Abstract
Although PIK3CA mutations and phosphorylated mTOR (p-mTOR) expression are two main events on PI3K/Akt/mTOR pathway, limited data has reported their roles in triple negative breast cancer (TNBC). Thus, we evaluated the associations of these two biomarkers and clinicopathological characteristics and prognosis in large Chinese TNBC patients. Immunohistochemistry (IHC) analysis was used to assess p-mTOR expression level in 218 TNBC patients. Direct sequencing was applied to detect the most important hotspot regions in exons 9 and 20 of PIK3CA gene. In the TNBC cohort, mutations were identified in 11.5% cases which were associated with basal-like subtype. The somatic point mutations were independently associated with worse overall survival (HR=0.400, 95% CI: 0.193-0.830, P=0.014). As for p-mTOR expression, 47.7% of the tumors were positive and the staining was shown in cytoplasm, nuclear and perinuclear areas. There were significant differences observed in tumor size, lymph node status, and clinical stage between p-mTOR positive and p-mTOR negative cases. Notably, we found a significant association between p-mTOR expression and PIK3CA mutations. Patients with p-mTOR staining also demonstrated shorter overall survival (HR=0.710, 95% CI: 0.514-0.980, P=0.037). Therefore, PIK3CA mutations and its downstream effector p-mTOR expression were two important regulators for activating the PI3K/Akt/mTOR pathway. Both of them could be served as adverse prognostic biomarkers and may contribute to the targeted therapy for TNBC patients with poor outcome.
Keywords: PIK3CA, p-mTOR, triple negative breast cancer, prognosis
Introduction
Triple negative breast cancer (TNBC) lacks the expression of estrogen-receptor (ER), progesterone-receptor (PR) and epidermal growth factor receptor 2 (HER2), accounting for approximately 16% of all BC [1,2]. A high proportion of TNBC comprises the basal-like breast cancer (BLBC) subtype which has been extensively characterized on the basis of gene expression profiling [3]. However, it is becoming increasingly common and facility to identify this phenotype based on the immunohistochemistry (IHC) staining of basal-like markers. BLBC specifically shows IHC expression of epidermal growth factor receptor (EGFR) and/or cytokeratin 5/6 (CK5/6), and is prone to have a poorer prognosis and higher recurrence than non-BLBC as reported [4]. The deficiency of target therapies and prognostic factors in TNBC patients is a critical clinical issue at present. Approximately 20% TNBC patients with family history carry an inherited BRCA1 or BRCA2 mutation. On the contrary, the sporadic TNBC patients do not have such genetic alterations [5]. The aggressive behavior and poor outcome of sporadic population might be controlled mainly by other genetic and molecular changes.
Phosphoinositide 3-kinase (PI3K)/Akt/mammalian target of Rapamycin (mTOR) (PAM) signaling pathway is a complicated intracellular pathway that participates in the regulation of cancer cell survival, metabolism, proliferation and differentiation [6]. PI3K heterodimer belongs to class IA of PI3K family that consists of two subunits: regulatory subunit (p85) and catalytic subunit (p110). PIK3CA is located on chromosome 3q26.3 which encodes p110α catalytic subunit of class IA of PI3K [7,8]. mTOR is a serine/threonine kinase that serve as a key downstream regulator on the pathway. The phosphorylation of mTOR (p-mTOR) stands for the hyperactivation of the molecule that can eventually lead to increased protein synthesis, cell growth and tumor development [9]. Emerging data has revealed that high PI3K pathway activity is the most frequent event in BLBC/TNBC when compared with TP53 and RB1 pathway. Among the aberrant activation pathway, PIK3CA mutations and downstream p-mTOR expression are two major events on the pathway [10]. Furthermore, Chen et al [11] recently demonstrated that PI3K/mTOR dual inhibitor BEZ235 and Trichostatin A (TSA) result in synergistic growth inhibition and apoptosis of BC cell lines. Hence, we postulated that the PIK3CA mutations and p-mTOR expression might serve as two critical regulators and have a role in sporadic TNBC.
Based on the former reports, we systematically evaluated the mutation type of PIK3CA and staining forms of p-mTOR in a large cohort study of 218 TNBC population. We also analyzed the associations of these two key markers with clinicopathological features and prognostic impacts, which might have clinical significance for further development of drug combinations in the treatment of TNBC patients.
Materials and methods
Patients and samples
A total of 218 patients who underwent surgery and diagnosed with TNBC were selected for this retrospective study during the period of January 1, 1999 - December 31, 2008 in the Department of Pathology, Cancer Hospital, Peking Union Medical College, Chinese Academy of Medical Sciences, Beijing, China. The inclusion criteria were as follows: primary operable breast cancer, no family history for breast or ovary cancer, no prior radiotherapy and chemotherapy before surgery, mastectomies, or lumpectomies specimens with sufficient tissue. Clinicopathological information was available for all the patients, including age, tumor size, grade, lymph node metastasis, tumor embolus, subtype, and prognosis. Survival data were last updated on August 24, 2014, and the median follow-up period was 77.25 months (range 2.13-168.47 months).
