Abstract
HSP90 (heat shock protein 90) is an ATP-dependent molecular chaperone involved in a proper folding and maturation of hundreds of proteins. HSP90 is abundantly expressed in cancer, including melanoma. HSP90 client proteins are the key oncoproteins of several signaling pathways controlling melanoma development, progression and response to therapy. A number of natural and synthetic compounds of different chemical structures and binding sites within HSP90 have been identified as selective HSP90 inhibitors. The majority of HSP90-targeting agents affect N-terminal ATPase activity of HSP90. In contrast to N-terminal inhibitors, agents interacting with the middle and C-terminal domains of HSP90 do not induce HSP70-dependent cytoprotective response. Several inhibitors of HSP90 were tested against melanoma in pre-clinical studies and clinical trials, providing evidence that these agents can be considered either as single or complementary therapeutic strategy. This review summarizes current knowledge on the role of HSP90 protein in cancer with focus on melanoma, and provides an overview of structurally different HSP90 inhibitors that are considered as potential therapeutics for melanoma treatment.
Keywords: Apoptosis, Chaperone, HSP70, HSP90 inhibitors, Melanoma, Targeted therapy
Introduction
The number of patients diagnosed with melanoma increases every year. In 2019, over 90,000 of new melanoma cases have been estimated to be diagnosed in United States alone [1]. Environmental factors, especially UV exposure, familiar history and genetic factors are amongst major causes of melanoma [2–4]. The most common mutations are found in genes encoding components of the RAS/RAF/MEK/ERK (MAPK) signaling pathway, and they lead to a constitutive activity of this cascade [5]. Algorithms for current treatment of melanoma patients include vemurafenib, dabrafenib and encorafenib, targeting mutated BRAF (B-RAF proto-oncogene, serine/threonine kinase); trametinib, cobimetinib and binimetinib that inhibit the activity of MEK1/2 (mitogen-activated protein kinase kinase), as well as immune checkpoint inhibitors including nivolumab and pembrolizumab binding to PD-1 (programmed cell death protein 1) and ipilimumab inhibiting CTLA-4 (cytotoxic T-lymphocyte antigen 4) [6–9]. However, available therapies have several limitations. Melanoma cells develop resistance towards BRAF and MEK inhibitors through a number of genetic and epigenetic mechanisms [10–13]. Resistance emerges through upregulation of expression of mutated BRAF, alternative splicing of BRAF transcript, secondary BRAF mutations, mutations in genes encoding MEK1/2 and RAS, reactivation of COT (cancer osaka thyroid oncogene) activity, dimerization of CRAF (RAF-1 proto-oncogene, serine/threonine kinase), which all can lead to the hyperactivation or recovery of the MAPK pathway activity. In addition, a loss of functional PTEN (phosphatase and tensin homolog) and enhanced activity of the PI3K (phosphatidylinositol 3-kinase)/AKT/mTOR (mechanistic target of rapamycin kinase) pathway, a suppression of BIM (BCL-2 interacting mediator of cell death), a loss of STAG2 (stromal antigen 2) or STAG3 (stromal antigen 3) that are the subunits of cohesion complex, an increase in cyclin D1 level, enhanced expression of several microRNAs, and expression of resistance-associated genes including AXL, PDGFRB (platelet-derived growth factor receptor beta) and EGFR (epidermal growth factor receptor) have been demonstrated to contribute to resistance of melanoma cells. In addition to cell-intrinsic mechanisms, growth factors derived from stromal cells and hypoxia can modulate melanoma cell sensitivity to targeted drugs [10–12, 14–16], and long-term therapy with BRAFV600E inhibitor can develop resistance to other drugs including dacarbazine [17]. Resistance to immunotherapy can also emerge, and melanoma cells resistant to PD-1 inhibitors show upregulation of receptors VISTA (V-domain Ig suppressor of T cell activation) and TIM-3 (T-cell immunoglobulin and mucin domain-containing 3), as well as acquisition of mutations in genes encoding JAK1 (Janus kinase 1), JAK2 (Janus kinase 2) and B2 M (beta-2-microglobulin), which results in reduced sensitivity to T-cell mediated killing [18, 19]. Therefore, there is still an urgent need for alternative therapeutic approaches for melanoma treatment, and new targets need to be identified.
HSP90 (heat shock protein 90) that is one of the crucial mediators of cellular physiology [20], is also recognized as a key facilitator of cancer cell survival [21]. This review delineates recent advances in our understanding of HSP90 function in melanoma cells and provides information on HSP90 inhibitors as potential drugs for melanoma treatment.
HSP90: structure and regulation of activity
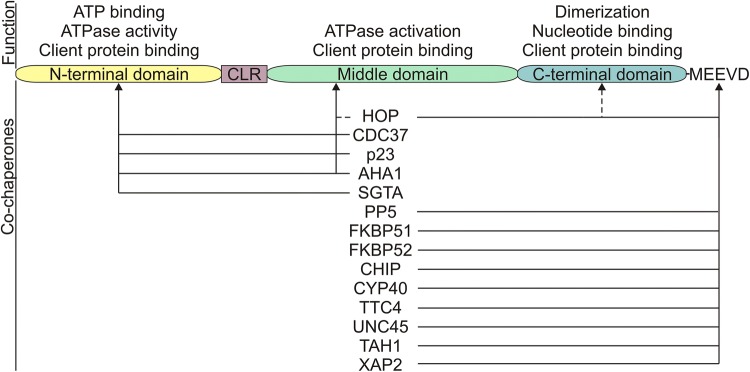
The HSP90 family of proteins includes the cytosolic HSP90α, HSP90β and HSP90 N isoforms, ER (endoplasmic reticulum)-residing member GRP94 (glucose-regulated protein 94) and mitochondrial protein TRAP1 (tumor necrosis factor receptor-associated protein 1) [22]. HSP90β is constitutively expressed, whereas HSP90α is induced in response to stress [23]. HSP90 homologues share conserved domains including an N-terminal domain (NTD; ~ 25 kDa), a middle-domain (MD; ~ 35 kDa), a C-terminal domain (CTD; ~ 10 kDa) and a flexible charged linker between NTD and MD [24–26]. In addition, HSP90α and HSP90β possess C-terminal Met-Glu-Glu-Val-Asp (MEEVD) motif [22]. HSP90 domains play specific roles (Fig. 1). NTD predominantly exerts ATPase activity, MD assists in activation of ATPase activity, whereas CTD is responsible for HSP90 dimerization, which is essential for its chaperone function [22, 27]. In addition, HSP90 domains serve as binding sites for the client proteins and co-chaperones (Fig. 1).
Fig. 1.
