Abstract
Introduction
Identification of adverse events and determination of their seriousness ensures timely detection of potential patient safety concerns. Adverse event seriousness is a key factor in defining reporting timelines and is often performed manually by pharmacovigilance experts. The dramatic increase in the volume of safety reports necessitates exploration of scalable solutions that also meet reporting timeline requirements.
Objective
The aim of this study was to develop an augmented intelligence methodology for automatically identifying adverse event seriousness in spontaneous, solicited, and medical literature safety reports. Deep learning models were evaluated for accuracy and/or the F1 score against a ground truth labeled by pharmacovigilance experts.
Methods
Using a stratified random sample of safety reports received by Celgene, we developed three neural networks for addressing identification of adverse event seriousness: (1) a binary adverse-event level seriousness classifier; (2) a classifier for determining seriousness categorization at the adverse-event level; and (3) an annotator for identifying seriousness criteria terms to provide supporting evidence at the document level.
Results
The seriousness classifier achieved an accuracy of 83.0% in post-marketing reports, 92.9% in solicited reports, and 86.3% in medical literature reports. F1 scores for seriousness categorization were 77.7 for death, 78.9 for hospitalization, and 75.5 for important medical events. The seriousness annotator achieved an F1 score of 89.9 in solicited reports, and 75.2 in medical literature reports.
Conclusions
The results of this study indicate that a neural network approach can provide an accurate and scalable solution for potentially augmenting pharmacovigilance practitioner determination of adverse event seriousness in spontaneous, solicited, and medical literature reports.
Key Points
| Volume, complexity, and time constraints of adverse event reporting are overwhelming the pharmacovigilance workforce. New solutions are needed to support these activities to meet global regulatory timelines. |
| We developed several augmented intelligence approaches to support the correct identification and classification of seriousness, a key factor in adverse reporting, in various document types. |
| Our deep learning models were trained using an extensive data set that captured deep institutional pharmacovigilance practitioner knowledge. |
Introduction
Marketing authorization holders and sponsors are required to collect and collate any safety event that is associated with a drug’s use regardless of whether it is drug related [1–3]. Under the remit of a company’s product safety department, adverse events (AEs) are processed and compiled into individual case safety reports (ICSRs). Each ICSR comprises all of the relevant reported data detailing one, or many, AEs [2, 4]. Currently each report is reviewed, compiled, and formatted for submission to the appropriate health authority using labor-intensive processes relying on a team of pharmacovigilance (PV) experts. The need for expertise combined with the dramatic increase in the volume of safety reports [5, 6], and their complexity, presents a challenge for meeting serious AE reporting timelines.
There are many components that need to be reviewed, assessed, and validated within each ICSR. Within a report, all events must each be documented as serious or not. An AE is considered serious when the patient outcome falls under any of the following seven categories: death, life threatening, hospitalization or hospital prolongation, disability, congenital anomaly, intervention required to prevent impairment, or an important medical event [7].
Accurate seriousness evaluation and the specific associated seriousness criteria of an AE are integral to ensuring the appropriate reporting of AEs to global health authorities within compliance of defined reporting timelines. Currently, the evaluation of whether a report is serious, and which specific seriousness criteria relate to a specific AE, is conducted by a PV professional. This assessment requires considerable training and expertise to accurately assess if and how the AEs identified are serious in nature.
In response to the growing number of safety reports, many efforts in recent years have been made to automate the identification of AEs using annotation and text-mining methods in a variety of source document types such as electronic health records, clinical notes, and social media [8–17]. Other groups have employed classification approaches for identifying text from similar sources as AEs [18–29]. However, few efforts have focused on the classification of identified AEs with regard to their type, severity, seriousness, or causality [30–33].
Here, we present a state-of-the-art method that applies cognitive technologies to accurately determine seriousness from unstructured information in ICSRs at the case level, and identify seriousness categories at the AE level, in spontaneous, solicited, and medical literature safety cases.
Methods
Scope
The aim of this study was to develop a cognitive deep learning methodology for accurate identification and classification of AE seriousness in spontaneous, solicited, and medical literature safety reports. Analyses were applied to English-language text data but otherwise unfiltered beyond the representation of the data in the sample. Seriousness determination from structured information (e.g., checkbox in CIOMS forms) using optical character recognition and template-based form understanding technology was outside the scope of this paper.
Data Collection and Management
A stratified random sample of 22,932 AE cases (including all associated versions and documents) was taken from a data set containing spontaneous or post-marketing (PM), solicited (SD), and medical literature (ML) AE reports received by Celgene between 2015 and 2016 (over 168,000 cases). Post-marketing reports included all spontaneous reports received during the same time period. Solicited cases included those from clinical trials, registries, post-approval named patient use programs, patient support and disease management programs, patient surveys, or information gathered through efficacy and compliance programs [34]. The medical literature is continually reviewed for any presented abstracts or published manuscripts that mention Celgene’s medicinal products; thus, they represent the medical literature cases.