PCR and direct sequencing of PIK3CA
DNA extraction from formalin-fixed and paraffin-embedded (FFPE) TNBC tissue samples, with minimum of 75% malignant cells, was performed according to the manufacturer’s instructions (ZEESAN, Xiamen, China). The PCR amplifications targeting PIK3CA exons 9 and 20 were conducted. Primers were designed using the Primer Design Tool from NCBI. The primers were as follows: 9-F: GTATTTGCTTTTTCTGTAAATCATCTG, 9-R: CATGCTGAGATCAGCCAAATTC; 20-F: CTCTGGAATGCCAGAACTAC, 20-R: ATGCTGTTTAATTGTGTGGAAG. The amplified products were sequenced using an ABI 3730XL sequencer and the sequencing profiles were analyzed by Mutation Surveyor software v4.0.8 (SoftGenetics Corporation, America), in comparison with the corresponding reference sequence (NM_006218) to identify mutations.
Tissue microarray (TMA) and IHC assay
TMA contained two tumor cores and one normal core of 1.0 mm diameter which were taken from each tumor sample based on hematoxylin and eosin (H&E) staining. The accurate definition of TNBC could be simply summarized as follows: ≤ 1% expression of ER and PR as determined by IHC, and that HER2 either 0-1+ by IHC, or 2+ with fluorescence in situ hybridization (FISH) negative. Tumors were considered as CK5/6 or EGFR positive when more than 5% of tumor cells were positively stained [12]. Staining for p-mTOR were performed using the monoclonal rabbit anti-p-mTOR (Ser2448) (49F9) antibody (Cell Signaling, Danvers, MA, USA) at a dilution of 1:100. Briefly, the slides were deparaffinized and implemented antigen retrieval in a steam cooker for 1.5 minutes with citric acid, pH 6.0 (ZSGB-BIO Co. Ltd, Beijing, China). The primary monoclonal antibodies were incubated at a room temperature for 1.5 h, and universal secondary antibody (DAKO) was applied for 15 min. Diaminobenzidine was used as chromogen and slides were counterstained with hematoxylin before mounting. p-mTOR expression was scored on a range from grade 0 to 2+ according to the previous study [13]: 0: no staining; 1+: 5-50% with weak intensity or 1-10% with moderate to strong intensity; 2+: > 50% with weak intensity or > 10% with moderate to strong intensity. These scores indicated, 0: negative; Score 1: low expression; Score 2: high expression; Both 1 and 2 scores were regarded as positive in the statistical analysis.
Statistical analysis
Statistical analysis was performed using the SPSS 17.0 software package. The associations between these two markers and clinicopathological parameters were analyzed using the Pearson’s χ2-test. The overall survival time was described by Kaplan-Meier method and Cox proportional hazard regression analysis from the date of surgery till the date of death from BC. All P values were two-tailed and statistical significance was considered to be P < 0.05.
Results
Sample cohort and clinical parameters
With regard to the complete cohort, a total of 218 TNBC patients could be further divide into 134 (61.5%) cases of BLBC and 84 (38.5%) cases of non-BLBC, based on IHC staining for EGFR and CK5/6 (Figure 1). The median age of all patients at the time of diagnosis was 50 years, with an age range of 24 to 82 years. According to the WHO Classification of Breast Tumors, 196 (89.9%) patients were histologically classified as invasive carcinomas of no special type (IC-NST, including 78 cases of grade 2 and 118 cases of grade 3) and 2 (0.9%) of the cases had invasive lobular carcinoma (ILC). Tumors numbering 20 (9.2%) had carcinoma of other histologic types such as medullary carcinoma (n=13), metaplastic carcinoma (n=4), secretory carcinoma (n=1), and adenoid cystic carcinoma (n=2).
Figure 1.
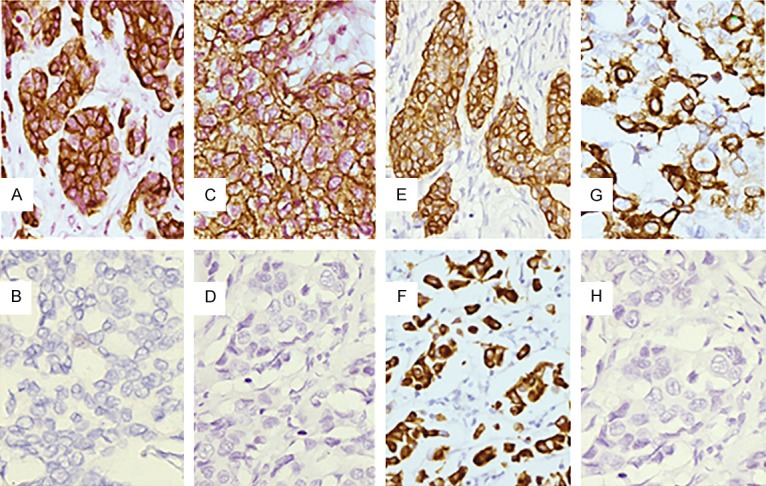
A. Cytoplasmic immunostaining of CK5/6; B. Negative control for CK5/6 by omission of the primary antibody; C. Membranous immunostaining of EGFR; D. Negative control for EGFR by omission of the primary antibody; E. Cytoplasmic immunostaining of p-mTOR; F. Nuclear immunostaining of p-mTOR; G. Perinuclear immunostaining of p-mTOR; H. Negative control for p-mTOR by omission of the primary antibody. (Magnification 400×).
