Abstract
Polycystic ovary syndrome (PCOS) is one of the most frequently encountered endocrine malfunctions. The etiology of PCOS is complex and unclear. We aimed to investigate the role of three common SNPs (rs1800896, rs1800871 and rs1800872) of IL-10 in the development of PCOS in a Chinese population. We recruited 360 patients with PCOS and 360 healthy controls in this study. SNP genotyping of IL-10 rs1800896, rs1800871 and rs1800872 was implemented in a 384-well plate format on the Sequenom MassARRAY® System (Sequenom, San Diego, USA). Individuals carrying the GG genotype of rs1800872 were associated with an increased risk of PCOS when compared with the AA genotype (OR = 3.04, 95% CI = 1.62-5.69). No linkage disequilibrium was observed among IL-10 rs1800896, rs1800871 and rs1800872. The C-T-A (OR = 1.53, 95% CI = 1.05-2.35) haplotype indicated an increased risk of PCOS, while the A-C-G (OR = 0.72, 95% CI = 0.53-0.98) showed a reduced risk of PCOS. In summary, this study firstly estimates the association between IL-10 polymorphisms and haplotype and PCOS risk in the Chinese population.
Keywords: Polycystic ovary syndrome, IL-10, polymorphism, haplotype
Introduction
Polycystic ovary syndrome (PCOS) is one of the most frequently encountered endocrine malfunctions. About 7.1-11.2% of women aged 12-44 years are suffering from PCOS in China [1]. The etiology of PCOS is complex and unclear. Many environmental factors are involved in the development of PCOS, such as obesity, adrenal dysfunction, as well as plastic-packaged food and drinking alcohol [2,3]. However, the incidence of PCOS greatly varied across different populations even when they are exposure to the same environmental risk factors, suggesting that hereditary factors could influence the pathogenesis of PCOS. An increasing number of studies have indicated that many genetic factors, such as paraoxonase 1, methylenetetrahydrofolate reductase C677T and A1298C, adiponectin, interleukin (IL)-1A and IL-6, play an important role in the risk of PCOS [4-7].
Immune dysregulation is an important factor in the development of PCOS, and chronic inflammation might be an critical underlying mechanism for PCOS risk [8]. Cytokines are important immunomodulatory proteins for controlling or regulating the cell activity and function in the immune system [9]. Low level of chronic inflammation and imbalance between pro- and anti-inflammatory cytokines have been considered to be involved in the pathogenesis of PCOS [10]. IL-10, encoded by a gene located on chromosome 1 (1q31-1q32), is an immunoregulatory cytokine produced by Th2 cells, regulatory T cells, and monocytes/macrophages. This anti-inflammatory cytokine can inhibit the synthesis of other cytokines, such as IL-6 and interferon-γ in T cells [11]. Single nucleotide polymorphisms (SNPs) in the promoter regions of IL-10 can influence the function and expression of protein, and thus affects susceptibility to diseases. However, only a few studies reported the relationship between IL-10 polymorphisms and risk of polycystic ovary syndrome in Caucasian [12-15], and no study reported their association in Chinese. Therefore, we aimed to investigate the relationship between three common SNPs (rs1800896, rs1800871 and rs1800872) of IL-10 and risk of PCOS in a Chinese population.
Material and methods
Subjects
We recruited a total of 360 patients with PCOS from the Department of Reproductive Medicine, Luoyang Center Hospital Affiliated to Zhengzhou University and the First Affiliated Hospital of Zhengzhou University between October 2014 and October 2015. The diagnosis of PCOS was based on the criteria of Rotterdam polycystic ovary syndrome consensus in 2004. Patients who had prior history of late onset adrenal cortex hyperplasia, 21-hydroxyulase deficiency, Cushing’s syndrome and premature ovarian failure were excluded.
Simultaneously, 360 healthy controls were randomly collected from the outpatient’s clinics and health examination centers of the two hospitals. All controls were diagnosed to be free of PCOS by B ultrasonic examination and sexual hormone examination. The exclusion criteria were patients with irregular menstrual periods, endocrine diseases and ovarian-related diseases. Written informed consents were obtained from all participants before enrollment. Implement of this study was approved by the ethics committee of Luoyang Center Hospital Affiliated to Zhengzhou University and the First Affiliated Hospital of Zhengzhou University.