A random sample was selected using stratification by the Medical Dictionary for Regulatory Activities (MedDRA®; http://www.MedDRA.org) code representation, distinct products, and AE seriousness classification to ensure a distribution that represented the diversity of case features. Positive linkage between AEs and seriousness was confirmed for all randomly selected reports prior to annotation by PV subject matter experts to establish the reference for all experiments. The random sample was divided into a training and test set for all experiments. Table 1 contains the statistics of the training data set.
Table 1.
Breakdown of report statistics for the training set
| Total number of cases | 22,932 (12,207 PM, 7,512 SD, 3,213 ML) |
| Total number of documents | 26,256 |
| By type | |
| Post-marketing | 13,083 |
| Solicited | 10,098 |
| Medical literature | 3075 |
| AE seriousness pairs, serious/non-serious | 48,118/25,076 |
| By seriousness classification | |
| Hospitalization | 26,019 |
| Important medical event | 15,149 |
| Death | 6955 |
| Disability | 13 |
| Congenital anomaly | 0 |
| Required intervention (devices) | 0 |
| Life threatening | 48 |
| Number of therapy areas covered | 3 (oncology, hematology, immunology) |
| Number of suspect drugs covered | PM = 23, SD = 237, ML = 32 |
| Number of unique adverse events covereda | PM (14,330), SD (8294), ML (3590) |
AE adverse event, ML medical literature, PM spontaneous reports, SD solicited reports
aCalculated using reported term
Study Design
Our cognitive approach was able to perform three tasks. First, determining if the AEs in a case were serious (yes vs. no). Second, identifying the seriousness criteria associated with each AE. Finally, we annotated the specific terms relating to seriousness classification for a case.
We developed a recurrent neural network (RNN) for the binary assessment of AE-level classification as serious or not. To classify each AE to a seriousness category, we developed a separate RNN. Finally, we used a bi-directional long short-term memory (LSTM) annotator, which identified terms pertaining to seriousness categories in all types of cases. These annotated entities provided human reviewers with focal points for their review as well as additional evidence to consider for determining seriousness categorization.
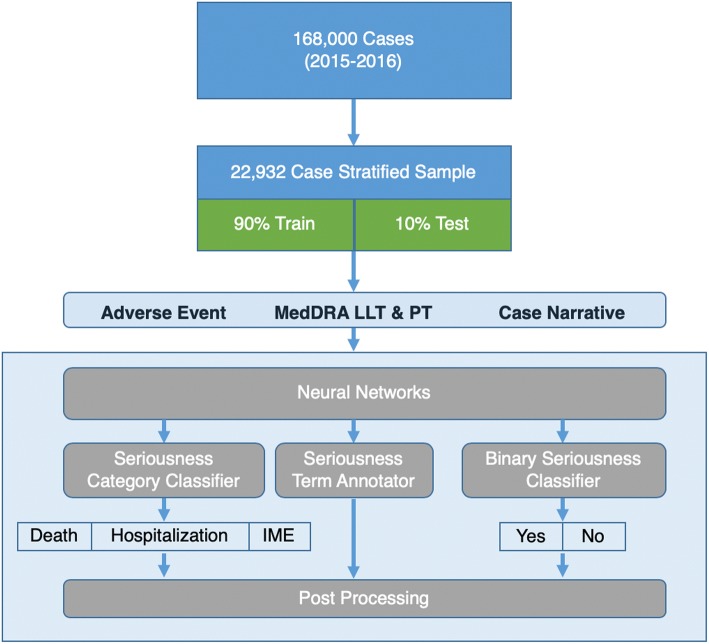
Figure 1 describes the study design used to test the accuracy of our RNN seriousness classifiers. The RNN binary seriousness classifier was applied to the narrative section of all report types (PM, SD, ML) to determine if the AE in the report was serious or non-serious. Reports containing AEs classified as serious were then further analyzed using a second RNN classifier to determine which of the categories of seriousness correspond to each AE in the report. Given the limitations of training data available resulting from Celgene’s therapy portfolio, only three of the seven seriousness categories (hospitalization, disability, and death) were evaluated. The neural networks referenced only the unstructured text within the source document, thus tick boxes commonly used on AE reporting forms such as CIOMS and MedWatch were not a factor in determining seriousness.
Fig. 1.
Study design. A stratified sample of 20,000 cases was derived from 2 years of safety data. Three neural networks were trained using 90% of the stratified sample and each was tested against the remaining 10% of the sample as depicted in the neural network architecture. IME important medical event, LLT lowest level term, MedDRA Medical Dictionary for Regulatory Activities, PT preferred term
The training set for the seriousness classifiers consisted of 26,256 documents (13,083 PM; 10,098 SD; 3075 ML), which were randomly selected from a stratified sample. Additionally, 2716 reports (1324 PM, 1045 SD, 347 ML) were randomly selected from the stratified sample as a test set as indicated in Table 2.
Table 2.