PIK3CA mutations and patients’ characteristics
Among the 218 samples, PIK3CA mutations were found in 25 (11.5%) TNBC patients. Mutations in helical (exon 9) and kinase (exon 20) domains were present in 10 (4.6%) and 15 (6.9%) cases, respectively. No cases had mutations in both the exons. The most frequent mutation was H1047R (6.9%), representing 100% of the exon 20 mutations. The exon 9 hotspot mutation was E545A (3.7%), corresponding to 80% of the exon 9 mutations. Other common helical mutations included were E542V and E545K which were found only in one case, respectively (Figure 2; Table 1). As shown in Table 2, there is no significant association of PIK3CA mutations with age, menopausal status, size, lymph node metastasis, tumor grade, type, embolus, and clinical stage. However, it is worth noting that patients with gene mutations were more frequently detected in BLBC than in non-BLBC (84% versus 16%, P=0.014).
Figure 2.
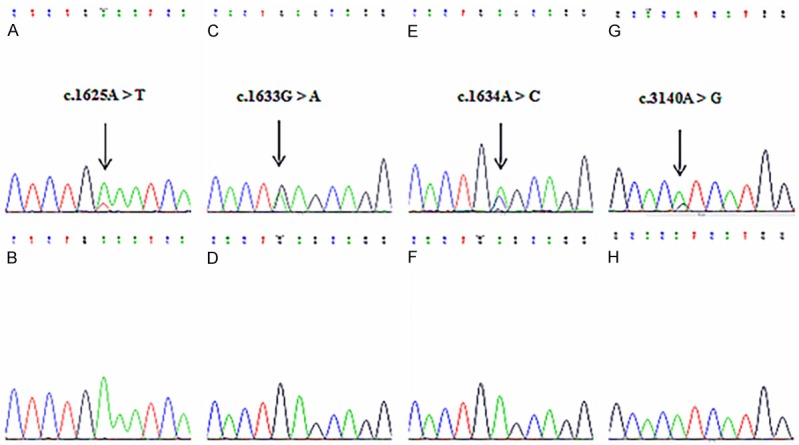
Mutational analysis of PIK3CA exons 9 and 20 in TNBC. A and B. Mutant sequence E542V (A1625T) and corresponding wild-type sequence; C and D. Mutant sequence E545K (G1633A) and corresponding wild-type sequence; E and F. Mutant sequence E545A (A1634C) and corresponding wild-type sequence; G and H. Mutant sequence H1047R (A3140G) and corresponding wild-type sequence.
Table 1.
PIK3CA mutations in TNBC patients
| Exon | Nucleotide | Amino acid | Domain | Cases |
|---|---|---|---|---|
| 9 | c.1625A>T | p.E542V | Helical | 1 |
| 9 | c.1633G>A | p.E545K | Helical | 1 |
| 9 | c.1634A>C | p.E545A | Helical | 8 |
| 20 | c.3140A>G | p.H1047R | Kinase | 15 |
Table 2.
PIK3CA mutations and clinicopathological characteristics of 218 TNBC patients
| Parameters | Status | Total | PIK3CA mutations | P value | |
|---|---|---|---|---|---|
|
| |||||
| Mutated (%) | Wildtype (%) | ||||
| Age | < 50 | 105 | 14 (56.0) | 91 (47.2) | NS |
| ≥ 50 | 113 | 11 (44.0) | 102 (52.8) | ||
| Menopausal status | Pre-menopausal | 172 | 20 (80.0) | 152 (78.8) | NS |
| Post-menopausal | 46 | 5 (20.0) | 41 (21.2) | ||
| Size | ≤ 2 | 73 | 10 (40.0) | 63 (32.6) | NS |
| > 2 | 145 | 15 (60.0) | 130 (67.4) | ||
| LNM | Present | 79 | 12 (48.0) | 67 (34.7) | NS |
| Absent | 139 | 13 (52.0) | 126 (65.3) | ||
| Tumor grade | G2 | 78 | 10 (40.0) | 68 (35.2) | NS |
| G3 | 118 | 11 (44.0) | 107 (55.4) | ||
| Tumor type | IC-NST | 196 | 21 (84.0) | 175 (90.7) | NS |
| ILC | 2 | 0 (0) | 2 (1.0) | ||
| Other | 20 | 4 (16.0) | 16 (8.3) | ||
| Tumor embolus | Yes | 24 | 5 (20.0) | 19 (9.8) | NS |
| No | 194 | 20 (80.0) | 174 (90.2) | ||
| Basal-like subtype | Yes | 134 | 21 (84.0) | 113 (58.5) | 0.014 |
| No | 84 | 4 (16.0) | 80 (41.5) | ||
| Clinical stage | 1 and 2 | 177 | 19 (76.0) | 158 (81.9) | NS |
| 3 | 41 | 6 (24.0) | 35 (77.8) | ||
Abbreviations: NS, not significant; IC-NST, invasive carcinoma of no special type; LNM, lymph nodes metastasis; ILC, invasive lobular carcinoma.
