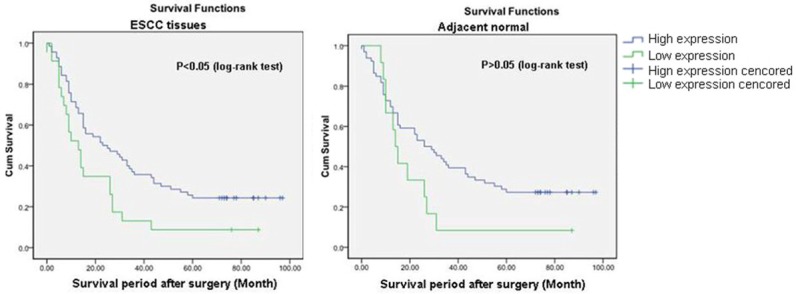
Overall survival curves of patients based on Occludin expression within ESCC and adjacent tissues.
In addition, we also performed multivariate analysis. All the significant factors in the univariate analysis were included in further multivariate analysis. It was indicated that expression of Occludin in ESCC tissues was an independent factor to predict OS of the patients (Table 5).
Table 5.
Multivariate Cox regression analysis predicting survival within ESCC patients
| RC | SE | Wald | DF | P value | RR | 95% CL of RR | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| LL | UL | |||||||
| Low expression of Occludin in ESCC | 0.537 | 0.267 | 4.066 | 1 | 0.044 | 1.763 | 0.214 | 2.886 |
| T stage | 0.209 | 0.392 | 0.286 | 1 | 0.593 | 1.233 | 0.572 | 2.658 |
| Lymph nodes metastasis | -0.180 | 0.633 | 0.081 | 1 | 0.777 | 0.836 | 0.242 | 2.890 |
| TNM stage | 0.88 | 0.668 | 1.735 | 1 | 0.188 | 2.412 | 0.651 | 8.941 |
RC: Regression coefficient; SE: Standard error; DF: Degree of freedom; RR: Relative risk; CL: Confidence level; LL: lower limit; UL: upper limit. Bold black indicated the significance of the factor for predicting prognosis of ESCC patients.
Discussion
Occludin, together with claudins, functions as a selective “gate and fence” in epithelial and endothelial cell layers. Besides, a number of studies illustrated Occludin was also important for carcinogenesis and cancer invasion. The purpose of this study was to figure out the relations between Occludin expression and clinicopathological findings in ESCC. We demonstrated that Occludin expression was down-regulated in ESCC as compared with adjacent normal tissues and Loss of Occludin was associated with poor prognosis.
The expression pattern of tight junction proteins was able to distinguish from different types of carcinoma, which was potential for diagnosis. Loss of Occludin expression might be one phenotypic feature distinguishing diffuse-type gastric carcinoma from intestinal-type gastric carcinoma [22]. Cldn1 expression was reported to be stronger in premalignant stages (e.g., carcinoma in situ of breast and cervical intraepithelial neoplasia), while a significant decrease was found in invasive carcinoma (e.g., invasive breast and cervical cancer) [23,24]. In addition, the expression pattern of tight junction proteins could be used to differentiate carcinoma from non-neoplasia, Several studies reported Cldn3 and Cldn4 were over-expressed in ovarian and uterine serous papillary carcinoma as compared with normal ovarian cells and endometrium, which showed potential diagnostic markers for neoplasia [25,26].
Over-expression of tight junction proteins would be expected to increase cell adhesion and consequently suppress invasion and motility of tumor cells. However, in certain cancers, over-expression of tight junction proteins might be related to carcinogenesis. The expression of Occludin was significantly increased in hepatocellular carcinoma and urothelial carcinoma as compared with non-neoplastic tissues and normal controls [27,28]. Cldn1 protein was found to be increased in colon carcinoma, which was identified as a potential target for β-catenin/Tcf signaling [29,30]. Cldn3 and Cldn4 were over-expressed in ovarian carcinoma [26]. In uterine serous papillary carcinoma, the mRNA levels for Cldn3 and Cldn4 were higher in carcinoma as compared with normal endometria [25]. Several additional reports confirmed these two Occludins were also increased in other cancers, such as breast, prostate and pancreatic cancers [9]. These results suggested tight junction proteins including Occludin might involve in regulation of signaling pathway, which were not associated with the formation of tight junctions [31].