Schematic representation of HSP90 protein domain structure. Functions of each domain and HSP90-interacting co-chaperones with their binding sites are shown. Dashed lines represent alternative binding sites. AHA1: activator of HSP90 ATPase protein 1; ATP: adenosine triphosphate; CDC37: cell division cycle 37; CHIP: carboxyl terminus of HSP70-interacting protein; CLR: charged linker region; CYP40: cyclophilin 40; FKBP51: FK506-binding protein 5; FKBP52: FK506-binding protein 4; HOP: homeodomain-only protein; PP5: protein phosphatase 5; SGTA: small glutamine rich tetratricopeptide repeat-containing alpha; TAH1: telomere-associated homeobox-containing protein 1; TTC4: tetratricopeptide repeat domain 4; UNC45: smooth muscle cell-associated protein 1; XAP2: HBV X-associated protein 2
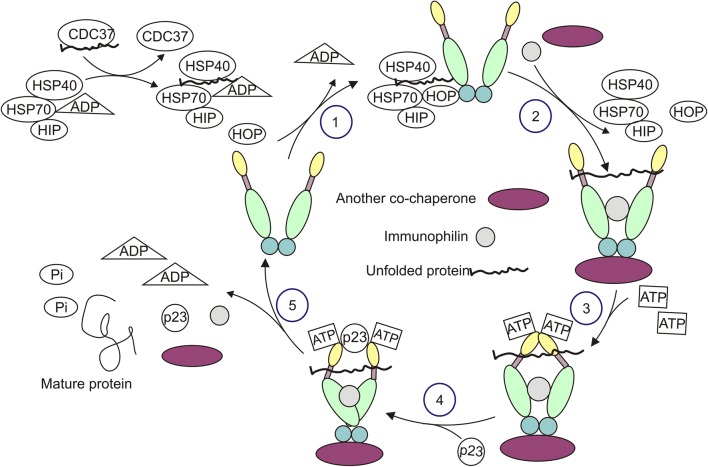
HSP90 contributes to folding and remodeling of proteins, assists in assembly of multi-protein complexes, and enables ligand binding to receptors [28]. HSP90 activity depends on the cooperation with co-chaperones and immunophilins, and is associated with highly dynamic conformational rearrangements during the chaperone cycle [26] (Fig. 2). In the absence of ATP, HSP90 predominantly adopts a V-shaped open conformation. ATP binding to NTD triggers closed state of HSP90, which is preceded by an intermediate steps involving the contribution of the N-terminal “lid” region. Closed state of HSP90 is crucial for ATP hydrolysis as it involves a reposition of a catalytic loop in the middle domain to activate ATPase activity in N-terminal domain. When HSP90 reaches the fully closed state, ATP undergoes hydrolysis which is followed by a disassembly of a multi-protein complex. ATPase cycle of HSP90 is symmetric as both ATP molecules are disrupted simultaneously. In addition, conformational cycle of HSP90 similarly appears in the presence and absence of the client proteins [28–30]. Co-chaperones substantially regulate HSP90 function as they diversely modulate HSP90 chaperone cycle and act as adaptors for specific client recruitment [22]. Regulation of HSP90 activity can be additionally modulated at the transcriptional level and through posttranscriptional modifications. Transcriptional regulation of HSP90 expression is mainly controlled by HSFs (heat shock factors), which bind to HSE (heat shock element) located in the promoter region of HSP90 [31, 32]. HDAC6 (histone deacetylase 6)-mediated regulation of HSP90 stability, posttranslational modifications of cytosolic HSP90 that include phosphorylation, acetylation, methylation, ubiquitylation and S-nitrosylation have been extensively discussed elsewhere [30, 33–35].
Fig. 2.
Exemplary chaperone cycle of HSP90. The consecutive steps are marked with numbers. Unfolded client protein of HSP90 is transferred from CDC37 to HSP70/HSP40/HIP/ADP complex, and becomes attached to HSP90 in an open conformation with assistance of HOP (1). Then, other co-chaperones and immunophilins are bound to the HSP90 homodimer, while HSP40, HSP70, HIP and HOP being released (2). Binding of ATP to the N-terminal domain of HSP90 switches the protein from an open to close conformation (3). Subsequently, p23 is attached (4), which is followed by ATP hydrolysis, and the release of mature protein, co-chaperones and immunophilins as well as conformational change of HSP90 (5). HSP40: heat shock protein 40; HIP: Hsc70-interacting protein
HSP90 in melanoma
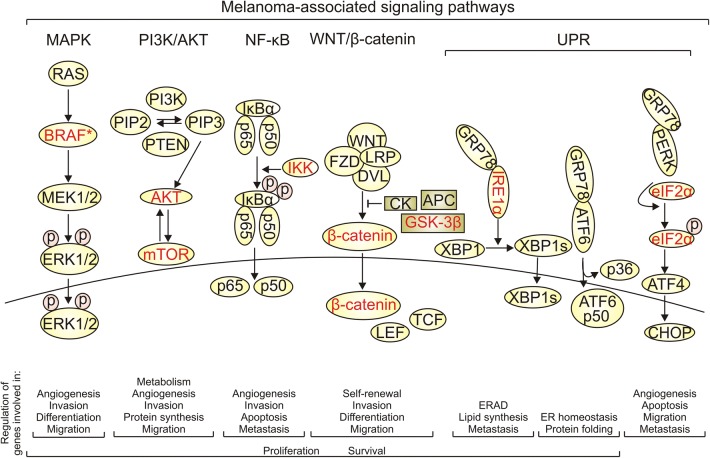
HSP90 is usually overexpressed in cancer [28]. While HSP90 level is low in benign melanocytic nevi, it increases during melanoma progression [36]. Consequently, a high level of HSP90 was assessed in more than 50% of melanoma tumors [37, 38]. Although high HSP90 expression is not a predictive factor for patient survival, HSP90 level significantly correlated with the Clark level and increased Breslow depth in primary melanomas [36]. Despite intracellular presence of HSP90, it is also identified on melanoma cell surface suggesting that HSP90 might be an immunorelevant target [39, 40]. It has been shown that a membrane-bound HSP90 facilitates immune clearance of dying cells [41]. In addition, HSP90 can be secreted into the extracellular space [42]. HSP90 was detected in serum of melanoma patients at significantly higher levels than in healthy controls [43]. As extracellular HSP90 can promote cell motility and angiogenesis [44], serum level of HSP90 might be considered as a putative biomarker of melanoma progression. HSP90 controls folding and maturation of more than 200 proteins, and up-to-date list of HSP90 clients and interactors is available at the Picard’s Lab website [https://www.picard.ch/downloads/Hsp90interactors.pdf]. HSP90 plays a multifactorial role in melanoma. HSP90 isoform was found in melanoma-derived exosomes and was considered as a part of ‘education’ program for bone marrow cells creating a pre-metastatic niche for melanoma cells [45]. Formation of a triple HSP90/HIF-1α/BCL-2 (B-cell CLL/lymphoma 2) complex results in stabilization of HIF-1α (hypoxia-inducible factor 1) under hypoxic conditions [46]. In addition, melanoma development and progression are substantially dependent on several key signaling pathways, all including essential oncoproteins identified as HSP90 client proteins (Fig. 3). MAPK signaling pathway is responsible for melanoma cell proliferation, differentiation, survival, invasion and angiogenesis [9, 47]. This signaling cascade is constitutively active in the majority of melanomas as a result of genetic alterations in BRAF, RAS or NF1 (neurofibromin 1) [48–50]. Most frequent mutations are found in BRAF (40–60% of melanoma patients) and NRAS (15–20%). Mutations in BRAF are mainly associated with a substitution of valine in codon 600, and valine can be substituted with either glutamic acid or lysine in up to 90% and 10–20% of patients harboring mutation in BRAF, respectively [5, 48]. Interestingly, HSP90 is required for folding of a protein product of mutated BRAF, whereas wild-type BRAF is not stabilized by HSP90 in cutaneous melanoma cells [51, 52]. By increasing intracellular protein load, oncogenic MAPK signaling broadly affects UPR (unfolded protein response) pathways involved in cell fate decision during prolonged ER stress [53, 54]. It has been also demonstrated that sustained activity of IRE1α (inositol-requiring enzyme 1 alpha) and ATF6 (activating transcription factor 6) promotes adaptation of melanoma cells to proteotoxic stress [55], which supports cancer progression [56]. HSP90 also contributes to the regulation of PI3 K/AKT signaling, which is involved in melanoma cell proliferation, migration and survival [57, 58], and it is often activated in melanoma cells resistant to BRAF and MEK inhibitors [59]. HSP90 may also control cell metabolism and protein synthesis through a cross-talk between PI3 K/AKT cascade and mTOR [57, 60, 61] as both AKT and mTOR are HSP90 client proteins (Fig. 3). IKKs (IκB kinases), other HSP90 client proteins, control NF-κB (nuclear factor kappa B) activation [62]. NF-κB signaling pathway is constitutively active in melanoma cells, where it regulates expression of genes involved in apoptosis, cell cycle, invasion and angiogenesis [63]. Activation of NF-κB is also associated with emergence of resistance to BRAF inhibitors [64–66]. WNT (Wingless-type)/β-catenin signaling pathway takes part in melanoma development, cell self-renewal and migration [67, 68]. It has been also shown that ABCB5 (ATP-binding cassette, sub-family B, member 5) contributes to WNT-dependent expression of CXCL8 (C-X-C motif chemokine ligand 8) encoding interleukin-8 to support a slow-cycling and chemoresistant phenotype of melanoma cells [69]. In addition, the enhanced activity of WNT signaling pathway was found in melanospheres [70]. AXL, another HSP90-interacting protein relevant for melanoma [71], has been identified as a driver of resistance to targeted drugs in melanoma [72], and a regulator of metastasis-promoting phenotype of melanoma cells [73].
Fig. 3.