p-mTOR expression and patients’ characteristics
A total of 104 (47.7%) tumors were considered to be positively stained for p-mTOR. The protein expression could be detected in the cytoplasm (38.1%, 83/218), the nuclear (7.8%, 17/218) and perinuclear (1.8%, 4/218) areas respectively. Typical staining patterns for p-mTOR were shown in Figure 1. As can be seen in Table 3, p-mTOR was more frequent in patients with lymph node metastasis than in those without lymph node metastasis; in tumors with size > 2 cm rather than in tumors with size ≤ 2 cm; and in advanced stage (3) compared with early stage (1 and 2). In contrast, p-mTOR was unrelated to age, menopausal status, tumor grade, type, embolus, and basal-like subtype. Particularly, p-mTOR-expressing tumors were more frequently detected in PIK3CA mutant than in PIK3CA wild-type patients. Moreover, we also noted a significant association between the protein present in the nuclear or perinuclear area and lymph node metastasis and PIK3CA mutations.
Table 3.
Relationship of p-mTOR expression with clinicopathological characteristics in 218 TNBC patients
| Characteristis | Status | Positive for p-mTOR | Positive for Cyto p-mTOR | Positive for Nuclear or PN p-mTOR | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| No (%) | P value | No (%) | P value | No (%) | P value | ||
| Age | < 50 | 47 (44.8) | NS | 38 (36.2) | NS | 9 (8.6) | NS |
| ≥ 50 | 57 (50.4) | 45 (39.8) | 12 (10.6) | ||||
| Menopausal status | Pre-menopausal | 78 (45.3) | NS | 62 (36.0) | NS | 16 (9.3) | NS |
| Post-menopausal | 26 (56.5) | 21 (45.7) | 5 (10.9) | ||||
| Size | ≤ 2 | 27 (37.0) | 0.025 | 22 (30.1) | NS | 5 (6.8) | NS |
| > 2 | 77 (53.1) | 61 (42.1) | 16 (11.0) | ||||
| LNM | Present | 46 (58.2) | 0.019 | 32 (40.5) | NS | 14 (17.7) | 0.002 |
| Absent | 58 (41.7) | 51 (36.7) | 7 (5.0) | ||||
| Tumor grade | G2 | 36 (46.2) | NS | 28 (35.9) | NS | 8 (10.3) | NS |
| G3 | 58 (49.2) | 47 (39.8) | 11 (9.4) | ||||
| Tumor type | IC-NST | 94 (48.0) | NS | 75 (38.3) | NS | 19 (9.7) | NS |
| ILC | 1 (50.0) | 1 (50.0) | 0 (0) | ||||
| Other | 9 (45.0) | 7 (35.0) | 2 (10.0) | ||||
| Tumor embolus | Yes | 15 (62.5) | NS | 10 (41.7) | NS | 5 (20.8) | NS |
| No | 89 (45.9) | 73 (37.6) | 16 (8.3) | ||||
| Basal-like subtype | Yes | 63 (47.0) | NS | 50 (37.3) | NS | 13 (9.7) | NS |
| No | 41 (48.8) | 33 (39.3) | 8 (9.5) | ||||
| Clinical stage | 1 and 2 | 77 (43.5) | 0.010 | 63 (35.6) | NS | 14 (7.9) | NS |
| 3 | 27 (65.9) | 20 (48.8) | 7 (17.1) | ||||
| PIK3CA | Mutant | 18 (72.0) | 0.010 | 12 (48.0) | NS | 6 (24.0) | 0.020 |
| Wild-type | 86 (44.6) | 71 (36.8) | 15 (7.8) | ||||
Abbreviations: NS, not significant; IC-NST, invasive carcinoma of no special type; LNM, lymph nodes metastasis; ILC, invasive lobular carcinoma; Cyto, Cytoplasmic; PN, perinuclear.
PIK3CA mutation and p-mTOR expression on prognosis
Patients with PIK3CA mutations or p-mTOR expression had significantly shorter overall survival by Kaplan-Meier analysis (P=0.001 and P=0.001, respectively; Figure 3A and 3B). Univariate and multivariate analysis of 218 TNBC patients revealed that PIK3CA mutation and p-mTOR expression were independent predictive factors for worse prognosis (HR=0.400, 95% CI: 0.193-0.830, P=0.014 and HR=0.710, 95% CI: 0.514-0.980, P=0.037, respectively), and also of lymph node metastasis (HR=0.253, P < 0.001; Table 4).
Figure 3.