In certain tumors, loss of Occludin was associated with carcinogenesis. A frequent complete loss of Occludin was observed in cutaneous squamous cell carcinoma as compared with the precursor lesions and sun-exposed skin, which might result in a decrease of epithelial cell-cell adhesion and a reduction of susceptibility to apoptosis [32]. Loss of Occludin protein was also observed in gallbladder adenocarcinoma [15], poorly differentiated carcinoma from stomach and colon [16,33,34] and cholangiocarcinoma [35] as compared with adjacent normal tissues and specific benign lesions. Tight junction was important for barrier function and cell polarity. Disruption of tight junction manifested by loss of Occuldin lead to destruction of barrier function and loss of cellular polarity, resulting in abnormal diffusion of nutrients and other factors necessary for survival and growth of malignant cells [36-38]. Loss of cell-to-cell adhesion triggered dissociation of cancer cells from primary nests, which was a crucial step for cancer progression and metastasis [9]. It was reported breast cancer with bone metastasis had significantly lower occludin expression as compared with that without metastasis, especially for breast ductal cancer [14]. A study gained the similar result that Occludin mRNA was significantly decreased in metastatic breast cancer as compared with normal breast tissues [17]. Although weak expression of Occludin protein was examined in normal hepatocytes, down-regulation of Occudin mRNA was reported in metastasis as compared with normal hepatic tissues [39]. In our present study, Loss of Occludin protein and down-regulation of Occludin mRNA were markedly observed in ESCC as compared with adjacent normal tissues.
We also reported the potential role of Occludin in clinico-pathological correlation of ESCC. Loss of Occludin was only associated with advanced tumor grade in our study. Other studies also suggested the association of Occludin expression with clinico-pathological parameters. It was reported loss of Occludin had a clear relationship with increased invasion, reduced adhesion and metastasis in breast cancer, which indicated loss of Occludin lead to complex changes in cellular phenotype of human cancers [14,17]. In gallbladder carcinoma, loss of Occludin was also associated with advanced tumor grade, a maximal tumor size >2 cm, lymph node metastasis and invasion to regional tissues [15]. Similarly, Occludin expression was significantly decreased in gastric carcinoma with undifferentiated-type [33]. On the one hand, loss of Occludin might lead to loss of cell-cell adhesion, driving the motility of cancer cells and the release of cancer cells from primary nests, conferring invasive and metastatic properties [9]. On the other, loss of Occludin also might lead to poor differentiation of cancer cells by influencing signaling pathways regulating cellular proliferation, differentiation and apoptosis, showing invasive and metastatic features [11]. However, some studies reported no association between Occludin expression and clinicopathological characteristics in cervical adenocarcinoma [40], hepatocellular carcinoma [27], ovarian carcinoma [41] and squamous cell carcinoma of tongue [41]. The discrepancy was not cleared now.
Many patients suffered from advanced ESCC with lymph node metastasis and regional invasion at the time of diagnosis, as it was difficult to detect this lesion at an early stage [42]. Clinicopathological characteristics, e.g., degree of differentiation, mass location, depth of invasion and lymph node status were associated with staging and grading [43], which were crucial parameters for predicting survival and prognosis of tumors [44]. It was also urgent to identify some novel and potential prognostic markers. Occludin might be a potential prognostic factor for ESCC manifested by regulating cell-cell adhesiveness and associating with frontal invasion and metastasis. For example, a negative prognostic effect of Occludin low-expression was reported in malignant gallbladder lesions [15] and brain tumors [45], showing a favorable-prognostic potential of Occludin. However, Occludin expression did not show any prognostic effects in some tumors, e.g., hepatocellular carcinoma [27], gastric carcinoma [33] and squamous cell carcinoma of the tongue [46]. The reasons for differential prognostic effects of Occludin in different cancers were unknown. The discrepancy was related to the possibility that functional implications of Occludin differed among organs.
In conclusion, this study determined the prognostic value of Occludin expression and loss of Occludin was found to be associated with poor prognosis in ESCC. Therefore, we considered Occludin might be a novel and potential indicator of prognosis in ESCC.
Acknowledgements
This work was supported by National Natural Science Foundation of China (NSFC) 81400590.