Major melanoma-associated signaling pathways, and their roles in melanoma. Proteins identified as direct HSP90 clients are depicted in red. *only BRAF mutants but not wild-type protein are reported as HSP90 clients in cutaneous melanoma. APC: adenomatous polyposis coli; CK: creatine kinase; DVL: dishevelled; ERAD: endoplasmic reticulum-associated protein degradation; FZD: frizzled; GSK-3β: glycogen synthase kinase 3 beta; IKK: IκB kinase; LEF: lymphoid enhancer-binding factor 1; LRP: low density lipoprotein receptor-related protein; PIP2: phosphatidylinositol biphosphate; PIP3: phosphatidylinositol(3,4,5)trisphosphate; TCF: T-cell factor
HSP90 inhibitors
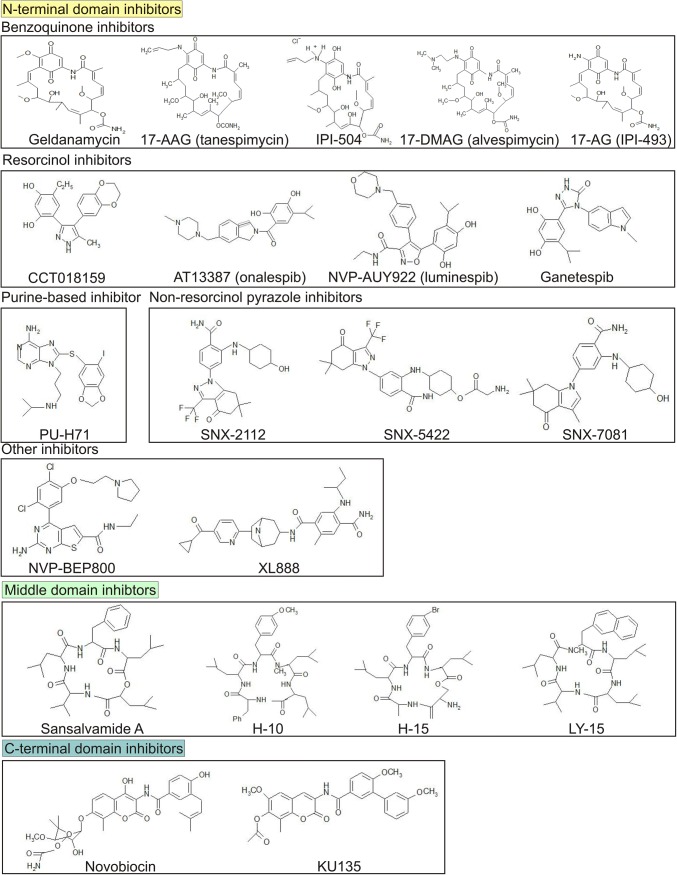
The role of HSP90 in melanoma development and progression makes this protein a promising therapeutic target. The rationale for targeting HSP90 is supported not only by a high level of this protein in cancer cells, but also by cancer cell-selective formation of HSP90 multi-chaperone complexes exerting a high ATPase activity [74]. In addition, HSP90 has been identified as a crucial regulator of melanoma cell phenotype, and inhibition of HSP90 has substantially affected both commercially available and primary melanoma cell lines [75], also those resistant to currently available therapeutics [76]. A number of natural and synthetic compounds of different chemical structures and binding sites have been identified as selective HSP90 inhibitors (Fig. 4). N-terminal domain inhibitors act by disrupting the interaction between ATP and ATP-binding pocket, and they restrain HSP90 in the ADP-bound state that leads to ubiquitylation and subsequent proteasomal degradation of the client proteins [77]. In turn, C-terminal domain inhibitors destabilize the chaperone complex and induce a release of co-chaperones and degradation of client proteins [78, 79]. Inhibitors of the middle domain of HSP90 directly or allosterically disrupt interactions between HSP90 and C-terminal binding proteins [80].
Fig. 4.
HSP90 inhibitors exerting anti-melanoma activity. Compounds were classified based on their binding sites and similarity in chemical structure
N-Terminal domain inhibitors
Benzoquinone inhibitors
Geldanamycin is a benzoquinone, ansamycin antibiotic of natural origin isolated from Streptomyces hygroscopicus, and exerts a potent activity against different types of cancer cells by competing with ATP for binding to the N-terminal domain of HSP90 [81]. However, substantial hepatotoxicity and unsatisfactory solubility of geldanamycin [82, 83] enforced the research to develop geldanamycin derivatives. 11-methoxy-17-formyl-17-demethoxy-18-O-21-O-dihydrogeldanamycin was isolated from another strain of Streptomyces hygroscopicus (A070101), and exerted a marked cytotoxicity against cancer cells including melanoma [84]. In addition, several 17-substituted semi-synthetic derivatives of geldanamycin exerted a promising activity while being less toxic against normal cells. The methoxy substituent of benzoquinone moiety in geldanamycin was replaced by an allyloamine group (17-AAG; tanespimycin), dimethyl-aminoethylamine group (17-DMAG; alvespimycin) or changed for an amine group (17-AG, IPI-493) [81]. Moreover, 17-AAG hydrochloride (IPI-504) was obtained by a reduction of benzoquinone moiety in 17-AAG to hydrochinon [85].
Activity of ansamycin HSP90 inhibitors was broadly studied in melanoma cells. It was demonstrated that 17-AAG induced degradation of BRAFV600E and other BRAF mutants, but not wild-type BRAF in cutaneous melanoma cells [51, 52]. However, HSP90 inhibition by 17-AAG or 17-DMAG affected wild-type BRAF in uveal melanoma cell lines [86]. Consequently, geldanamycin derivatives inactivated MAPK signaling that was shown as reduced level of phosphorylated ERK1/2 [86–89]. This effect was similarly observed in melanoma cells harboring mutation in NRAS [89, 90]. 17-DMAG also decreased levels of phospho-ERK1/2 and phospho-AKT under hyperthermic conditions [90]. As a result, activity of HSP90 inhibitors was associated with downregulation of cyclin D1 and inhibition of cell proliferation [86, 87]. Geldanamycin derivatives differed in a cytostatic potential as shown for 17-DMAG that was more effective at inhibiting melanoma cell proliferation than 17-AAG [90]. In addition, 17-AAG inhibited vemurafenib-mediated paradoxical activation of ERK1/2 [91]. It was also demonstrated that geldanamycin and its analogs induced cell death [89, 90, 92]. 17-AAG and 17-DMAG induced apoptosis associated with the activation of caspase-9, caspase-2 and caspase-7, and PARP (poly-ADP ribose polymerase) cleavage [90]. 17-AG was more potent in caspase-3/7 activation than geldanamycin in patient-derived melanoma cells [89], although geldanamycin exerted lower IC50 values for anti-clonogenic activity than 17-AG [93]. 17-AG-mediated apoptosis was associated with attenuation of cytoprotective IRE1α-XBP1s (spliced X-Box binding protein 1) axis in melanoma cells harboring either BRAFV600E or NRASQ61R variant [89].
In addition to cytostatic and cytotoxic effects of ansamycin HSP90 inhibitors, these compound can affect melanoma cell phenotype. 17-AAG induced differentiation by increasing protein levels of tyrosinase and PMEL/gp100 (premelanosome protein/glycoprotein 100) in both BRAF-mutated and wild-type BRAF melanoma cell lines [87]. In another study, 17-AAG increased expression of DCT (dopachrome tautomerase) and TYRP1 (tyrosinase-related protein 1) encoding pigmentation-related proteins, and elevated melanin production in melanoma cells [91]. Additionally, 17-AAG increased glycerophosphocholine levels which was coupled with an elevated content of cytoplasmic mobile lipid droplets and enhanced fatty acid signaling suggesting that HSP90 inhibition can also broadly affect metabolism of melanoma cells [87].
Geldanamycin-related HSP90 inhibitors upregulate co-chaperones and stress-related response genes that can affect cell sensitivity to these drugs. Geldanamycin, 17-AAG, 17-DMAG and 17-AG induced expression of HSP70 (heat shock protein 70) [87, 89, 94–96], however, this effect was transient and silenced already after 22 h in melanoma cells exposed to 17-AG [89]. HSP70 upregulation can be the outcome of HSF-1 (heat shock factor 1) activation to induce expression of genes encoding heat shock proteins [97]. Accumulation of HSP70 reduced the extent of cell death induction in response to HSP90 inhibition [98]. In addition, 17-AG upregulated ER-located chaperone protein GRP78 (glucose-regulated protein 78) in a manner similar to the induction of HSP70 [89].