DNA extraction and genotyping methods
Three ml venous whole blood samples were collected and stored in ethylenediaminetetraacetic acid tubes. Tubes and plates with reagents are lightly vortexed and centrifuged before use. Genomic DNA was extracted using the TIANamp blood DNA kit (Tiangen Biotech, Beijing, China) according to the instructions. SNP genotyping analysis was implemented in a 384-well plate format on the Sequenom MassARRAY® System (Sequenom, San Diego, USA). Primers for polymerase chain reaction (PCR) amplification and single base extension assays were designed using Sequenom Assay Design 3.1 software. PCR reactions for genotyping IL-10 rs1800896, rs1800871 and rs1800872 were performed in 5 μL are performed, including 2.8 HPLC grade water, 0.5 μl of 10× PCR buffer with 20 mM MgCl2, 0.4 μl of 25 mM MgCl2, 0.1 μl of 25 mM dNTP Mix, 1.0 μl of 0.5 μM Primer Mix, 0.2 μl Sequenom PCR enzyme (5 U/μl). Then the sample was extended in the SAP and iPLEX reaction. The samples were finally analyzed with MALDI-TOF MS.
Statistical analysis
Categorical variables were displayed as percentages of the total, and continuous variables were shown as mean ± standard deviation (SD). The demographic and lifestyle characteristics between patients with PCOS and controls were compared by Chi-square (x2) test. The Hardy-Weinberg equilibrium (HWE) of IL-10 rs1800896, rs1800871 and rs1800872 was tested by x2 test with one degree of freedom. The relationship between IL-10 rs1800896, rs1800871 and rs1800872 and PCOS risk was estimated by conditional logistic regression analysis. The results were shown by odds ratio (OR) and 95% confident intervals (95% CI). Linkage disequilibrium and haplotype analysis was calculated by SHEsis software (http://analysis.bio-x.cn/myAnalysis.php). SPSS Statistics for Windows, version 20.0 (IBM Corp, Armonk, NY, USA) was used to perform the statistical analysis. P value < 0.05 was set to be statistical significant.
Results
Compared to the controls, patients with PCOS have higher levels of FPG (t = 3.73, P < 0.001), FINS (t = 2.40, P < 0.001), HOMA-IR (t = 3.44, P < 0.001), FSH (t = 4.30, P < 0.001), LH (t = 4.78, P < 0.001), P (t = 3.61, P < 0.001) and T (t = 6.29, P < 0.001) values (Table 1).
Table 1.
Demographic, lifestyle and environmental characteristics of included patients with PCOS and controls
| Variables | Patients (%) N = 360 | Controls (%) N = 360 | x2 or t value | P value |
|---|---|---|---|---|
| Age, years | 26.43±4.34 | 26.71±4.34 | -0.86 | 0.39 |
| Age of menarche, years | 13.38±2.01 | 13.61±1.93 | -1.58 | 0.12 |
| Weight, kg | 57.81±7.28 | 57.69±6.49 | 0.23 | 0.82 |
| Height, m | 1.60±0.06 | 1.61±0.07 | -1.59 | 0.11 |
| BMI, kg/m2 | 22.57±3.30 | 22.29±2.79 | 1.22 | 0.22 |
| FPG, mmol/L | 5.38±1.26 | 5.04±1.19 | 3.73 | < 0.001 |
| FINS, µIU/mL | 15.66±4.93 | 14.75±5.28 | 2.40 | < 0.05 |
| HOMA-IR | 3.85±1.73 | 3.41±1.72 | 3.44 | < 0.05 |
| Smoking | ||||
| No | 353 (98.60) | 349 (96.94) | 0.91 | 0.34 |
| Yes | 7 (1.94) | 11 (3.06) | ||
| Drinking | ||||
| No | 295 (81.94) | 288 (80.00) | 0.44 | 0.51 |
| Yes | 65 (18.06) | 72 (20.00) | ||
| Family history of diabetes mellitus | ||||
| No | 300 (83.33) | 311 (86.39) | 1.31 | 0.25 |
| Yes | 60 (16.67) | 49 (13.61) | ||
| FSH | 4.53±2.54 | 3.68±2.77 | 4.30 | < 0.001 |
| LH | 13.76±6.94 | 11.38±6.43 | 4.78 | < 0.001 |
| E2 | 91.82±21.85 | 94.66±24.14 | -1.65 | 0.10 |
| P | 8.93±3.28 | 8.05±3.26 | 3.61 | < 0.001 |
| T | 0.65±0.24 | 0.54±0.24 | 6.29 | < 0.001 |
The AA, AG and GG genotype distributions of rs1800872 showed significant differences between patients with PCOS and controls (x2 = 18.85, P < 0.001) (Table 2). However, no significant differences were found in the genotype frequencies of IL-10 rs1800896 and rs1800871. The genotype distributions of the three SNPs were in line with the HWE. We observed that individuals carrying the GG genotype of rs1800872 were associated with higher risk of PCOS when compared with the AA genotype (OR = 3.04, 95% CI = 1.62-5.69).