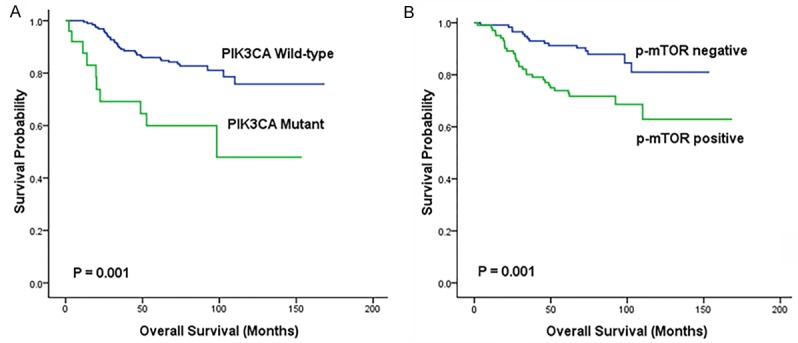
Kaplan-Meier curves for overall survival according to PIK3CA mutation (A) and p-mTOR expression (B) in TNBC patients. (n=218).
Table 4.
PIK3CA mutations and p-mTOR expression status in 218 TNBC patients
| Variable | Univariate HR (95% CI) | P value | Multivariate HR (95% CI) | P value |
|---|---|---|---|---|
| Tumor size | 0.394 (0.183-0.846) | 0.017 | 0.553 (0.244-1.254) | 0.156 |
| Lymph node metastasis | 0.227 (0.122-0.423) | < 0.001 | 0.253 (0.121-0.529) | < 0.001 |
| Clinical stage | 0.353 (0.191-0.650) | 0.001 | 1.332 (0.626-2.832) | 0.457 |
| PIK3CA mutations | 0.614 (0.450-0.837) | 0.002 | 0.400 (0.193-0.830) | 0.014 |
| P-mTOR expression | 0.324 (0.160-0.655) | 0.002 | 0.710 (0.514-0.980) | 0.037 |
Discussion
In the TNBC cohort, 11.5% cases were identified with PIK3CA mutations which were a little lower than the COSMIC databases with a total mutation frequency of 13%. The possible reason for the slight difference might be due to different sequencing methods as reported previously [14]. Other causes could be the differences of patient cohorts, sample preservation and methods used for DNA isolation [15]. Somatic mutations of PIK3CA are clustered into the helical domain (exon 9, commonly E545K and E542K) and the kinase domain (exon 20, commonly H1047R) [16]. The major mutations of our study were present in exon 20 (60% vs 40% in exon 9), entirely in the hot spots of H1047R. Wallin et al [17] detected that PIK3CA-H1047R mutation promoted PI3K pathway activity and induced obvious epithelial-mesenchymal transition (EMT) as well as invasive phenotype, in comparison with the isogenic wild-type mammary epithelial cells. A previous study from the Peruvian BC patients showed that E545A was the main mutation site in exon 9 (70%, 7/10) rather than E545K or E542K in usual [18], which were in accordance with our findings. It is possible that heterogeneous feature of TNBC may lead to various mutated types of oncogene in diverse ethnic population. Although there is no significant association between PIK3CA mutations and most clinical parameters, we found that PIK3CA mutations were statistically correlated with BLBC (P=0.014) which has not been reported in large TNBC cohort before. It has become clear that BLBC often induced unfavorable outcomes [4]. Thus, we assume that the relation of basal markers with oncogene mutations in TNBC can generate more aggressive biological behaviors and worse prognostic effects than wild-type.
The effect of PIK3CA mutations on the prognosis of BC remains unclear. Deng et al [19] suggested that the role of gene mutations with outcomes should be assessed according to different molecular subtypes. It has been reported that PIK3CA mutations are mostly found in the luminal group of BC, specifically in luminal A (49% vs 32% in luminal B) which was defined by higher expression of ER and classical ER-dependent genes [20]. Actually, the association of PIK3CA mutations and favorable clinical outcomes was mainly restricted to high ER positivity (luminal A) tumors, especially in the context of endocrine treatment [21,22]. On the contrary, luminal B tumors were often resistant to endocrine therapy with lower ER levels. The hyperactive GFR/PI3K signaling pathway in this tumor type usually lead to a pejorative effect on survival time [23]. Additionally, patients with PIK3CA mutations in HER2 overexpression subtype also possessed an adverse effect on the outcome after treatment with trastuzumab [24]. Our study confirmed that PIK3CA mutations could be served as an independent poor prognostic factor for overall survival in TNBC patients. From the above conclusions, specific BC samples based on different molecular subtypes which also include other genomic alterations (such as ER, HER2, EGFR, and so on) may contribute to the controversial results of PIK3CA mutations on prognosis.
Everolimus, as an oral inhibitor of mTOR, is the only US Food and Drug Administration (FDA)-approved drug for patients with hormone-receptor-positive, HER2-negative locally advanced or metastatic BC [25]. Nearly half of the TNBCs (47.7%) in our data showed positive immunostaining for p-mTOR, indicating a potential role of mTOR inhibitors in TNBC targeted therapy. Most of the previous studies regarded cytoplasm as the only positive form for p-mTOR expression without referring to other staining patterns [26-28]. Furthermore, few in-depth studies have been implemented on its expression in TNBC patients. Our study found that the staining form of the protein was characterized in the cytoplasm, nucleus and perinuclear area. Moreover, there were no significant differences when identified tumors into high and low expression group in our cases. Thus, we consider any of the three staining forms as positive regardless of the staining intensity when analysis was performed. The p-mTOR positive group was related to bigger tumor size (≥ 2 cm), lymph node metastasis and advanced stage (3), all of which stand for aggressive biological behaviors and result in poor outcome.