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Pennathur A, Farkas A, Krasinskas AM, Ferson PF, Gooding WE, Gibson MK, Schuchert MJ, Landreneau RJ, Luketich JD. Esophagectomy for T1 esophageal cancer: outcomes in 100 patients and implications for endoscopic therapy. Ann Thorac Surg. 2009;87:1048–1054. doi: 10.1016/j.athoracsur.2008.12.060. discussion 1054-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 5.Oshima T, Miwa H. Gastrointestinal mucosal barrier function and diseases. J Gastroenterol. 2016;51:768–778. doi: 10.1007/s00535-016-1207-z. [DOI] [PubMed] [Google Scholar]
- 6.Sawada N. Tight junction-related human diseases. Pathol Int. 2013;63:1–12. doi: 10.1111/pin.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akimoto T, Takasawa A, Murata M, Kojima Y, Takasawa K, Nojima M, Aoyama T, Hiratsuka Y, Ono Y, Tanaka S, Osanai M, Hasegawa T, Saito T, Sawada N. Analysis of the expression and localization of tight junction transmembrane proteins, claudin-1, -4, -7, occludin and JAM-A, in human cervical adenocarcinoma. Histol Histopathol. 2016;31:921–931. doi: 10.14670/HH-11-729. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Tully O, Ngo B, Zitin M, Mullin JM. Epithelial tight junctional changes in colorectal cancer tissues. ScientificWorldJournal. 2011;11:826–841. doi: 10.1100/tsw.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding L, Lu Z, Lu Q, Chen YH. The claudin family of proteins in human malignancy: a clinical perspective. Cancer Manag Res. 2013;5:367–375. doi: 10.2147/CMAR.S38294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soini Y. Tight junctions in lung cancer and lung metastasis: a review. Int J Clin Exp Pathol. 2012;5:126–136. [PMC free article] [PubMed] [Google Scholar]
- 11.Takano K, Kojima T, Sawada N, Himi T. Role of tight junctions in signal transduction: an update. EXCLI J. 2014;13:1145–1162. [PMC free article] [PubMed] [Google Scholar]
- 12.Martin TA, Mason MD, Jiang WG. Tight junctions in cancer metastasis. Front Biosci (Landmark Ed) 2011;16:898–936. doi: 10.2741/3726. [DOI] [PubMed] [Google Scholar]
- 13.Tsukita S, Yamazaki Y, Katsuno T, Tamura A. Tight junction-based epithelial microenvironment and cell proliferation. Oncogene. 2008;27:6930–6938. doi: 10.1038/onc.2008.344. [DOI] [PubMed] [Google Scholar]
- 14.Martin TA, Jordan N, Davies EL, Jiang WG. Metastasis to bone in human cancer is associated with loss of occludin expression. Anticancer Res. 2016;36:1287–1293. [PubMed] [Google Scholar]
- 15.Xiong L, Wen Y, Miao X, Yang Z. Expressions of cell junction regulatory proteins and their association with clinicopathologic parameters in benign and malignant gallbladder lesions. Am J Med Sci. 2011;342:388–394. doi: 10.1097/MAJ.0b013e31821e12af. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, Zhang S, Zhen K, Wang XF. [Relationship between the expression of occludin and tumor genesis and development in the gastric carcinoma] . Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2010;26:1008–1011. [PubMed] [Google Scholar]
- 17.Martin TA, Mansel RE, Jiang WG. Loss of occludin leads to the progression of human breast cancer. Int J Mol Med. 2010;26:723–734. doi: 10.3892/ijmm_00000519. [DOI] [PubMed] [Google Scholar]