Table 2.
Genotype distributions of IL-10 rs1800896, rs1800871 and rs1800872 and their association with PCOS risk
| Genotype | Patients | % | Controls | % | x2 | P | x2 for HWE | P for HWE | Adjusted OR (95% CI)1 | P |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1800896 | ||||||||||
| AA | 148 | 41.11 | 155 | 43.06 | 1.0 (Ref.) | - | ||||
| AC | 179 | 49.72 | 175 | 48.61 | 0.91 (0.66-1.25) | 0.55 | ||||
| CC | 33 | 9.17 | 30 | 8.33 | 0.35 | 0.84 | 4.11 | 0.055 | 0.83 (0.49-1.42) | 0.50 |
| rs1800871 | ||||||||||
| CC | 134 | 37.22 | 140 | 38.79 | 1.0 (Ref.) | - | ||||
| CT | 166 | 46.11 | 159 | 44.24 | 1.02 (0.73-1.41) | 0.93 | ||||
| TT | 60 | 16.67 | 61 | 16.97 | 0.29 | 0.87 | 1.87 | 0.187 | 1.05 (0.65-1.69) | 0.99 |
| rs1800872 | ||||||||||
| AA | 147 | 40.83 | 185 | 51.48 | 1.0 (Ref.) | - | ||||
| AG | 164 | 45.56 | 157 | 43.64 | 0.99 (0.73-1.36) | 0.97 | ||||
| GG | 49 | 13.61 | 18 | 4.88 | 18.85 | < 0.001 | 0.09 | 0.76 | 3.04 (1.62-5.69) | 0.001 |
Adjusted for age, BMI, drinking and T value.
Linkage disequilibrium did not find among IL-10 rs1800896, rs1800871 and rs1800872 (Figure 1 and Table 3). Eight haplotypes (frequency > 0.03 in either the patients with cerebral infarction or controls) were selected into finally haplotype analysis, and the C-T-A (OR = 1.53, 95% CI = 1.05-2.35) haplotype displayed an increased risk of PCOS, while the A-C-G (OR = 0.72, 95% CI = 0.53-0.98) showed a reduced risk of PCOS (Table 4).
Figure 1.
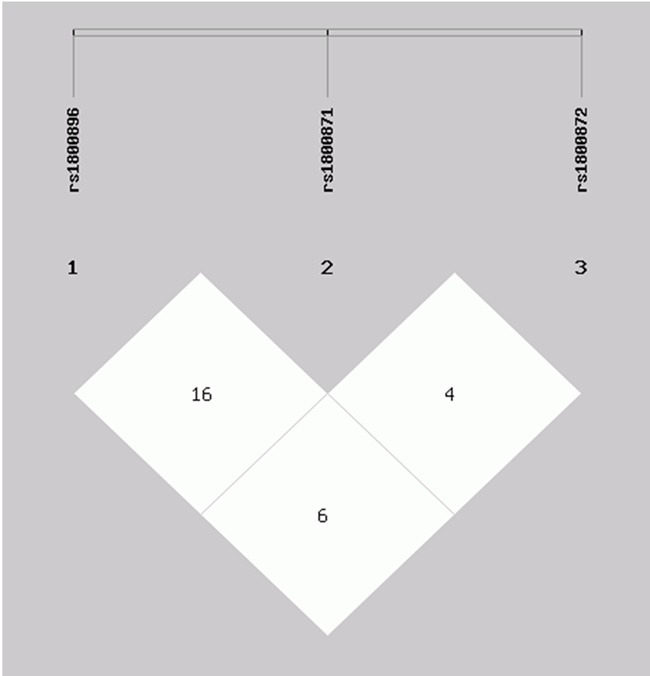
Linkage disequilibrium tests for rs1800896, rs1800871 and rs1800872.