Breakdown of test set report statistics
| Data type | PM | SD | ML |
|---|---|---|---|
| Total number of reports | 1324 | 1045 | 347 |
| AE seriousness pairs, serious/non-seriousa | 763/1565 | 2615/660 | 811/668 |
| By seriousness classification | |||
| Hospitalization | 207 | 1837 | 184 |
| Important medical event | 485 | 330 | 507 |
| Death | 71 | 448 | 120 |
| Disability | 0 | 0 | 0 |
| Congenital anomaly | 0 | 0 | 0 |
| Required intervention (devices) | 0 | 0 | 0 |
| Life threatening | 0 | 0 | 0 |
AE adverse event, ML medical literature, PM spontaneous reports, SD solicited reports
aCalculated using the Medical Dictionary for Regulatory Activities code
Classifier Structure
Three neural networks were required in our methodology for determining seriousness: (1) an RNN classifier to determine the seriousness or non-seriousness of each AE; (2) an additional RNN classifier to determine seriousness categorization for each AE; and (3) a bi-directional LSTM (Bi-LSTM) deep neural network annotator to identify seriousness terms; all methods require subsequent human review and use the concatenated narrative report, AE, and the MedDRA® preferred term as input. All approaches relied on the use of word embeddings created using the Glove algorithm [35] with the PubMed corpus [36].
We have used LSTM for classification tasks and a Bi-LSTM neural network with a conditional random field [37] for annotation tasks based on the simple intuition of the structure of the tasks. Typical classification requires analysis of the whole input sequence to generate the label. Long short-term memory does this by interpreting the vector representation of the last word of the input, which essentially encodes the information of the whole sequence of words. Annotation tasks, however, need to make decisions for each word while reading the input sentence, and the accuracy of such decisions can be improved by knowing the semantics of the words on both sides of the word in focus. Hence, a Bi-LSTM network is more appropriate for these instances as it has the capability to provide the semantics of both sides of the input text with regard to a given word in the input.
Recurrent Neural Network for Seriousness Classification
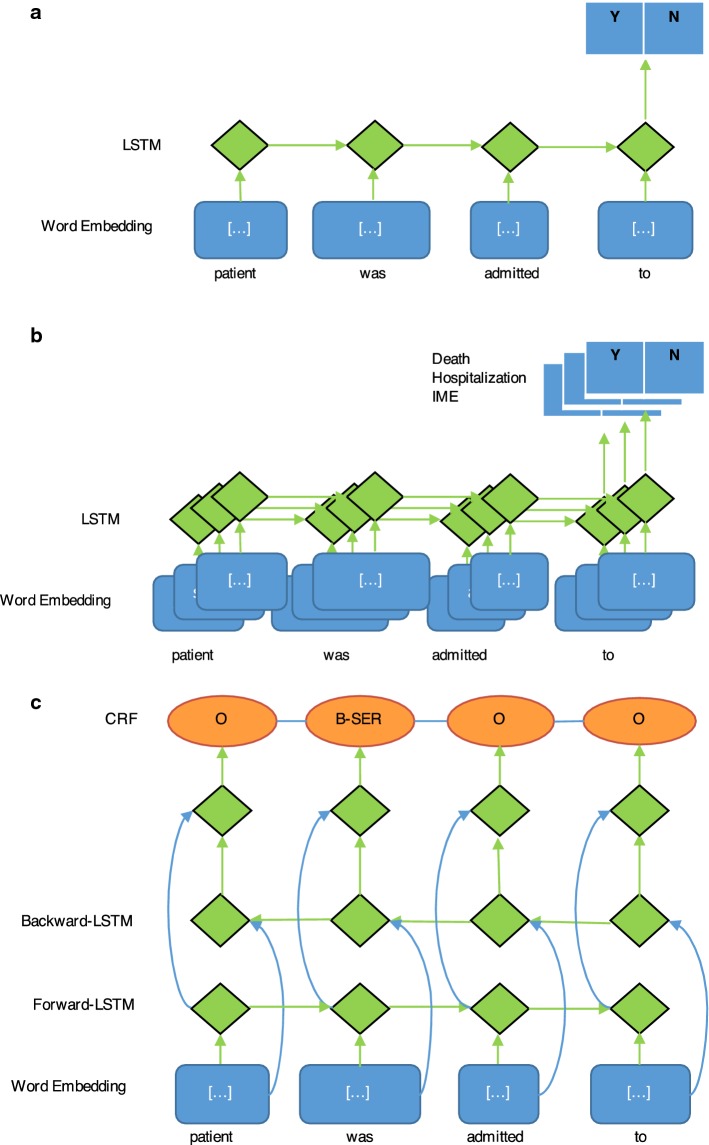
We developed a neural network inspired by Lipton et al. [38] to determine whether each AE in our sample should be classified as serious or non-serious. Case-report-level seriousness is a simple UNION of seriousness criteria from each AE in a report. If any one of the AEs is serious, the whole case report is considered serious. The classifier used as inputs the report narrative, AEs identified in the report, and the MedDRA® lowest level term and preferred terms corresponding to the identified AE. We created a binary classifier using an LSTM deep neural network to determine seriousness = yes/no for each AE. Figure 2a describes the structure of the classifier.