Cell sensitivity to ansamycin inhibitors of HSP90 was also associated with expression of NQO1 encoding NAD(P)H:quinone oxidoreductase 1 that converted these compounds to hydroquinone to enhance their activity by increasing hydrogen bonding [99]. It was reported that NQO1P187S variant exerted diminished activity compared with wild-type NQO1 [99], but genetic alterations affecting a His80 residue in this protein could compensate for P187S substitution [100]. Loss of NQO1 expression and acquisition of NQO1P187S variant contributed to the development of resistance to 17-AAG [101]. Accordingly, sensitivity of melanoma cells to 17-AAG was related to NQO1 overexpression [102]. In NQO1low melanoma cells, combination of 17-AAG and cisplatin exerted cooperation, which was driven by cisplatin-mediated induction of reactive oxygen species and up-regulation of NQO1 [102]. Interestingly, melanoma cells harboring P187S variant of NQO1 were susceptible to 17-AG-induced apoptosis, although the occurrence of cell death was delayed compared with melanoma cells harboring wild-type NQO1 [89]. This could result from similar affinity of quinone and hydroquinone forms of 17-AG to HSP90 [103] as NQO1-independent cell response to 17-AG was also reported in another study [104].
While the molecular effects of 17-AAG in melanoma cells are diverse, no objective clinical response to this drug was reported in a phase II trial in patients with metastatic melanoma [105]. In addition, several adverse effects of grade 2 and 3 severity were reported, including nausea, vomiting and fatigue [105]. Geldanamycin derivatives cooperated with other drugs or therapeutic approaches. Combination of 17-AAG and tipifarnib, a farnesyltransferase inhibitor, was cooperatively cytotoxic against melanoma cell lines derived from advanced stage tumors, but not against cells from radial growth phase melanoma [106]. Drug combination induced apoptosis through mitochondrial pathway as evidenced by increase in caspase-3 and caspase-9 activation, and DNA fragmentation [106]. By degrading HIF-1α protein, 17-AAG synergistically cooperated with glucose analog (2-DG) and imiquimod (a ligand for Toll-like receptor 7/8) to induce cell apoptosis and inhibit melanoma tumor growth in vivo [107]. Combination of 17-AAG and PI3 K inhibitor diminished ERK1/2 and AKT phosphorylation, and exerted enhanced anti-melanoma activity than either drug used alone [88]. Geldanamycin [108] and 17-DMAG cooperated with hyperthermia to more potently inhibit melanoma cell proliferation and increase the number of apoptotic and necrotic cells in a time-dependent manner [90]. 17-AG enhanced activity of MAPK pathway inhibitors, vemurafenib and trametinib, in induction of apoptosis that might result from concurrent inhibition of IRE1α and ERK1/2 activities [89]. Interestingly, it was also demonstrated that prolonged treatment with 17-AAG could develop resistance to a spectrum of structurally distinct inhibitors of HSP90, and acquired resistance could be overcome by HDAC inhibitors [109].
Resorcinol inhibitors
CCT018159
CCT018159 is a synthetic 3,4-diarylpyrazole resorcinol inhibitor of N-terminal ATPase activity of HSP90 [110]. Lack of a benzoquinone moiety in CCT018159 may determine lower hepatotoxicity than this observed for ansamycin inhibitors of HSP90 [111]. CCT018159 displayed a number of similar activities compared with geldanamycin and geldanamycin derivatives including induction of HSP70 expression [111], depletion of melanoma-related oncoproteins such as BRAFV600E, CRAF, CDK4 (cyclin-dependent kinase 4), ERBB2 (receptor tyrosine protein kinase ERBB2) [111], attenuation of ERK1/2 activity [87, 111] and upregulation of genes involved in melanoma cell differentiation [87]. In addition, CCT018159 caused a substantial accumulation of melanoma cells in the G1 phase of cell cycle, and induced apoptosis [87, 111]. Melanoma cell response to CCT018159 was independent of NQO1 expression and the level of P-glycoprotein/ABCB1 (ATP binding cassette subfamily B member 1) involved in drug efflux [111].
AT13387 (onalespib)
AT13387 is a long-acting inhibitor that interacts with the N-terminal ATPase catalytic site of HSP90. AT13387 exerted high activity against cancer cells addicted to several oncoproteins including receptor tyrosine kinases such as EGFR, ERBB2, c-MET (hepatocyte growth factor receptor) and FLT3 (Fms-related tyrosine kinase 3) [112]. AT13387 also depleted HSP90 client proteins including CRAF, BRAFV600E and AKT in a concentration-dependent manner [112, 113] leading to attenuation of MAPK and AKT signaling pathways, also in a three-dimensional model of melanoma [113]. Notably, molecular effects of AT13387 activity were still visible 48 h after drug wash out [112]. AT13387 at low nanomolar concentration efficiently inhibited melanoma cell proliferation compared with other types of cancer cells [112], induced apoptosis and delayed tumor growth when used either alone or in combination with vemurafenib [113]. Notably, AT13387 delayed the emergence of resistance to vemurafenib in vitro and in vivo. In addition, melanoma cells resistant to vemurafenib or resistant to a combination of BRAF and MEK inhibitors were sensitive to AT13387 [113]. Similarly to other N-terminal HSP90 inhibitors, AT13387 induced expression of chaperones, including HSP70 [112, 113]. In a phase I study, AT13387 was tolerable in patients with advanced solid tumors and had acceptable safety profile. Cardiotoxicity and unfavorable hepatotoxicity observed for ansamycin HSP90 inhibitors were not observed in this study [114]. Importantly, AT13387 was shown to cross the blood–brain barrier [115]. AT13387 is currently evaluated in combination with dabrafenib and trametinib in a phase I clinical trial (Table 1).
Table 1.
Active clinical trials evaluating the efficacy of HSP90 inhibitors in patients either with melanoma or other malignancies. HSP90 inhibitors that have shown anti-melanoma activity in preclinical studies were included. Data were extracted from https://clinicaltrials.gov
| HSP90 inhibitor | Additional drugs | Major inclusion criteria | Phase | Identifier |
|---|---|---|---|---|
| On-going clinical trials evaluating the efficacy of HSP90 inhibitors in melanoma patients | ||||
| XL888 | Vemurafenib | BRAFV600E/K mutation; AJCC stage IIIB, IIIC, IV; unresectable | 1 | NCT01657591 |
| XL888 | Vemurafenib + cobimetinib | BRAFV600 mutation; unresectable AJCC stage IV, IIIB or IIIC | 1 | NCT02721459 |
| AT13387 | Dabrafenib + trametinib | BRAFV600E/K mutation; metastatic or unresectable | 1 | NCT02097225 |
| Other active clinical trials on HSP90 inhibitors with known anti-melanoma properties | ||||
| AT13387 | – | different lymphomas | 2 | NCT02572453 |
| AT13387 | Paclitaxel | Breast cancer triple-negative breast carcinoma | 1 | NCT02474173 |
| AT13387 | AT7519 M | Solid tumors | 1 | NCT02503709 |
| AT13387 | Olaparib | Unresectable solid tumors fallopian tube/ovarian carcinoma triple-negative breast carcinoma | 1 | NCT02898207 |
| AT13387 | Cisplatin | Squamous cell carcinoma | 1 | NCT02381535 |
| AT13387 | Erlotinib | Lung non-small cell carcinoma | 1/2 | NCT02535338 |
| ganetespib | Crizotinib | Lung cancer | 1 | NCT01579994 |
| ganetespib | – | Lung cancer | 1/2 | NCT01590160 |
| ganetespib | Niraparib carboplatin | Fallopian tube/ovarian carcinoma primary peritoneal carcinoma | 2 | NCT03783949 |
| ganetespib | – | Breast cancer | 2 | NCT01042379 |
| NVP-AUY922 | – | Gastrointestinal stromal tumor | 2 | NCT01389583 |
| NVP-AUY922 | Alpelisib or capmatinib or ceritinib or binimetinib | Adenocarcinoma lung cancer squamous cell lung carcinoma | 2 | NCT02276027 |
| PU-H71 | – | Non-Hodgkin’s lymphoma myeloma solid malignancy | 1 | NCT01269593 |
| PU-H71 | Ruxolitinib | Myelofibrosis | 1 | NCT03373877 |
| PU-H71 | – | Metastatic solid tumor lymphoma | 1 | NCT01393509 |
| PU-H71 | Nab-paclitaxel | Metastatic breast cancer | 1 | NCT03166085 |
| PU-H71 | – | Myelofibrosis | 1 | NCT03935555 |
| XL888 | Pembrolizumab | Colorectal and pancreatic cancer | 1 | NCT03095781 |
NVP-AUY922 (luminespib)
NVP-AUY922 is a resorcinol isoxazole amide compound with a high affinity to HSP90 [116]. NVP-AUY922 affected proliferation and inhibited colony-forming capacity of melanoma cells. NVP-AUY922 decreased protein level of cyclin D1, and decreased activity of ERK1/2 and NF-κB signaling pathway [117]. In addition to inducing apoptosis, NVP-AUY922 elevated LC3II/LC3I (microtubule-associated proteins 1A/1B light chain 3B) ratio in a time-dependent manner indicating activation of autophagy [117]. Conflicting results of NVP-AUY922 activity on melanoma tumor growth and metastasis were published [116, 117]. NVP-AUY922 induced expression of HSP70, GRP78 and DDIT3 (DNA damage-inducible transcript 3) encoding CHOP, thereby increasing endoplasmic reticulum stress and activating unfolded protein response in melanoma cells [117]. Co-treatment with PFT-μ (2-phenylethynesulphonamide), which acted as a dual inhibitor of HSP70 and autophagy, showed a synergistic anti-melanoma activity both in vitro and in vivo probably by deregulating redox balance [117].