Table 3.
Linkage disequilibrium tests for rs1800896, rs1800871 and rs1800872
| D’ | r2 | |||
|---|---|---|---|---|
|
|
||||
| rs1800871 | rs1800872 | rs1800871 | rs1800872 | |
| rs1800896 | 0.161 | 0.066 | 0.009 | 0.004 |
| rs1800871 | 0.042 | 0.001 | ||
Table 4.
Haplotype analysis of the correlation between IL-10 rs1800896-rs1800871-rs1800872 and PCOS risk
| Haplotype | Patients | % | Controls | % | OR (95% CI)1 | P value |
|---|---|---|---|---|---|---|
| A-C-A | 190 | 26.39 | 171 | 23.75 | 1.15 (0.91-1.46) | 0.25 |
| A-C-G | 80 | 11.11 | 106 | 14.72 | 0.72 (0.53-0.98) | 0.04 |
| A-T-A | 142 | 19.72 | 150 | 20.83 | 0.93 (0.72-1.21) | 0.59 |
| A-T-G | 50 | 6.94 | 47 | 6.53 | 1.08 (0.72-1.63) | 0.71 |
| C-C-A | 124 | 17.22 | 112 | 15.56 | 1.13 (0.85-1.49) | 0.40 |
| C-C-G | 47 | 6.53 | 61 | 8.47 | 0.75 (0.50-1.11) | 0.15 |
| C-T-A | 51 | 7.08 | 35 | 4.86 | 1.53 (1.05-2.35) | 0.04 |
| C-T-G | 36 | 5.00 | 38 | 5.28 | 0.95 (0.60-1.52) | 0.83 |
Global result: Total control = 720, total case = 720; Global chi2 = 10.51, pearson’s P value = 0.16.
Discussion
Genetic factors, such as SNPs, have been considered as parts of the etiology of diseases. In the current study, we observed that the GG genotype of rs1800872 was associated with PCOS susceptibility, and the C-T-A and A-C-G haplotypes could affect the development of PCOS.
IL-10 is an important anti-inflammatory factor, with roles in the regulation of immune functions and inflammatory processes, in addition to being associated with the development of several kinds of diseases. IL-10 level is very low in normal healthy human tissues, but its cellular secretion significantly increases during microbial infection and autoimmune diseases [16,17]. Such increased IL-10 secretion is an endogenous response to an excess of inflammatory factors, and may aid in eliminating inflammation [18]. Single nucleotide polymorphisms of genes encoding inflammatory factors may influence an individual’s cytokine production level and reaction intensity, and are associated with disease development [19]. Many previous studies have examined associations of IL-10 polymorphisms with the susceptibility to several diseases related to inflammation, such as metabolic syndrome, multiple sclerosis and chronic and aggressive periodontitis [20-22].
To date, only a few studies reported the association between IL-10 genetic polymorphisms and risk of PCOS, and the results are inconsistent. Talaat RM et al. performed a study in 61 patients with PCOS and 80 healthy controls in a Egyptian population, and investigated the influence of IL-10 serum levels and genetic polymorphisms in the risk of PCOS [12]. They found that IL-10 -1082 GG and -819 TT genotype could be regarded as risk factors for PCOS, and IL-10 levels were significantly lower in PCOS patients than those in normal controls [12]. Vural P et al. performed a study with 97 PCOS patients and 95 healthy control women in a Turkish population, and they reported no significant association between IL-10 -1082 genetic polymorphisms and the occurrence and metabolic abnormalities in PCOS in a Turkish population [14]. Karadeniz M et al. also performed a study in 91 young with PCOS and 74 healthy control women in a Turkish population, and showed that IL-10 genetic polymorphism had no effect on the risk of PCOS [15]. In our study, we firstly observed a significant relationship between IL-10 rs1800871 polymorphism and development of PCOS, indicating that this gene polymorphism could influence the risk of this disease. Moreover, we found that the C-T-A and A-C-G haplotypes constructed with rs1800896-rs1800871-rs1800872 differed significantly between PCOS patients and controls, suggesting that the haplotype could be a genetic marker for PCOS. The C-T-A and A-C-G haplotypes may change the activity of IL-10, and consequently affect pathogenesis of PCOS.