Fig. 2.
Model architectures. Neural network architectures for the a binary seriousness classifier, b seriousness category classifier, and c seriousness term annotator. B-SER beginning of seriousness term, CRF conditional random field, IME important medical event, LSTM long short-term memory, O other
Recurrent Neural Network for Seriousness Categorization
To determine which seriousness categories pertain to each report, we trained an RNN also based on Lipton et al. [38], utilizing the same inputs as above, but limited to train on only cases classified as serious. The representation of seriousness criteria in the training data resulted in the ability to train three of the seven seriousness criteria (death, hospitalization, and important medical events). We created a combined set of binary classifiers using an LSTM deep neural network to determine death = yes/no, hospitalization = yes/no, and IME = yes/no for each AE. Figure 2b describes the structure of the seriousness categorization classifiers.
Long Short-Term Memory Neural Network to Annotate Seriousness Category Terms
We created a Bi-LSTM deep neural network with a conditional random field seriousness annotator to analyze the narrative text of reports. The output of the annotator included identified terms regarding seriousness and was used for human review to support seriousness category determination. Annotated terms are further normalized in post-processing to map to the seriousness sub-categories to augment human review. Figure 2c describes the structure of the seriousness term annotator.
Performance Analysis
Performance of all neural networks was assessed against the manually annotated ground truth: for seriousness classification, accuracy was used; for seriousness category classification and the seriousness annotator, F1 score was used. For comparisons to alternate approaches, standard methods were used to develop random forest [39, 40] and support-vector machine [41–43] algorithms using the same PM training and test data utilized for neural network development and testing.
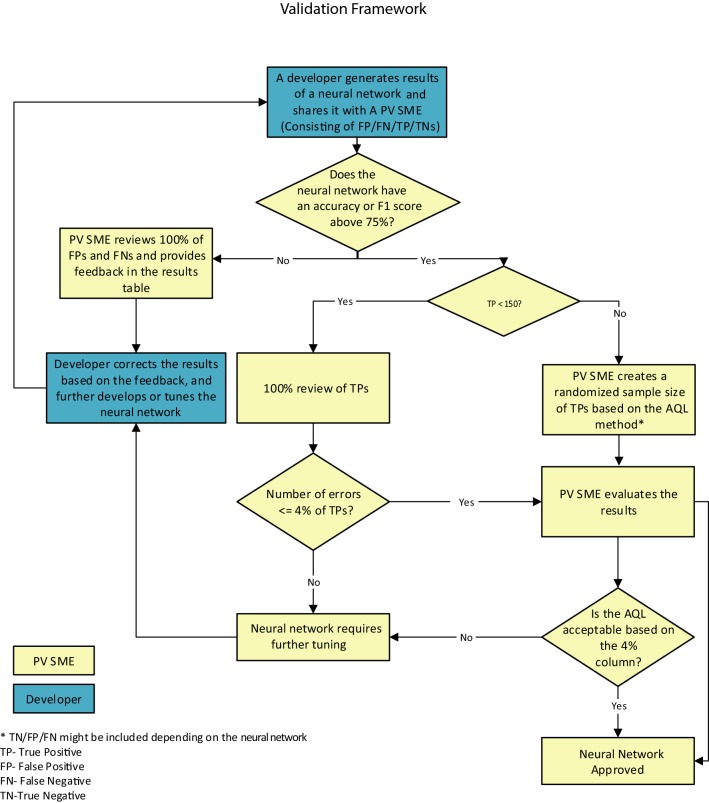
Although PV is a highly regulated space, there are currently no thresholds defined by regulators for validating neural networks for use within PV. We therefore established that an acceptable performance target for our neural networks would be an F1 score or an accuracy of 75.0 or higher. To affirm the F1 score or accuracy, we derived a sampling approach that was based on the Z1.4 standard developed by the American National Standards Institute/American Society for Quality [44], or acceptable quality level (AQL) method. The AQL method defines the maximum number of errors allowed to accept a set of outputs. In our research, we defined our outputs as true positives. We set our quality threshold to 96% because the models were performing at an F1 score of 75.0 and above, and PV SMEs had been integrated into the development process. Further detail can be found in Fig. 3, which details the AQL process for PV neural networks and has been adapted with permission from its original source [45].
Fig. 3.