Ganetespib
Ganetespib and its prodrug, STA-1474, are water soluble compounds that bind to N-terminal domain of HSP90, and exert anti-cancer activity [118–120]. Ganetespib destabilized MAPK signaling in melanoma cells by diminution of HSP90 client proteins including CRAF and BRAFV600E leading to attenuation of MEK1/2 and ERK1/2 activity in a concentration-dependent manner, while not affecting BRAF protein level in wild-type BRAF melanoma cells and melanocytes [121, 122]. In addition, ganetespib reduced expression of EGFR, IGF1R (insulin-like growth factor 1 receptor) and MET, inhibited AKT activity [121, 123], and upregulated HSP70 [123]. Ganetespib induced melanoma cell cycle arrest in G2, G1 and G2/M phases in a cell line-dependent manner [121]. This was associated with reduction of CDK1 (cyclin-dependent kinase 1) [121, 123], CDK2 (cyclin-dependent kinase 2) and CDK4 expression, and diverse alterations in protein levels of p27Kip1 (cyclin-dependent kinase inhibitor 1B), p21Cip1 (CDK-interacting protein 1) and cyclins [121]. In addition to cytostatic effect, ganetespib activated caspase-3 and caspase-7 leading to apoptosis in melanoma cells [121, 122]. Ganetespib-induced apoptosis was associated with decrease in protein levels of anti-apoptotic proteins including survivin, BCL-2, BCL-XL (B-cell lymphoma-extra-large) and MCL-1 (myeloid cell leukemia 1), although elevated levels of these proteins were reported in certain cell lines exposed to ganetespib [121]. Ganetespib inhibited tumor growth in mice xenografts [122], significantly potentiated the tumor growth inhibitory effect of BRAF and MEK inhibitors, and overcame mechanisms of primary and acquired resistance of melanoma cells to BRAF inhibitors [121, 122]. Ganetespib exerted similar activity in melanoma cells of different genetic subtypes including those harboring both BRAF and NRAS as wild-types [121]. More recently, ganetespib was demonstrated to potentiate anti-tumor effect of immunotherapy as it sensitized melanoma cells to T-cell-mediated killing by upregulating interferon response genes, IFIT1, IFIT2, IFIT3 (interferon-induced protein with tetratricopeptide repeats 1–3) in vitro and in vivo [124]. In a clinical trial on patients with metastatic uveal melanoma stage IV, progression-free survival was 1.6 months and 1.8 months in patient cohorts that received 200 mg of ganetespib weekly and 150 mg of the drug twice a week, respectively [125]. However, ganetespib was poorly tolerated in uveal melanoma patients as it evoked a significant gastrointestinal toxicity, including increased aspartate aminotransferase and alanine aminotransferase activity, nausea, vomiting and diarrhea [125].
Purine-based inhibitor
PU-H71 is a purine-scaffold inhibitor of HSP90 that exerts a broader accessibility to a greater number of undimerized HSP90 conformations than geldanamycin, and PU-H71 activity is less affected by phosphorylation of HSP90 [126]. PU-H71 exerted higher selectivity in targeting HSP90-oncoprotein complexes than several other N-terminal inhibitors of HSP90 [127]. PU-H71 was shown to induce ER stress and activate UPR pathway involving upregulation of DDIT3 expression. This was followed by loss of mitochondrial membrane potential and activation of caspase-3, culminating in induction of apoptosis in melanoma cells [128]. Interestingly, these effects were also reported in cancer cell lines of different origin, but not in normal fibroblasts and tissues [128, 129]. Selectivity towards cancer cells was also observed for PU-H71-dependent radiosensitization [130]. More recently, a first-in-human study revealed that PU-H71 was well tolerated in patients with refractory solid tumors, and exerted no dose-limiting toxicity with predominantly grade 1 adverse effects [131].
Non-resorcinol pyrazole inhibitors
SNX-2112 is an N-terminal domain binding HSP90 inhibitor containing the 2-aminobenzamide scaffold [132]. SNX-2112 more potently inhibited melanoma cell proliferation than 17-AAG [133, 134], and arrested cells in G0/G1 phase of cell cycle in a dose-dependent manner [133]. SNX-2112 induced a time-dependent degradation of HSP90 client proteins crucial for melanoma cell maintenance including AKT, IKKα, BRAF and glycogen synthase kinase 3 beta (GSK-3β) [133]. SNX-2112 also induced apoptosis in melanoma cells [132, 134], which was associated with an activation of caspase-3, caspase-7 and caspase-8, and PARP cleavage. SNX-2112 induced a time-dependent release of cytochrome c from mitochondria and upregulated pro-apoptotic protein BIM, simultaneously leading to down-regulation of BCL-2, BCL-XL and XIAP (X-linked inhibitor of apoptosis protein). In addition, SNX-2112 inhibited AKT/mTOR/p70S6K (ribosomal protein S6 kinase) pathway to induce autophagy [134]. Two other SNX-2112-related agents were tested in melanoma. SNX-5422, a SNX-2112 prodrug, is rapidly metabolized to SNX-2112 by enzymatic hydrolysis, and exerted promising activity in phase I trials involving patients with solid tumors including melanoma [135, 136]. Most adverse effects were grade 1 or 2 in severity, although few events of grade 3 such as diarrhea, non-septic arthritis and thrombocytopenia were also reported [135, 136]. In SNX-7081, another SNX-2112 derivative, a side chain indazole was replaced with indole. SNX-7081 inhibited cancer cell proliferation more efficiently than its parent drug. Both compounds were highly selective towards cancer cells [132].
NVP-BEP800
NVP-BEP800 is a fully synthetic N-terminal inhibitor of HSP90 exerting activity against a number of cancer cell lines at nanomolar concentrations. NVP-BEP800 induced degradation of several melanoma oncoproteins including ERBB2, CRAF, BRAFV600E and AKT [137]. NVP-BEP800 exhibited good bioavailability after oral administration. Pharmacokinetic analysis revealed its short half-life of less than 2 h in plasma, and selective retention in tumor cells with half-life of more than 16 h. Notably, no hepatotoxicity was observed in the preclinical studies [137].