Two limitations should be considered in this study. First, a case-control study design was used, and questionnaires relied on subjects’ memories, which may result in recall bias. Second, patients and controls were selected from only two hospitals, and they may not be sufficiently to present other populations. Therefore, selection bias may not be avoided in this study.
In conclusion, this study firstly estimates the association between IL-10 polymorphisms and haplotypes and PCOS risk in the Chinese population. Its findings suggest that IL-10 could be a biomarker for the risk of PCOS. Further research employing a greater number of study subjects is greatly required to corroborate our results.
Acknowledgements
We thank the great help and funding from Scientific and Technological Project in Henan Province (201602359).
Disclosure of conflict of interest
None.
References
- 1.Zhuang J, Liu Y, Xu L, Liu X, Zhou L, Tang L, Kang D, Guo W, He M, Yang F, Qiu D. Prevalence of the polycystic ovary syndrome in female residents of Chengdu, China. Gynecol Obstet Invest. 2014;77:217–223. doi: 10.1159/000358485. [DOI] [PubMed] [Google Scholar]
- 2.Merkin SS, Phy JL, Sites CK, Yang D. Environmental determinants of polycystic ovary syndrome. Fertil Steril. 2016;106:16–24. doi: 10.1016/j.fertnstert.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Liu XF, Liu Y, Xu LZ, Zhou LL, Tang LL, Zhuang J, Li TT, Guo WQ, Hu R, Qiu DS, Han DW. Environmental risk factors for women with polycystic ovary syndrome in china: a population-based case-control study. J Biol Regul Homeost Agents. 2014;28:203–211. [PubMed] [Google Scholar]
- 4.Gu HF, Mou M, Liang ZG, Sun C, Ren XY, Xiao YB. The association between paraoxonase 1 gene polymorphisms and polycystic ovarian syndrome. Cell Mol Biol (Noisy-le-grand) 2016;62:44–47. [PubMed] [Google Scholar]
- 5.Wu JB, Zhai JF, Yang J. Role of methylenetetrahydrofolate reductase C677T and A1298C polymorphisms in polycystic ovary syndrome risk. Genet Mol Res. 2016;15 doi: 10.4238/gmr15048570. [DOI] [PubMed] [Google Scholar]
- 6.Sun X, Wu X, Duan Y, Liu G, Yu X, Zhang W. Family-based association study of rs17300539 and rs12495941 polymorphism in adiponectin gene and polycystic ovary syndrome in a Chinese population. Med Sci Monit. 2017;23:78–84. doi: 10.12659/MSM.901944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eser B, Islimye Taskin M, Hismiogullari AA, Aksit H, Bodur AS. The effects of IL-1A and IL-6 genes polymorphisms on gene expressions, hormonal and biochemical parameters in polycystic ovary syndrome. J Obstet Gynaecol. 2017;37:358–362. doi: 10.1080/01443615.2016.1256966. [DOI] [PubMed] [Google Scholar]
- 8.Qin L, Xu W, Li X, Meng W, Hu L, Luo Z, Wang Y, Luo S, Li S. Differential expression profile of immunological cytokines in local ovary in patients with polycystic ovarian syndrome: analysis by flow cytometry. Eur J Obstet Gynecol Reprod Biol. 2016;197:136–141. doi: 10.1016/j.ejogrb.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Urbanek M. The genetics of the polycystic ovary syndrome. Nat Clin Pract Endocrinol Metab. 2007;3:103–111. doi: 10.1038/ncpendmet0400. [DOI] [PubMed] [Google Scholar]