Theoretically, mutations in PIK3CA can result in the hyperactivation of the PAM pathway which then transformed PIP2 into PIP3. High levels of PIP3 lead to phosphorylation of Akt, which showed an impact on the cancer cell cycling, survival and growth [7]. Akt can activate mTOR directly by phosphorylation at S2448 or indirectly, by phosphorylation and inactivation of tuberous sclerosis complex 2 (TSC2). When TSC2 loses its function, the GTPase Rheb is maintained in its GTP-bound state, allowing for increased activation of mTOR [29]. Hence, overexpression of p-mTOR (active form of mTOR) represents aberrant activation of this pathway and results in increased protein synthesis, proliferation and anti-apoptosis via its action on substrate 40S ribosomal protein S6 kinase 1 (S6K1) and eukaryotic initiation factor 4E binding protein (4EBP1) [30]. The expression of p-mTOR was expressed more frequently in PIK3CA mutant patients compared to PIK3CA wild-type patients. To some extent, our finding reflects the activated associations between upstream and downstream effectors on the pathway. PIK3CA mutations have been shown to be associated with mTOR inhibitor sensitive in both cell lines and clinical studies [31,32]. Therefore, it can be speculated from our findings that TNBC patients harboring both PIK3CA mutations and p-mTOR expression are more likely to respond to mTOR inhibitors than those with either alteration alone.
According to our IHC results, a considerable number of tumors were present in the nucleus in spite of the small proportion of nuclear expression. Previous studies have revealed that nuclear location of the protein could mediate transcription of rDNA and tDNA which are vital for protein synthesis and lead to an increased proliferation in the BC cell lines [33,34]. In our research, a high proportion of mTOR phosphorylation in nuclear form possessed lymph node metastasis (85.7%, 12/14) and PIK3CA mutations (83.3%, 5/6), which might indicate that nuclear p-mTOR staining induced a more aggressive phenotype in TNBC. For perinuclear staining, Walsh et al [35] confirmed that it was linked to low grade tumors and found more frequently in non-TNBC than TNBC (37% vs 4%) patients, which was similar to our findings with only 4 cases showing the perinuclear pattern. To sum up the above conclusions, along with our findings, the expression form of p-mTOR is actually variable in BC with different meaning on specific subtypes. Although cytoplasmic positive is the main form, we believe that the nuclear p-mTOR plays a significant role in the progression of TNBC and potentially triggers an negative impact on the disease behaviors of patients. On the contrary, the perinuclear pattern is typically present in non-TNBC and appears to have an effect on tumors without triple-negative features.
The expression of p-mTOR induced different disease behaviors and outcomes through particular mechanisms in special subtypes of BC. Zhou et al [36] demonstrated the association of higher mTOR phosphorylation with significantly shorter disease-free survival (P < 0.01), and could lead to enhanced sensitivity to mTOR inhibitors in ErbB2-overexpressing cells. In contrast, one study found that p-mTOR expression was independently associated with longer disease-free survival and overall survival in luminal BC [26]. The protein expression has an unfavorable impact on survival in our TNBC patients. Zhang et al [37] found that mTOR inhibitor testing in TNBC xenografts showed significant tumor growth inhibition. Our finding emphasized the potential prognostic value for TNBC patients who were treated with mTOR inhibitor.
In summary, our findings indicated that PIK3CA mutations and p-mTOR expression were common genetic-molecular events of the activated PAM pathway in TNBC. Both of the two markers were independently associated with poor overall survival and patients who had both oncogene mutations and downstream protein expression seem to benefit from the PAM pathway inhibitors. While, clinical trials are required in future to clarify these two biomarkers as suitable and reliable drug targets.
Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosure of conflict of interest
None.