Acceptable quality level (AQL) process for pharmacovigilance (PV) neural networks. This process depicts the framework for the validation of neural networks leveraging the AQL method. It was customized in a manner to accommodate for the inherent needs of PV. The validation process begins once the developer generates the results of a neural network and creates an excel output of the true positive (TP), false positive (FP), false negative (FN), and true negatives (TNs). If the F1 score or accuracy is below the 75% threshold, the PV subject matter expert (SME) reviews 100% of the FP and FN results and reports any trends in errors and results of the review to the developer for further training. If the F1 score or accuracy is above 75%, the PV SME reviews the TP results to ensure the neural network is performing at the F1 score or accuracy claimed. For our purposes, if the number of TPs was less than 150, the PV SME would perform a 100% review of TPs to ensure the system result matches the safety database entry and is indeed a TP, as it was within the work capacity of the team. If there were more than 150 TPs, the PV SME would randomize the TPs, select the appropriate AQL sample of TPs, and then review the results. For both instances, if the TP error rate was ≤ 4%, then the neural network was deemed passed, and if not, it was sent back to the developer for further training
Results
Recurrent Neural Network for Seriousness Classification
To test our automated seriousness classifier’s ability to accurately predict AE seriousness, we assessed the classifier’s predictions against the human ground truth determinations. Table 3 shows the results of this testing yielded equal to or greater than 83.0% accuracy in all report types.
Table 3.
Performance of neural networks
| Source data type | Seriousness classification (accuracy) | Seriousness categorization (F1 score) | Annotation of seriousness category terms (F1 score) |
|---|---|---|---|
| Post-marketing | 83.0% (precision = 0.95, recall = 0.74) |
Death—0.78 (precision = 0.88, recall = 0.70) Hospitalization—0.79 (precision = 0.84, recall = 0.74) IME—0.76 (precision = 0.81, recall = 0.72) |
NC |
| Solicited reports | 92.9% (precision = 0.87, recall = 0.87) | NC | 0.90 (precision = 0.88, recall = 0.91) |
| Medical literature | 86.3% (precision = 0.83, recall = 0.82) | NC | 0.75 (precision = 0.62, recall = 0.96) |
IME important medical event, NC not calculated
Recurrent Neural Network for Seriousness Categorization
We hypothesized that our method could be extended to identifying the seriousness categories corresponding to each AE in our reports. To test this, we used the same training data to develop an RNN classifier focused on classification of AEs in reports to seriousness categories. In initial tests, we encountered two issues: (1) because our model is designed to match seriousness categories to specific AEs, the large distance between AEs and seriousness terms (average of over 500 words) in long reports (typically in SD and ML reports) limited the classifier’s ability to connect an AE to seriousness terms; (2) our data set contained sufficient training data for classifying only three of the seven seriousness criteria—death, hospitalization, and IME as identified from the unstructured text within the source document. This imbalance in seriousness category representation within our sample reflects the composition of Celgene’s therapy portfolio and risk management programs, which dictate the variety and frequency of other categories of seriousness. As a result, the seriousness category classifier was only applied to PM reports, yielding the F1 score results depicted in Table 3 of 77.7 for death, 78.9 for hospitalization, and 75.5 for IME.
To observe if there were any cascade effects from combining the binary classifier into our multi-category classifier approach, we integrated the binary classification as a separate category in the multi-category classifier. After integration, we observed the binary classification (new category “not serious”) F1 score increase by 0.6–1.3 (post-marketing = 84.3, solicited reports = 93.5, medical literature = 87.0), whereas the remaining category classification F1 scores were very similar to the values in Table 3 (less than a 0.2 difference).
To understand how our deep learning classifiers performed compared to other methods, we trained two additional classifiers using random forests [39, 40] and support-vector machines [41–43]. Table 4 depicts the results generated by these two algorithms in the PM data set.
Table 4.
Analysis of alternate algorithm performance on post-marketing data
| Algorithm | Seriousness classification (accuracy) | Seriousness categorization (F1 score) |
|---|---|---|
| Random forests | 81.2% (precision = 0.89, recall = 0.71) |
Death—0.59 (precision = 0.53, recall = 0.66) Hospitalization—0.74 (precision = 0.78, recall = 0.70) IME—0.76 (precision = 0.84, recall = 0.69) |
| Support-vector machine | 82.3% (precision = 0.94, recall = 0.72) |
Death—0.80 (precision = 0.92, recall = 0.71) Hospitalization—0.75 (precision = 0.79, recall = 0.71) IME – 0.82 (precision = 0.87, recall = 0.77) |
IME important medical event
Long Short-Term Memory Neural Network to Annotate Seriousness Category Terms
Next, we trained a Bi-LSTM deep neural network seriousness annotator to help facilitate human review of seriousness determination. This annotator performed with an F1 score of 89.9 in SD reports, and 75.2 in ML reports, as shown in Table 3.
Example Model Analysis
An example from the data set of our models working in concert to determine the seriousness of each AE, categorization of serious AEs, and annotation of potential seriousness terms is provided in Table 5.
Table 5.