XL888
XL888 is an orally bioavailable inhibitor of HSP90 exerting selectivity for this chaperone protein over almost 30 kinases [138]. A high-throughput analysis of XL888-mediated perturbations in cell cycle distribution revealed that XL888 activity might depend on the mutation status of driver oncogenes. XL888 induced G2/M phase accumulation of melanoma cells harboring unmutated BRAF, RAS and EGFR, and homozygous P72R variant of p53 [139]. In turn, the presence of a homozygous BRAFV600E variant was predominantly associated with accumulation of XL888-treated melanoma cells in G1 phase of cell cycle [139]. Cytostatic effect of XL888 activity was accompanied with diminution of cell cycle-related protein levels including WEE1 (WEE1 G2 checkpoint kinase), CHK1 (checkpoint kinase 1), CDK1 and CDK4 [138]. In addition, XL888 diminished protein levels of ARAF (ARAF proto-oncogene, serine/threonine kinase) and CRAF leading to attenuation of ERK1/2 activity, and decreased activity of AKT and S6 kinases [138]. XL888 upregulated BIM and BAX (BCL-2 associated X protein) expression while decreasing protein level of MCL-1 that resulted in apoptosis induction in NRAS-mutant melanoma cells. MCL1 overexpression, however, prevented from XL888-induced apoptosis [138]. Notably, XL888 efficiently exerted similar effects in three-dimensional spheroid cultures, and in a mouse xenograft model of melanoma with milder effect on the activity of MAPK signaling pathway but retaining its inhibitory potential on CDK4 and WEE1 expression, and activity of AKT and S6 [138]. In a panel of NRAS-mutated melanoma cell lines, XL888 caused degradation of IGF-1Rβ, PDGFR-β, c-MET and VEGFR1 (vascular endothelial growth factor receptor (1), although it surprisingly increased VEGFR2 (vascular endothelial growth factor receptor (2) level in one cell line [140]. XL888 inhibited cell proliferation also in melanoma cells with intrinsic and acquired resistance to BRAF inhibitors that were dependent on different mechanisms including either overexpression of cyclin D1, PDGFR-β or COT, or NRAS mutation [141]. XL888 efficiently reduced levels of ARAF, CRAF and cyclin D1, and inhibited AKT, ERK1/2 and S6 activity in resistant melanoma cells. XL888 restored nuclear localization of FOXO3a (forkhead box O3) that was followed by upregulation of BIM expression and diminution of MCL-1 level in vemurafenib-resistant cell lines leading to cleavage of caspase-3 and loss of mitochondrial membrane potential [141]. Interestingly, XL888 arrested vemurafenib-sensitive melanoma cells in G1 phase of cell cycle, but G2/M cell accumulation was predominantly reported in matched vemurafenib-resistant cells exposed to XL888 [141]. XL888 decreased number of non-melanoma skin lesions developed as a result of paradoxical MAPK pathway activation in patients treated with vemurafenib. Similarly, XL888 suppressed ERK1/2 activity in NRAS-mutant melanoma cell lines exposed to vemurafenib, and this effect was associated with down-regulation of CRAF [142]. In addition, XL888-mediated inhibition of HSP90 was accompanied with induction of a compensatory mechanism involving HSP70 upregulation as shown in both in vitro and in vivo models of melanoma. HSP70 induction was similarly observed in drug-naïve and resistant melanoma cells [138, 140, 141]. In a clinical study, XL888 in combination with vemurafenib displayed a tolerable side-effect profile and promising activity in melanoma patients with BRAFV600E-mutant tumors. Objective responses were reported in 15/20 patients, including 3 complete and 12 partial responses. The most common adverse effects of grade 3 and 4 were rash and diarrhea, and cutaneous squamous cell carcinomas was developed in 14% of patients [143]. XL888 resistance mechanism involving CDK2 was identified in melanoma cells, and CDK2 expression was dependent on MITF (microphthalmia-associated transcription factor) [144]. XL888 is currently evaluated in phase I clinical trials (Table 1).
Middle domain inhibitors
Sansalvamide A and sansalvamide A derivatives
Sansalvamide A (San A) is a cyclic pentapeptide isolated from the marine fungi Fusarium sp. [80, 145]. Sansalvamide A binds to N-terminal fragment of the middle domain of HSP90, and exerts the ability to allosterically disrupt the interactions of C-terminal binding co-chaperones and client proteins [80]. Interestingly, Di-Sansalvamide A (Di-San A), a dimerized derivative of San A, was found to bind C-middle domain of HSP90 suggesting that Di-San A physically prevents from binding of C-terminal binding clients [80]. Three San A-derived compounds, H-10, H-15 and LY-15 were investigated as potential HSP90 inhibitors in melanoma cells. These agents inhibited proliferation in melanoma cell lines in a concentration- and time-dependent manner [146–148]. Additionally, LY-15 and H-10 induced mitochondrial pathway of apoptosis associated with activation of caspase-3 and caspase-9, but not caspase-8 [147, 148]. LY-15 increased BAX and diminished BCL-2 protein levels, and inhibited cell migration [148]. H-15 increased melanin production and upregulated TYR (tyrosinase) expression suggesting that H-15 was capable of inducing differentiation in melanoma cells [146].
C-Terminal domain inhibitors
Novobiocin and novobiocin derivative
Novobiocin is an aminocoumarin antibiotic that showed a dose-dependent anti-proliferative effect in melanoma cells, and increased activity of NADPH: cytochrome c reductase and γ-glutamyltranspeptidase [149, 150]. C-terminal domain of HSP90 was identified as a novobiocin-HSP90 interaction surface [151, 152]. Novobiocin induced degradation of HSP90 client proteins including ERBB2, CRAF, mutated p53 and SRC (proto-oncogene tyrosine-protein kinase SRC), although only when used at relatively high concentrations [152]. To improve activity, novobiocin analogues were synthesized with several structural modifications. In one of them, KU135, the coumarin core was modified, and the noviose sugar was replaced with a methylated phenol that can participate in hydrogen bonding [153]. KU135 arrested melanoma cells in the G2/M phase of cell cycle by increasing the phosphorylation of CDC25C (cell division cycle 25C) at Ser216 and diminishing cyclin B level in contrast to novobiocin that did not influence the level of both proteins [154]. Moreover, KU135 reduced melanoma cell viability more potently than novobiocin and N-terminal inhibitor, 17-AAG. KU135-induced apoptosis was associated with dissipation of mitochondrial membrane potential that led to the release of cytochrome c, activation of caspase-8, caspase-9 and caspase-3, and PARP cleavage [154]. Interestingly, KU135 induced AKT phosphorylation after short incubation, but AKT activity was attenuated after 48 h. In addition, KU135 inhibited ERK1/2 activity that might be related to the reduction of HSP90 client proteins, BRAF and CRAF [154]. Unlike N-terminal inhibitors, KU135 did not affect HSP27 (heat shock protein 27), HSP70 and GRP94 expression, and activity of HSF-1 [154].
Conclusions
Cancer cells are under constant stress due to the presence of mutant proteins and rapid cell proliferation that affects the control of proteostasis, and in turn elevates cell dependence on HSP90. HSP90 is a promising therapeutic target in melanoma as HSP90 clients have been identified among melanoma-associated oncoproteins involved in determination of cell phenotype and response to drugs. Several HSP90 inhibitors exerting anti-melanoma activity in pre-clinical in vitro and in vivo studies are currently evaluated in clinical trials (Table 1). Further research is, however, necessary to more precisely define unique isoforms or conformational preferences for particular HSP90 inhibitors. Limited reports addressing this issue suggest that substantial differences in both client and drug preferences can exist for HSP90α and HSP90β isoforms, and indicate that geldanamycin and ganetespib bind to HSP90β with greater affinity than to HSP90α [155]. Recent advance showing that activity of HSP90 inhibitors can be monitored by using [11C]NMS-E973 as a PET tracer to both quantify HSP90 level in vivo and to determine HSP90 occupancy after treatment with HSP90 inhibitors [123] has provided a tool for in-depth discoveries.
Acknowledgements
This study was funded by National Science Centre, Grant No. 2014/15/B/NZ7/00947.