- 10.Xiong YL, Liang XY, Yang X, Li Y, Wei LN. Low-grade chronic inflammation in the peripheral blood and ovaries of women with polycystic ovarian syndrome. Eur J Obstet Gynecol Reprod Biol. 2011;159:148–150. doi: 10.1016/j.ejogrb.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 11.D’Andrea A, Aste-Amezaga M, Valiante NM, Ma X, Kubin M, Trinchieri G. Interleukin 10 (IL-10) inhibits human lymphocyte interferon gamma-production by suppressing natural killer cell stimulatory factor/IL-12 synthesis in accessory cells. J Exp Med. 1993;178:1041–1048. doi: 10.1084/jem.178.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talaat RM, Mohamed YA, Mohamad EH, Elsharkawy M, Guirgis AA. Interleukin 10 (- 1082 G/A) and (- 819 C/T) gene polymorphisms in Egyptian women with polycystic ovary syndrome (PCOS) Meta Gene. 2016;9:254–258. doi: 10.1016/j.mgene.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H, Yu K, Yang Z. Associations between TNF-α and interleukin gene polymorphisms with polycystic ovary syndrome risk: a systematic review and meta-analysis. J Assist Reprod Genet. 2015;32:625–634. doi: 10.1007/s10815-015-0449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vural P, Degirmencioglu S, Saral NY, Akgul C. Tumor necrosis factor alpha (-308), interleukin-6 (-174) and interleukin-10 (-1082) gene polymorphisms in polycystic ovary syndrome. Eur J Obstet Gynecol Reprod Biol. 2010;150:61–65. doi: 10.1016/j.ejogrb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Karadeniz M, Erdogan M, Zengi A, Tamsel S, Berdeli A, Saygili F, Yilmaz C. Polymorphism of the interleukin-10 gene in polycystic ovary syndrome. Int J Immunogenet. 2008;35:119–123. doi: 10.1111/j.1744-313X.2007.00746.x. [DOI] [PubMed] [Google Scholar]
- 16.Rutz S, Ouyang W. Regulation of interleukin-10 expression. Adv Exp Med Biol. 2016;941:89–116. doi: 10.1007/978-94-024-0921-5_5. [DOI] [PubMed] [Google Scholar]
- 17.Rutz S, Ouyang W. Polymorphism of the interleukin-10 gene in polycystic ovary syndrome. Polymorphism of the interleukin-10 gene in polycystic ovary syndrome. Curr Opin Immunol. 2011;23:605–612. doi: 10.1016/j.coi.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 18.Nakahata T, Tsuji K. [Function, molecular structure and gene expression regulation of interleukin-10 (IL-10)] . Nihon Rinsho. 1992;50:1827–1832. [PubMed] [Google Scholar]
- 19.Mera-Ramirez A, Castillo A, Orobio Y, Gomez MA, Gallego-Marin C. Screening of TNFalpha, IL-10 and TLR4 single nucleotide polymorphisms in individuals with asymptomatic and chronic cutaneous leishmaniasis in Colombia: a pilot study. BMC Infect Dis. 2017;17:177. doi: 10.1186/s12879-017-2281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maintinguer Norde M, Oki E, Ferreira Carioca AA, Teixeira Damasceno NR, Fisberg RM, Lobo Marchioni DM, Rogero MM. Influence of IL1B, IL6 and IL10 gene variants and plasma fatty acid interaction on metabolic syndrome risk in a cross-sectional population-based study. Clin Nutr. 2017 doi: 10.1016/j.clnu.2017.02.009. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Ben Fredj N, Aissi M, Ben Selma W, Mahmoud I, Nefzi F, Frih-Ayed M, Boukadida J, Aouni M. Association of the IL-10 receptor A536G (S138G) loss-of-function variant with multiple sclerosis in Tunisian patients. APMIS. 2017;125:444–451. doi: 10.1111/apm.12659. [DOI] [PubMed] [Google Scholar]
- 22.Silveira VR, Pigossi SC, Scarel-Caminaga RM, Cirelli JA, Rego R, Nogueira NA. Analysis of polymorphisms in Interleukin 10, NOS2A, and ESR2 genes in chronic and aggressive periodontitis. Braz Oral Res. 2016;30:e105. doi: 10.1590/1807-3107BOR-2016.vol30.0105. [DOI] [PubMed] [Google Scholar]