References
- 1.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, Prentice LM, Khattra J, Burleigh A, Yap D, Bernard V, McPherson A, Shumansky K, Crisan A, Giuliany R, Heravi-Moussavi A, Rosner J, Lai D, Birol I, Varhol R, Tam A, Dhalla N, Zeng T, Ma K, Chan SK, Griffith M, Moradian A, Cheng SW, Morin GB, Watson P, Gelmon K, Chia S, Chin SF, Curtis C, Rueda OM, Pharoah PD, Damaraju S, Mackey J, Hoon K, Harkins T, Tadigotla V, Sigaroudinia M, Gascard P, Tlsty T, Costello JF, Meyer IM, Eaves CJ, Wasserman WW, Jones S, Huntsman D, Hirst M, Caldas C, Marra MA, Aparicio S. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmadeka R, Harmon BE, Singh M. Triplenegative breast carcinoma: current and emerging concepts. Am J Clin Pathol. 2014;141:462–477. doi: 10.1309/AJCPQN8GZ8SILKGN. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tischkowitz M, Brunet JS, Begin LR, Huntsman DG, Cheang MC, Akslen LA, Nielsen TO, Foulkes WD. Use of immunohistochemical markers can refine prognosis in triple negative breast cancer. BMC Cancer. 2007;7:134. doi: 10.1186/1471-2407-7-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Afghahi A, Telli ML, Kurian AW. Genetics of triple-negative breast cancer: implications for patient care. Curr Probl Cancer. 2016;40:130–140. doi: 10.1016/j.currproblcancer.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zardavas D, Phillips WA, Loi S. PIK3CA mutations in breast cancer: reconciling findings from preclinical and clinical data. Breast Cancer Res. 2014;16:201. doi: 10.1186/bcr3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translation control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Jin T, Zhu K, Piao Y, Quan T, Quan C, Lin Z. PI3K/mTOR dual inhibitor BEZ235 and histone deacetylase inhibitor trichostatin a synergistically exert anti-tumor activity in breast cancer. Oncotarget. 2017;8:11937–11949. doi: 10.18632/oncotarget.14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu X, Shan L, Wang F, Wang J, Wang F, Shen G, Liu X, Wang B, Yuan Y, Ying J, Yang H. Hypermethylation of BRCA1 gene: implication for prognostic biomarker and therapeutic target in sporadic primary triple-negative breast cancer. Breast Cancer Res Tr. 2015;150:479–486. doi: 10.1007/s10549-015-3338-y. [DOI] [PubMed] [Google Scholar]
- 13.Ueng SH, Chen SC, Chang YS, Hsueh S, Lin YC, Chien HP, Lo YF, Shen SC, Hsueh C. Phosphorylated mTOR expression correlates with poor outcome in early-stage triple negative breast carcinomas. Int J Clin Exp Pathol. 2012;5:806–813. [PMC free article] [PubMed] [Google Scholar]
- 14.Arsenic R, Treue D, Lehmann A, Hummel M, Dietel M, Denkert C, Budczies J. Comparison of targeted next-generation sequencing and Sanger sequencing for the detection of PIK3CA mutations in breast cancer. BMC Clin Pathol. 2015;15:20. doi: 10.1186/s12907-015-0020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arsenic R, Lehmann A, Budczies J, Koch I, Prinzler J, Kleine-Tebbe A, Schewe C, Loibl S, Dietel M, Denkert C. Analysis of PIK3CA mutations in breast cancer subtypes. Appl Immunohistochem Mol Morphol. 2014;22:50–56. doi: 10.1097/pdm.0b013e318297afea. [DOI] [PubMed] [Google Scholar]
- 16.Janku F, Lee JJ, Tsimberidou AM, Hong DS, Naing A, Falchook GS, Fu S, Luthra R, Garrido-Laguna I, Kurzrock R. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS One. 2011;6:e22769. doi: 10.1371/journal.pone.0022769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallin JJ, Guan J, Edgar KA, Zhou W, Francis R, Torres AC, Haverty PM, Eastham-Anderson J, Arena S, Bardelli A, Griffin S, Goodall JE, Grimshaw KM, Hoeflich KP, Torrance C, Belvin M, Friedman LS. Active PI3K pathway causes an invasive phenotype which can be reversed or promoted by blocking the pathway at divergent nodes. PLoS One. 2012;7:e36402. doi: 10.1371/journal.pone.0036402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castaneda CA, Lopez-Ilasaca M, Pinto JA, Chirinos-Arias M, Doimi F. PIK3CA mutations in Peruvian patients with HER2-amplified and triple negative non-metastatic breast cancers. Hematol Oncol Stem Cell Ther. 2014;7:142–148. doi: 10.1016/j.hemonc.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Deng L, Chen J, Zhong XR, Luo T, Wang YP, Huang HF, Yin L, Qiu Y, Bu H, Lv Q, Zheng H. Correlation between activation of PI3K/AKT/mTOR pathway and prognosis of breast cancer in Chinese women. PLoS One. 2015;10:e120511. doi: 10.1371/journal.pone.0120511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu X, Osborne CK, Schiff R. Biology and therapeutic potential of PI3K signaling in ER+/HER2-negative breast cancer. Breast. 2013;22(Suppl 2):S12–S18. doi: 10.1016/j.breast.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis MJ, Lin L, Crowder R, Tao Y, Hoog J, Snider J, Davies S, DeSchryver K, Evans DB, Steinseifer J, Bandaru R, Liu W, Gardner H, Semiglazov V, Watson M, Hunt K, Olson J, Baselga J. Phosphatidyl-inositol-3-kinase alpha catalytic subunit mutation and response to neoadjuvant endocrine therapy for estrogen receptor positive breast cancer. Breast Cancer Res Treat. 