Example analysis by seriousness models
| Model inputs | Model outputs |
|---|---|
|
Narrative: patient was hospitalized for arrhythmia and passed away 3 days later from cardiac arrest AE: arrhythmia MedDRA® PT: arrhythmia; LLT: arrhythmia AE: cardiac arrest MedDRA® PT: cardiac arrest; LLT: cardiac arrest |
Binary seriousness classifier AE: arrhythmia = serious AE: cardiac arrest = serious Case = serious |
|
Seriousness category classifier AE: arrhythmia = hospitalization, IME AE: cardiac arrest = death | |
|
Annotator Patient was hospitalized for arrhythmia and passed away 3 days later from cardiac arrest |
AE adverse event, LLT lowest level term, MedDRA® Medical Dictionary for Regulatory Activities, PT preferred term, IME important medical event
Discussion
Our results show that AE seriousness can be determined with a high accuracy and/or F1 score at both the binary and subcategorization level, in various sources of unstructured document narratives, using an advanced neural network classification/annotator approach. We chose a neural network approach over other techniques such as support-vector machines and random forests because of the expected exponential growth in training data and the need for scalability based on the published prior art [46–48].
To the best of our knowledge, this is the first work of this type to be published and we are not aware of any other directly comparable work. Initial approaches applying natural language processing to AE seriousness using deep learning neural networks have been reported, but they are not comprehensive and focus on AE-level seriousness [32, 33]. Our work is differentiated from these studies by addressing the problem in a comprehensive manner through determination of seriousness at the AE level, combined with seriousness category classification and seriousness term annotation.
While the results of our modeling are notable, additional exploration of algorithmic techniques such as the use of Bi-LSTM in classification tasks may increase performance and should be pursued in future studies. Regardless of performance levels, however, it is clear that safety report seriousness classification will always require human confirmation. Under increasing report volumes, the PV workforce needs help to identify, from the massive number of reports, those that need immediate attention. Our models could potentially be used to assist human review by enabling the identification of documents containing serious AEs. In addition, our annotator model could be used to assist human review with quickly identifying the key text supporting seriousness classification within a report.
It should be noted that there are various limitations to our approach. First, our neural network approach was not able to categorize AE seriousness in case documents with lengthy case narrative sections. We were unable to train our networks to understand the on average 500-word span between AEs and seriousness criteria terms. To address this limitation, we developed a seriousness criteria annotator to accelerate human review of these larger documents. This illustrates the fact that any practical PV technology solution will likely require various methodologies, working in concert with human experts, to be successful.
Additional limitations of our method include the requirement for already identified AEs and MedDRA® terms as inputs for our classifier and the use of data from a single company’s drug portfolio, which may impact its generalizability. However, the method can be applied at additional companies using training data representative of that company’s AEs, seriousness categories, drug/indication portfolios, and conventions.
Despite these limitations, our methodology represents a state-of-the-art approach to highly accurate automated seriousness classification, at both the AE and case level, and suggests that the potential benefits hypothesized with the automation of seriousness determination—increased consistency and efficiencies enabling compliance with stringent reporting timelines despite increasing report volumes—may be demonstrated when evaluated in future studies. Of particular interest is the conduct of user studies to evaluate what level of improvement, if any, might be expected from employing this method in a prospective evaluation environment. The benefits demonstrated in any such studies will need to be considered with system implementations and business process improvements.
Conclusions
To our knowledge, our work is the first to demonstrate that deep learning can be applied to the evaluation of event and/or case seriousness classification within PV. Given the increasing demands on the PV workforce, approaches like deep learning need to be considered for supporting the volume, complexity, and time constraints of AE report processing. Augmentation of human review with deep learning is a viable approach to tackle these current challenges. We demonstrate that our deep learning algorithms were able to identify serious AEs, classify the seriousness of AEs, and annotate seriousness text in unstructured document narratives. If introduced in the PV case management process, we believe that our algorithms could positively impact the consistency and timeliness of reporting.
Acknowledgements
We thank Mark Widdowson for his insights in developing this study.
Compliance with Ethical Standards
Funding
Funding for this study was provided by Celgene Corporation.
Conflict of interest
Ramani Routray, Claire Abu-Assal, Hanqing Chen, Shenghua Bao, Van Willis, Sharon Hensley Alford, and Vivek Krishnamurthy were employed by IBM Watson Health at the time this research was conducted. Niki Tatarenko, Ruta Mockute, Bruno Assuncao, Sameen Desai, Salvatore Cicirello, Karolina Danysz, and Edward Mingle were employed by Celgene Corporation at the time this research was conducted and hold stock/stock options therein.
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Ramani Routray and Niki Tetarenko contributed equally to this work.
References
- 1.World Health Organization . The importance of pharmacovigilance: safety monitoring of medicinal products. Geneva: World Health Organization; 2002. [Google Scholar]
- 2.European Medicines Agency . Guideline on good pharmacovigilance practices. Annex I: definitions (Rev 4) Amsterdam: European Medicines Agency; 2017. [Google Scholar]
- 3.US Food and Drug Administration. Investigational new drug safety reporting. 21CFR31232. Silver Spring: U.S. Food and Drug Administration; 2017.
- 4.US Food and Drug Administration. Individual case safety reports. 2–18. https://www.fdagov/forindustry/datastandards/individualcasesafetyreports/defaulthtm. Accessed Dec 2018.