Abbreviations
- 17-AAG
17-N-Allylamino-17-demethoxygeldanamycin (tanespimycin)
- 17-AG
17-Aminogeldanamycin (IPI-493)
- 17-DMAG
17-Dimethylaminoethylamino-17-demethoxygeldanamycin (alvespimycin)
- BAX
BCL-2 associated X protein
- BCL-2
B-Cell CLL/lymphoma 2
- BCL-XL
B-Cell lymphoma-extra large
- BIM
BCL-2-interacting mediator of cell death
- BRAF
B-Raf proto-oncogene serine/threonine kinase
- c-MET
Hepatocyte growth factor receptor
- CDC37
Cell division cycle 37
- CDK1/2/4
Cyclin-dependent kinase 1/2/4
- COT
Cancer Osaka thyroid oncogene
- CRAF
RAF-1 proto-oncogene, serine/threonine kinase
- DDIT3
DNA damage-inducible transcript 3
- EGFR
Epidermal growth factor receptor
- EIF2α
Eukaryotic initiation factor 2
- ER
Endoplasmic reticulum
- ERK1/2
Extracellular signal-regulated kinase 1/2
- GRP78/BiP
Glucose-regulated protein 78/binding immunoglobulin protein
- GRP94
Glucose-regulated protein 94
- HIF-1α
Hypoxia-inducible factor 1
- HSF-1
Heat shock factor 1
- HSP70/90
Heat shock protein 70/90
- IRE1α
Inositol-requiring enzyme 1 alpha
- MCL-1
Myeloid cell leukemia-1
- MEK1/2
Mitogen-activated protein kinase 1/2
- mTOR
Mechanistic target of rapamycin kinase
- NF-κB
Nuclear factor-kappa B
- NRAS
Neuroblastoma RAS viral oncogene homolog
- NQO1
NAD(P)H:quinone oxidoreductase 1
- PARP1
Poly(ADP-ribose) polymerase 1
- PD-1
Programmed cell death protein-1
- PI3 K
Phosphatidylinositol 3-kinase
- UPR
Unfolded protein response
- WNT
Wingless-type
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Aleksandra Mielczarek-Lewandowska and Mariusz L. Hartman have contributed equally to this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Rastrelli M, Tropea S, Rossi CR, Alaibac M. Melanoma: epidemiology, risk factors, pathogenesis, diagnosis and classification. Vivo. 2014;28:1005–1011. [PubMed] [Google Scholar]
- 3.Vogan K. Pigmentation and skin-cancer risk. Nat Rev Genet. 2008;9:502. doi: 10.1038/nrg2409. [DOI] [Google Scholar]
- 4.Schadendorf D, Fisher DE, Garbe C, Gershenwald JE, Grob JJ, Halpern A, Herlyn M, Marchetti MA, McArthur G, Ribas A, Roesch A, Hauschild A. Melanoma. Nat Rev Dis Prim. 2015;1:15003. doi: 10.1038/nrdp.2015.3. [DOI] [PubMed] [Google Scholar]
- 5.Cheng L, Lopez-Beltran A, Massari F, MacLennan GT, Montironi R. Molecular testing for BRAF mutations to inform melanoma treatment decisions: a move toward precision medicine. Mod Pathol. 2018;31:24–38. doi: 10.1038/modpathol.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaufman HL, Margolin K, Sullivan R. Management of metastatic melanoma in 2018. JAMA Oncol. 2018;4:857–858. doi: 10.1001/jamaoncol.2018.0170. [DOI] [PubMed] [Google Scholar]
- 7.Domingues B, Lopes JM, Soares P, Pópulo H. Melanoma treatment in review. Immunotargets Ther. 2018;7:35–49. doi: 10.2147/ITT.S134842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seidel JA, Otsuka A, Kabashima K. Anti-PD-1 and Anti-CTLA-4 therapies in cancer: mechanisms of action, efficacy, and limitations. Front Oncol. 2018;8:86. doi: 10.3389/fonc.2018.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savoia P, Fava P, Casoni F, Cremona O. Targeting the ERK signaling pathway in melanoma. Int J Mol Sci. 2019;20:E1483. doi: 10.3390/ijms20061483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan RJ, Flaherty KT. Resistance to BRAF-targeted therapy in melanoma. Eur J Cancer. 2013;49:1297–1304. doi: 10.1016/j.ejca.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Almeida FV, Douglass SM, Fane ME, Weeraratna AT. Bad company: microenvironmentally mediated resistance to targeted therapy in melanoma. Pigment Cell Melanoma Res. 2019;32:237–247. doi: 10.1111/pcmr.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozar I, Margue C, Rothengatter S, Haan C, Kreis S. Many ways to resistance: how melanoma cells evade targeted therapies. Biochim Biophys Acta Rev Cancer. 2019;1871:313–322. doi: 10.1016/j.bbcan.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Czyz M, Sztiller-Sikorska M, Gajos-Michniewicz A, Osrodek M, Hartman ML. Plasticity of drug-naïve and vemurafenib- or trametinib-resistant melanoma cells in execution of differentiation/pigmentation program. J Oncol. 2019;2019:1697913. doi: 10.1155/2019/1697913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gajos-Michniewicz A, Czyz M. Role of miRNAs in melanoma metastasis. Cancers. 2019;11:E326. doi: 10.3390/cancers11030326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres-Collado AX, Knott J, Jazirehi AR. Reversal of resistance in targeted therapy of metastatic melanoma: lessons learned from vemurafenib (BRAF(V600E)-specific inhibitor) Cancers (Basel) 2018;10:E157. doi: 10.3390/cancers10060157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen CH, Kim SH, Trousil S, Frederick DT, Piris A, Yuan P, Cai L, Gu L, Li M, Lee JH, Mitra D, Fisher DE, Sullivan RJ, Flaherty KT, Zheng B. Loss of cohesin complex components STAG2 or STAG3 confers resistance to BRAF inhibition in melanoma. Nat Med. 2016;22:1056–1061. doi: 10.1038/nm.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erdmann S, Seidel D, Jahnke HG, Eichler M, Simon JC, Robitzki AA. Induced cross-resistance of BRAF(V600E) melanoma cells to standard chemotherapeutic dacarbazine after chronic PLX4032 treatment. Sci Rep. 2019;9:30. doi: 10.1038/s41598-018-37188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao J, Ward JF, Pettaway CA, Shi LZ, Subudhi SK, Vence LM, Zhao H, Chen J, Chen H, Efstathiou E, Troncoso P, Allison JP, Logothetis CJ, Wistuba II, Sepulveda MA, Sun J, Wargo J, Blando J, Sharma P. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat Med. 2017;23:551–555. doi: 10.1038/nm.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, Saco J, Homet Moreno B, Mezzadra R, Chmielowski B, Ruchalski K, Shintaku IP, Sanchez PJ, Puig-Saus C, Cherry G, Seja E, Kong X, Pang J, Berent-Maoz B, Comin-Anduix B, Graeber TG, Tumeh PC, Schumacher TN, Lo RS, Ribas A. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dezwaan DC, Freeman BC. HSP90: the Rosetta stone for cellular protein dynamics? Cell Cycle. 2008;7:1006–1012. doi: 10.4161/cc.7.8.5723. [DOI] [PubMed] [Google Scholar]
- 21.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 22.Hoter A, El-Sabban ME, Naim HY. The HSP90 family: structure, regulation, function, and implications in health and disease. Int J Mol Sci. 2018;19:E2560. doi: 10.3390/ijms19092560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen B, Zhong D, Monteiro A. Comparative genomics and evolution of the HSP90 family of genes across all kingdoms of organisms. BMC Genom. 2006;7:156. doi: 10.1186/1471-2164-7-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsutsumi S, Mollapour M, Prodromou C, Lee CT, Panaretou B, Yoshida S, Mayer MP, Neckers LM. Charged linker sequence modulates eukaryotic heat shock protein 90 (Hsp90) chaperone activity. Proc Natl Acad Sci USA. 2012;109:2937–2942. doi: 10.1073/pnas.1114414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verma S, Goyal S, Jamal S, Singh A, Grover A. Hsp90: friends, clients and natural foes. Biochimie. 2016 doi: 10.1016/j.biochi.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 26.Genest O, Wickner S, Doyle SM. Hsp90 and Hsp70 chaperones: collaborators in protein remodeling. J Biol Chem. 2019;294:2109–2120. doi: 10.1074/jbc.REV118.002806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutz DA, Luo Q, Freiburger L, Madl T, Ville KRI, Sattler M, Buchner J. A switch point in the molecular chaperone Hsp90 responding to client interaction. Nat Commun. 2018;9:1472. doi: 10.1038/s41467-018-03946-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schopf FH, Biebl MM, Buchner J. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol. 2017;18:345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 29.Li R, Albertini DF. The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat Rev Mol Cell Biol. 2013;14:141–152. doi: 10.1038/nrm3531. [DOI] [PubMed] [Google Scholar]