2010;119:379–390. doi: 10.1007/s10549-009-0575-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalinsky K, Jacks LM, Heguy A, Patil S, Drobnjak M, Bhanot UK, Hedvat CV, Traina TA, Solit D, Gerald W, Moynahan ME. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15:5049–5059. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- 23.Creighton CJ, Fu X, Hennessy BT, Casa AJ, Zhang Y, Gonzalez-Angulo AM, Lluch A, Gray JW, Brown PH, Hilsenbeck SG, Osborne CK, Mills GB, Lee AV, Schiff R. Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res. 2010;12:R40. doi: 10.1186/bcr2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cizkova M, Dujaric ME, Lehmann-Che J, Scott V, Tembo O, Asselain B, Pierga JY, Marty M, de Cremoux P, Spyratos F, Bieche I. Outcome impact of PIK3CA mutations in HER2-positive breast cancer patients treated with trastuzumab. Br J Cancer. 2013;108:1807–1809. doi: 10.1038/bjc.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jerusalem G, Mariani G, Ciruelos EM, Martin M, Tjan-Heijnen VC, Neven P, Gavila JG, Michelotti A, Montemurro F, Generali D, Simoncini E, Lang I, Mardiak J, Naume B, Camozzi M, Lorizzo K, Bianchetti S, Conte P. Safety of everolimus plus exemestane in patients with hormone-receptor-positive, HER2-negative locally advanced or metastatic breast cancer progressing on prior non-steroidal aromatase inhibitors: Primary results of a phase IIIb, openlabel, single-arm, expanded-access multicenter trial (BALLET) Ann Oncol. 2016;27:1719–1725. doi: 10.1093/annonc/mdw249. [DOI] [PubMed] [Google Scholar]
- 26.Beca F, Andre R, Martins DS, Bilhim T, Martins D, Schmitt F. P-mTOR expression is associated with better prognosis in luminal breast carcinoma. J Clin Pathol. 2014;67:961–967. doi: 10.1136/jclinpath-2014-202320. [DOI] [PubMed] [Google Scholar]
- 27.Bakarakos P, Theohari I, Nomikos A, Mylona E, Papadimitriou C, Dimopoulos AM, Nakopoulou L. Immunohistochemical study of PTEN and phosphorylated mTOR proteins in familial and sporadic invasive breast carcinomas. Histopathology. 2010;56:876–882. doi: 10.1111/j.1365-2559.2010.03570.x. [DOI] [PubMed] [Google Scholar]
- 28.Bostner J, Karlsson E, Pandiyan MJ, Westman H, Skoog L, Fornander T, Nordenskjold B, Stal O. Activation of Akt, mTOR, and the estrogen receptor as a signature to predict tamoxifen treatment benefit. Breast Cancer Res Treat. 2013;137:397–406. doi: 10.1007/s10549-012-2376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 30.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 31.Meric-Bernstam F, Akcakanat A, Chen H, Do KA, Sangai T, Adkins F, Gonzalez-Angulo AM, Rashid A, Crosby K, Dong M, Phan AT, Wolff RA, Gupta S, Mills GB, Yao J. PIK3CA/PTEN mutations and Akt activation as markers of sensitivity to allosteric mTOR inhibitors. Clin Cancer Res. 2012;18:1777–1789. doi: 10.1158/1078-0432.CCR-11-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez-Angulo AM, Blumenschein GJ Jr. Defining biomarkers to predict sensitivity to PI3K/Akt/mTOR pathway inhibitors in breast cancer. Cancer Treat Rev. 2013;39:313–320. doi: 10.1016/j.ctrv.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakarakos P, Theohari I, Nomikos A, Mylona E, Papadimitriou C, Dimopoulos AM, Nakopoulou L. Immunohistochemical study of PTEN and phosphorylated mTOR proteins in familial and sporadic invasive breast carcinomas. Histopathology. 2010;56:876–882. doi: 10.1111/j.1365-2559.2010.03570.x. [DOI] [PubMed] [Google Scholar]
- 34.Vazquez-Martin A, Oliveras-Ferraros C, Bernado L, Lopez-Bonet E, Menendez JA. The serine 2481-autophosphorylated form of mammalian Target of Rapamycin (mTOR) is localized to midzone and midbody in dividing cancer cells. Biochem Biophys Res Commun. 2009;380:638–643. doi: 10.1016/j.bbrc.2009.01.153. [DOI] [PubMed] [Google Scholar]
- 35.Walsh S, Flanagan L, Quinn C, Evoy D, McDermott EW, Pierce A, Duffy MJ. MTOR in breast cancer: differential expression in triplenegative and non-triple-negative tumors. Breast. 2012;21:178–182. doi: 10.1016/j.breast.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 36.Zhou X, Tan M, Stone HV, Klos KS, Lan KH, Yang Y, Yang W, Smith TL, Shi D, Yu D. Activation of the Akt/mammalian target of rapamycin/4E-BP1 pathway by ErbB2 overexpression predicts tumor progression in breast cancers. Clin Cancer Res. 2004;10:6779–6788. doi: 10.1158/1078-0432.CCR-04-0112. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Cohen AL, Krishnakumar S, Wapnir IL, Veeriah S, Deng G, Coram MA, Piskun CM, Longacre TA, Herrler M, Frimannsson DO, Telli ML, Dirbas FM, Matin AC, Dairkee SH, Larijani B, Glinsky GV, Bild AH, Jeffrey SS. Patientderived xenografts of triple-negative breast cancer reproduce molecular features of patient tumors and respond to mTOR inhibition. Breast Cancer Res. 2014;16:R36. doi: 10.1186/bcr3640. [DOI] [PMC free article] [PubMed] [Google Scholar]