- 5.Price J. Pharmacovigilance in crisis: drug safety at a crossroads. Clin Ther. 2018;40(5):790–797. doi: 10.1016/j.clinthera.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 6.US Food and Drug Administration Adverse Event Reporting System (FAERS) Public dashboard. Silver Spring: U.S. Food and Drug Administration; 2018. [Google Scholar]
- 7.US Food and Drug Administration. Postmarketing reporting of adverse drug experiences. 21CFR31480. Silver Spring: U.S. Food and Drug Administration; 2018.
- 8.Bollegala D, Maskell S, Sloane R, Hajne J, Pirmohamed M. Causality patterns for detecting adverse drug reactions from social media: text mining approach. JMIR Public Health Surveill. 2018;4(2):e51. doi: 10.2196/publichealth.8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eriksson R, Jensen PB, Frankild S, Jensen LJ, Brunak S. Dictionary construction and identification of possible adverse drug events in Danish clinical narrative text. J Am Med Inform Assoc. 2013;20(5):947–953. doi: 10.1136/amiajnl-2013-001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang SH, Lee S, Koo HK, Kim Y. Evaluation of a computer-based adverse-drug-event monitor. Am J Health Syst Pharm. 2008;65(23):2265–2272. doi: 10.2146/ajhp080122. [DOI] [PubMed] [Google Scholar]
- 11.LePendu P, Iyer SV, Bauer-Mehren A, Harpaz R, Mortensen JM, Podchiyska T, et al. Pharmacovigilance using clinical notes. Clin Pharmacol Ther. 2013;93(6):547–555. doi: 10.1038/clpt.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melton GB, Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries. J Am Med Inform Assoc. 2005;12(4):448–457. doi: 10.1197/jamia.M1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murff HJ, Forster AJ, Peterson JF, Fiskio JM, Heiman HL, Bates DW. Electronically screening discharge summaries for adverse medical events. J Am Med Inform Assoc. 2003;10(4):339–350. doi: 10.1197/jamia.M1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polepalli Ramesh B, Belknap SM, Li Z, Frid N, West DP, Yu H. Automatically recognizing medication and adverse event information from Food and Drug Administration’s adverse event reporting system narratives. JMIR Med Inform. 2014;2(1):e10. doi: 10.2196/medinform.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tinoco A, Evans RS, Staes CJ, Lloyd JF, Rothschild JM, Haug PJ. Comparison of computerized surveillance and manual chart review for adverse events. J Am Med Inform Assoc. 2011;18(4):491–497. doi: 10.1136/amiajnl-2011-000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Hripcsak G, Markatou M, Friedman C. Active computerized pharmacovigilance using natural language processing, statistics, and electronic health records: a feasibility study. J Am Med Inform Assoc. 2009;16(3):328–337. doi: 10.1197/jamia.M3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wunnava S, Qin X, Kakar T, Rundensteiner EA, Kong X. Bidirectional LSTM-CRF for adverse drug event tagging in electronic health records. Proc. Mach. Learn. Res. 2018;90:48–56. [Google Scholar]
- 18.Gurulingappa H, Mateen-Rajpu A, Toldo L. Extraction of potential adverse drug events from medical case reports. J. Biomed. Semant. 2012;3(1):15. doi: 10.1186/2041-1480-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarker A, Gonzalez G. Portable automatic text classification for adverse drug reaction detection via multi-corpus training. J. Biomed. Inform. 2015;53:196–207. doi: 10.1016/j.jbi.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adrover C, Bodnar T, Huang Z, Telenti A, Salathé M. Identifying adverse effects of HIV drug treatment and associated sentiments using Twitter. JMIR Public Health Surveill. 2015;1(2):e7. doi: 10.2196/publichealth.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comfort S, Perera S, Hudson Z, Dorrell D, Meireis S, Nagarajan M, et al. Sorting through the safety data haystack: using machine learning to identify individual case safety reports in social–digital media. Drug Saf. 2018;41(6):579–590. doi: 10.1007/s40264-018-0641-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eshleman R, Singh R. Leveraging graph topology and semantic context for pharmacovigilance through twitter-streams. BMC Bioinform. 2016;17(Suppl. 13):335. doi: 10.1186/s12859-016-1220-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tafti AP, Badger J, LaRose E, Shirzadi E, Mahnke A, Mayer J, et al. Adverse drug event discovery using biomedical literature: a big data neural network adventure. JMIR Med Inform. 2017;5(4):e51. doi: 10.2196/medinform.9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raja K, Patrick M, Elder JT, Tsoi LC. Machine learning workflow to enhance predictions of adverse drug reactions (ADRs) through drug-gene interactions: application to drugs for cutaneous diseases. Sci Rep. 2017;7(1):3690. doi: 10.1038/s41598-017-03914-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang G, Jung K, Winnenburg R, Shah NH. A method for systematic discovery of adverse drug events from clinical notes. J Am Med Inform Assoc. 2015;22(6):1196–1204. doi: 10.1093/jamia/ocv102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bian J, Topaloglu U, Yu F. Towards large-scale twitter mining for drug-related adverse events. SHB12 (2012). 2012;2012:25–32. doi: 10.1145/2389707.2389713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jagannatha AN, Yu H. Bidirectional RNN for medical event detection in electronic health records. Proc. Conf. Assoc. Comput. Linguist. N. Am. Chapter Meet. 2016;2016:473–482. doi: 10.18653/v1/n16-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanovsky G, Gruhl D, Mendes PN. Recognizing mentions of adverse drug reactions in social media using knowledge-infused recurrent models. In: Proceedings of the 15th conference of the European chapter of the association for computational linguistics, vol 1. Long papers; 2017. pp 142–51.