- 18.Hu N, Clifford RJ, Yang HH, Wang C, Goldstein AM, Ding T, Taylor PR, Lee MP. Genome wide analysis of DNA copy number neutral loss of heterozygosity (CNNLOH) and its relation to gene expression in esophageal squamous cell carcinoma. BMC Genomics. 2010;11:576. doi: 10.1186/1471-2164-11-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su H, Hu N, Yang HH, Wang C, Takikita M, Wang QH, Giffen C, Clifford R, Hewitt SM, Shou JZ, Goldstein AM, Lee MP, Taylor PR. Global gene expression profiling and validation in esophageal squamous cell carcinoma and its association with clinical phenotypes. Clin Cancer Res. 2011;17:2955–2966. doi: 10.1158/1078-0432.CCR-10-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q, Zhang N, Jia Z, Le X, Dai B, Wei D, Huang S, Tan D, Xie K. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69:3501–3509. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ajani JA, D’Amico TA, Almhanna K, Bentrem DJ, Besh S, Chao J, Das P, Denlinger C, Fanta P, Fuchs CS, Gerdes H, Glasgow RE, Hayman JA, Hochwald S, Hofstetter WL, Ilson DH, Jaroszewski D, Jasperson K, Keswani RN, Kleinberg LR, Korn WM, Leong S, Lockhart AC, Mulcahy MF, Orringer MB, Posey JA, Poultsides GA, Sasson AR, Scott WJ, Strong VE, Varghese TK Jr, Washington MK, Willett CG, Wright CD, Zelman D, McMillian N, Sundar H National comprehensive cancer network. Esophageal and esophagogastric junction cancers, version 1.2015. J Natl Compr Canc Netw. 2015;13:194–227. doi: 10.6004/jnccn.2015.0028. [DOI] [PubMed] [Google Scholar]
- 22.Soini Y, Tommola S, Helin H, Martikainen P. Claudins 1, 3, 4 and 5 in gastric carcinoma, loss of claudin expression associates with the diffuse subtype. Virchows Arch. 2006;448:52–58. doi: 10.1007/s00428-005-0011-6. [DOI] [PubMed] [Google Scholar]
- 23.Sobel G, Paska C, Szabo I, Kiss A, Kadar A, Schaff Z. Increased expression of claudins in cervical squamous intraepithelial neoplasia and invasive carcinoma. Hum Pathol. 2005;36:162–169. doi: 10.1016/j.humpath.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Morohashi S, Kusumi T, Sato F, Odagiri H, Chiba H, Yoshihara S, Hakamada K, Sasaki M, Kijima H. Decreased expression of claudin-1 correlates with recurrence status in breast cancer. Int J Mol Med. 2007;20:139–143. [PubMed] [Google Scholar]
- 25.Dhawan P, Singh AB, Deane NG, No Y, Shiou SR, Schmidt C, Neff J, Washington MK, Beauchamp RD. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest. 2005;115:1765–1776. doi: 10.1172/JCI24543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Y, Brannstrom M, Janson PO, Sundfeldt K. Differences in expression patterns of the tight junction proteins,claudin 1, 3, 4 and 5, in human ovarian surface epithelium as compared to epithelia in inclusion cysts and epithelial ovarian tumours. Int J Cancer. 2006;118:1884–1891. doi: 10.1002/ijc.21506. [DOI] [PubMed] [Google Scholar]
- 27.Bouchagier KA, Assimakopoulos SF, Karavias DD, Maroulis I, Tzelepi V, Kalofonos H, Kardamakis D, Scopa CD, Tsamandas AC. Expression of claudins-1, -4, -5, -7 and occludin in hepatocellular carcinoma and their relation with classic clinicopathological features and patients’ survival. In Vivo. 2014;28:315–326. [PubMed] [Google Scholar]
- 28.Nakanishi K, Ogata S, Hiroi S, Tominaga S, Aida S, Kawai T. Expression of occludin and claudins 1, 3, 4, and 7 in urothelial carcinoma of the upper urinary tract. Am J Clin Pathol. 2008;130:43–49. doi: 10.1309/U77A6BTEXVCA5D0E. [DOI] [PubMed] [Google Scholar]
- 29.Long H, Crean CD, Lee WH, Cummings OW, Gabig TG. Expression of clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in prostate cancer epithelium. Cancer Res. 2001;61:7878–7881. [PubMed] [Google Scholar]
- 30.Miwa N, Furuse M, Tsukita S, Niikawa N, Nakamura Y, Furukawa Y. Involvement of claudin-1 in the beta-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol Res. 2001;12:469–476. doi: 10.3727/096504001108747477. [DOI] [PubMed] [Google Scholar]
- 31.Rangel LB, Agarwal R, D’Souza T, Pizer ES, Alo PL, Lancaster WD, Gregoire L, Schwartz DR, Cho KR, Morin PJ. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res. 2003;9:2567–2575. [PubMed] [Google Scholar]