- 29.Tutubalina E, Nikolenko S. Combination of deep recurrent neural networks and conditional random fields for extracting adverse drug reactions from user reviews. J Healthc Eng. 2017;2017:9451342. doi: 10.1155/2017/9451342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dev S, Zhang S, Voyles J, Rao A. Automated classification of adverse events in pharmacovigilance. In: 2017 IEEE internationalc conference on bioinformatics and biomedicine (BIBM), Kansas City, MO; 2017. pp. 905–9. 10.1109/BIBM.2017.8217777. https://www.computer.org/csdl/api/v1/citation/asciitext/proceedings/12OmNx6g6nT/08217777.
- 31.Wang Y, Coiera E, Runciman W, Magrabi F. Using multiclass classification to automate the identification of patient safety incident reports by type and severity. BMC Med Inform Decis Mak. 2017;17(1):84. doi: 10.1186/s12911-017-0483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuwen L, Chen S, Zhang H, editors. Detecting potential serious adverse drug reactions using sequential pattern mining method. In: 2018 IEEE 9th international conference on software engineering and service science (ICSESS), Beijing; 23–25 Nov 2018.
- 33.Zhang S, Dev S, Voyles J, Rao AS, editors. Attention-based multi-task learning in pharmacovigilance. In: 2018 IEEE international conference on bioinformatics and biomedicine (BIBM), Madrid; 3–6 Dec 2018.
- 34.ICH. Post-approval safety data management: definitions and standards for expedited reporting E2D. Step 4 version. Geneva: International Council for Harmonisation; 2003.
- 35.Pennington J, Socher R, Manning C. Glove: global vectors for word representation. Stroudsburg: Association for Computational Linguistics; 2014. [Google Scholar]
- 36.Coordinators NR. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44(D1):D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Z, Xu W, Yu K. Bidirectional LSTM-CRF models for sequence tagging. Ithaca: ArXiv; 2015. [Google Scholar]
- 38.Lipton Z, Kale D, Elkan C, Wetzel R. Learning to diagnose with LSTM recurrent neural networks. Ithaca: ArXiv; 2015. [Google Scholar]
- 39.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi: 10.1023/a:1010933404324. [DOI] [Google Scholar]
- 40.Khalilia M, Chakraborty S, Popescu M. Predicting disease risks from highly imbalanced data using random forest. BMC Med Inform Decis Mak. 2011;11(1):51. doi: 10.1186/1472-6947-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20(3):273–297. doi: 10.1007/bf00994018. [DOI] [Google Scholar]
- 42.Go A, Bhayani R, Huang L. Twitter sentiment classification using distant supervision. CS224N Project Report; 2009. pp. 1–12. https://cs.stanford.edu/people/alecmgo/papers/TwitterDistantSupervision09.pdf.
- 43.Lilleberg J, Zhu Y, Zhang Y. Support vector machines and Word2vec for text classification with semantic features. New York: Institute of Electrical and Electronics Engineers; 2015. [Google Scholar]
- 44.Institute American National Standards. American Society for Quality . ANSI/ASQ Z1.4-2003 (R2013): sampling procedures and tables for inspection by attributes. Milwaukee: American Society for Quality; 2013. [Google Scholar]
- 45.Mockute R, Desai S, Perera S, Assuncao B, Danysz K, Tetarenko N, et al. Artificial intelligence within pharmacovigilance: a means to identify cognitive services and the framework for their validation. Pharm Med. 2019;33(2):109–120. doi: 10.1007/s40290-019-00269-0. [DOI] [PubMed] [Google Scholar]
- 46.Dos Santos C, Gatti de Bayser M, editors. Deep convolutional neural networks for sentiment analysis of short texts. In: Proceedings of COLING 2014, the 25th international conference on computational linguistics: technical papers; Dublin.
- 47.Jeeva M. The scuffle between two algorithms: neural network vs. support vector machine. 2018. https://medium.com/analytics-vidhya/the-scuffle-between-two-algorithms-neural-network-vs-support-vector-machine-16abe0eb4181. Accessed May 2019.
- 48.Zaghloul W, Lee S, Trimi S. Text classification: neural networks vs support vector machines. Ind Manag Data Syst. 2009;109(5):708–717. doi: 10.1108/02635570910957669. [DOI] [Google Scholar]