- 32.Rachow S, Zorn-Kruppa M, Ohnemus U, Kirschner N, Vidal-y-Sy S, von den Driesch P, Bornchen C, Eberle J, Mildner M, Vettorazzi E, Rosenthal R, Moll I, Brandner JM. Occludin is involved in adhesion, apoptosis, differentiation and Ca2+-homeostasis of human keratinocytes: implications for tumorigenesis. PLoS One. 2013;8:e55116. doi: 10.1371/journal.pone.0055116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohtani S, Terashima M, Satoh J, Soeta N, Saze Z, Kashimura S, Ohsuka F, Hoshino Y, Kogure M, Gotoh M. Expression of tight-junctionassociated proteins in human gastric cancer: downregulation of claudin-4 correlates with tumor aggressiveness and survival. Gastric Cancer. 2009;12:43–51. doi: 10.1007/s10120-008-0497-0. [DOI] [PubMed] [Google Scholar]
- 34.Kimura Y, Shiozaki H, Hirao M, Maeno Y, Doki Y, Inoue M, Monden T, Ando-Akatsuka Y, Furuse M, Tsukita S, Monden M. Expression of occludin, tight-junction-associated protein, in human digestive tract. Am J Pathol. 1997;151:45–54. [PMC free article] [PubMed] [Google Scholar]
- 35.Nemeth Z, Szasz AM, Somoracz A, Tatrai P, Nemeth J, Gyorffy H, Szijarto A, Kupcsulik P, Kiss A, Schaff Z. Zonula occludens-1, occludin, and E-cadherin protein expression in biliary tract cancers. Pathol Oncol Res. 2009;15:533–539. doi: 10.1007/s12253-009-9150-4. [DOI] [PubMed] [Google Scholar]
- 36.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 37.Matsuda Y, Semba S, Ueda J, Fuku T, Hasuo T, Chiba H, Sawada N, Kuroda Y, Yokozaki H. Gastric and intestinal claudin expression at the invasive front of gastric carcinoma. Cancer Sci. 2007;98:1014–1019. doi: 10.1111/j.1349-7006.2007.00490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez-Mariscal L, Betanzos A, Nava P, Jaramillo BE. Tight junction proteins. Prog Biophys Mol Biol. 2003;81:1–44. doi: 10.1016/s0079-6107(02)00037-8. [DOI] [PubMed] [Google Scholar]
- 39.Orban E, Szabo E, Lotz G, Kupcsulik P, Paska C, Schaff Z, Kiss A. Different expression of occludin and ZO-1 in primary and metastatic liver tumors. Pathol Oncol Res. 2008;14:299–306. doi: 10.1007/s12253-008-9031-2. [DOI] [PubMed] [Google Scholar]
- 40.Marzesco AM, Dunia I, Pandjaitan R, Recouvreur M, Dauzonne D, Benedetti EL, Louvard D, Zahraoui A. The small GTPase Rab13 regulates assembly of functional tight junctions in epithelial cells. Mol Biol Cell. 2002;13:1819–1831. doi: 10.1091/mbc.02-02-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Jin X, Lin D, Liu Z, Zhang X, Lu Y, Liu Y, Wang M, Yang M, Li J, Quan C. Clinicopathologic significance of claudin-6, occludin, and matrix metalloproteinases -2 expression in ovarian carcinoma. Diagn Pathol. 2013;8:190. doi: 10.1186/1746-1596-8-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goseki N, Koike M, Yoshida M. Histopathologic characteristics of early stage esophageal carcinoma. A comparative study with gastric carcinoma. Cancer. 1992;69:1088–1093. doi: 10.1002/cncr.2820690503. [DOI] [PubMed] [Google Scholar]
- 43.Akiyama H, Tsurumaru M, Kawamura T, Ono Y. Principles of surgical treatment for carcinoma of the esophagus: analysis of lymph node involvement. Ann Surg. 1981;194:438–446. doi: 10.1097/00000658-198110000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klimstra DS. Pathologic prognostic factors in esophageal carcinoma. Semin Oncol. 1994;21:425–430. [PubMed] [Google Scholar]
- 45.Park MW, Kim CH, Cheong JH, Bak KH, Kim JM, Oh SJ. Occludin expression in brain tumors and its relevance to peritumoral edema and survival. Cancer Res Treat. 2006;38:139–143. doi: 10.4143/crt.2006.38.3.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bello IO, Vilen ST, Niinimaa A, Kantola S, Soini Y, Salo T. Expression of claudins 1, 4, 5, and 7 and occludin, and relationship with prognosis in squamous cell carcinoma of the tongue. Hum Pathol. 2008;39:1212–1220. doi: 10.1016/j.humpath.2007.12.015. [DOI] [PubMed] [Google Scholar]