- 30.Sima S, Richter K. Regulation of the Hsp90 system. Biochim Biophys Acta Mol Cell Res. 2018;1865:889–897. doi: 10.1016/j.bbamcr.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 31.Ortner V, Ludwig A, Riegel E, Dunzinger S, Czerny T. An artificial HSE promoter for efficient and selective detection of heat shock pathway activity. Cell Stress Chaperon. 2015;20:277–288. doi: 10.1007/s12192-014-0540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prodromou C. Mechanisms of Hsp90 regulation. Biochem J. 2016;473:2439–2452. doi: 10.1042/BCJ20160005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mollapour M, Neckers L. Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochim Biophys Acta. 2012;1823:648–655. doi: 10.1016/j.bbamcr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sager RA, Woodford MR, Neckers L, Mollapour M. Detecting posttranslational modifications of Hsp90. Methods Mol Biol. 2018;1709:209–219. doi: 10.1007/978-1-4939-7477-1_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imai Y, Hirano M, Kobayashi M, Futami M, Tojo A. HDAC inhibitors exert anti-myeloma effects through multiple modes of action. Cancers. 2019;11:E475. doi: 10.3390/cancers11040475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarthy MM, Pick E, Kluger Y, Gould-Rothberg B, Lazova R, Camp RL, Rimm DL, Kluger HM. HSP90 as a marker of progression in melanoma. Ann Oncol. 2008;19:590–594. doi: 10.1093/annonc/mdm545. [DOI] [PubMed] [Google Scholar]
- 37.Shipp C, Weide B, Derhovanessian E, Pawelec G. Hsps are up-regulated in melanoma tissue and correlate with patient clinical parameters. Cell Stress Chaperon. 2013;18:145–154. doi: 10.1007/s12192-012-0363-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strickler AG, Vasquez JG, Yates N, Ho J. Potential diagnostic significance of HSP90, ACS/TMS1, and L-plastin in the identification of melanoma. Melanoma Res. 2014;24:535–544. doi: 10.1097/CMR.0000000000000115. [DOI] [PubMed] [Google Scholar]
- 39.Becker B, Multhoff G, Farkas B, Wild PJ, Landthaler M, Stolz W, Vogt T. Induction of Hsp90 protein expression in malignant melanomas and melanoma metastases. Exp Dermatol. 2004;13:27–32. doi: 10.1111/j.0906-6705.2004.00114.x. [DOI] [PubMed] [Google Scholar]
- 40.Girola N, Matsuo AL, Figueiredo CR, Massaoka MH, Farias CF, Arruda DC, Azevedo RA, Monteiro HP, Resende-Lara PT, Cunha RL, Polonelli L, Travassos LR. The Ig V(H) complementarity-determining region 3-containing Rb9 peptide, inhibits melanoma cells migration and invasion by interactions with Hsp90 and an adhesion G-protein coupled receptor. Peptides. 2016;85:1–15. doi: 10.1016/j.peptides.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 41.Zhu H, Fang X, Zhang D, Wu W, Shao M, Wang L, Gu J. Membrane-bound heat shock proteins facilitate the uptake of dying cells and cross-presentation of cellular antigen. Apoptosis. 2016;21:96–109. doi: 10.1007/s10495-015-1187-0. [DOI] [PubMed] [Google Scholar]
- 42.Eustace BK, Sakurai T, Stewart JK, Yimlamai D, Unger C, Zehetmeier C, Lain B, Torella C, Henning SW, Beste G, Scroggins BT, Neckers L, Ilag LL, Jay DG. Functional proteomic screens reveal an essential extracellular role for hsp90 alpha in cancer cell invasiveness. Nat Cell Biol. 2004;6:507–514. doi: 10.1038/ncb1131. [DOI] [PubMed] [Google Scholar]
- 43.Tas F, Bilgin E, Erturk K, Duranyildiz D. Clinical significance of circulating serum cellular Heat Shock Protein90 (HSP90) level in patients with cutaneous malignant melanoma. Asian Pac J Cancer Prev. 2017;18:599–601. doi: 10.22034/APJCP.2017.18.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hance MW, Nolan KD, Isaacs JS. The double-edged sword: conserved functions of extracellular hsp90 in wound healing and cancer. Cancers. 2014;6:1065–1097. doi: 10.3390/cancers6021065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, García-Santos G, Ghajar C, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trisciuoglio D, Gabellini C, Desideri M, Ziparo E, Zupi G, Del Bufalo D. Bcl-2 regulates HIF-1alpha protein stabilization in hypoxic melanoma cells via the molecular chaperone HSP90. PLoS ONE. 2010;5:e11772. doi: 10.1371/journal.pone.0011772. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Cheng Y, Zhang G, Li G. Targeting MAPK pathway in melanoma therapy. Cancer Metastasis Rev. 2013;32:567–584. doi: 10.1007/s10555-013-9433-9. [DOI] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas Network Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, Patch AM, Kakavand H, Alexandrov LB, Burke H, Jakrot V, Kazakoff S, Holmes O, Leonard C, Sabarinathan R, Mularoni L, Wood S, Xu Q, Waddell N, Tembe V, Pupo GM, De Paoli-Iseppi R, Vilain RE, Shang P, Lau LMS, Dagg RA, Schramm SJ, Pritchard A, Dutton-Regester K, Newell F, Fitzgerald A, Shang CA, Grimmond SM, Pickett HA, Yang JY, Stretch JR, Behren A, Kefford RF, Hersey P, Long GV, Cebon J, Shackleton M, Spillane AJ, Saw RPM, López-Bigas N, Pearson JV, Thompson JF, Scolyer RA, Mann GJ. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 50.Hartman ML, Sztiller-Sikorska M, Czyz M. Whole-exome sequencing reveals novel genetic variants associated with diverse phenotypes of melanoma cells. Mol Carcinog. 2019;58:588–602. doi: 10.1002/mc.22953. [DOI] [PubMed] [Google Scholar]
- 51.Da Rocha Dias S, Friedlos F, Light Y, Springer C, Workman P, Marais R. Activated B-RAF is an Hsp90 client protein that is targeted by the anticancer drug 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2005;65:10686–10691. doi: 10.1158/0008-5472.CAN-05-2632. [DOI] [PubMed] [Google Scholar]
- 52.Grbovic OM, Basso AD, Sawai A, Ye Q, Friedlander P, Solit D, Rosen N. V600E B-Raf requires the Hsp90 chaperone for stability and is degraded in response to Hsp90 inhibitors. Proc Natl Acad Sci USA. 2006;103:57–62. doi: 10.1073/pnas.0609973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corazzari M, Rapino F, Ciccosanti F, Giglio P, Antonioli M, Conti B, Fimia GM, Lovat PE, Piacentini M. Oncogenic BRAF induces chronic ER stress condition resulting in increased basal autophagy and apoptotic resistance of cutaneous melanoma. Cell Death Differ. 2015;22:946–958. doi: 10.1038/cdd.2014.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Croft A, Tay KH, Boyd SC, Guo ST, Jiang CC, Lai F, Tseng HY, Jin L, Rizos H, Hersey P, Zhang XD. Oncogenic activation of MEK/ERK primes melanoma cells for adaptation to endoplasmic reticulum stress. J Invest Dermatol. 2014;134:488–497. doi: 10.1038/jid.2013.325. [DOI] [PubMed] [Google Scholar]
- 55.Tay KH, Luan Q, Croft A, Jiang CC, Jin L, Zhang XD, Tseng HY. Sustained IRE1 and ATF6 signaling is important for survival of melanoma cells undergoing ER stress. Cell Signal. 2014;26:287–294. doi: 10.1016/j.cellsig.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 56.Sykes EK, Mactier S, Christopherson RI. Melanoma and the unfolded protein response. Cancers (Basel) 2016;8:E30. doi: 10.3390/cancers8030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davies MA. The role of the PI3 K-AKT pathway in melanoma. Cancer J. 2012;18:142–147. doi: 10.1097/PPO.0b013e31824d448c. [DOI] [PubMed] [Google Scholar]
- 58.Shannan B, Chen Q, Watters A, Perego M, Krepler C, Thombre R, Li L, Rajan G, Peterson S, Gimotty PA, Wilson M, Nathanson KL, Gangadhar TC, Schuchter LM, Weeraratna AT, Herlyn M, Vultur A. Enhancing the evaluation of PI3 K inhibitors through 3D melanoma models. Pigment Cell Melanoma Res. 2016;29:317–328. doi: 10.1111/pcmr.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Irvine M, Stewart A, Pedersen B, Boyd S, Kefford R, Rizos H. Oncogenic PI3 K/AKT promotes the step-wise evolution of combination BRAF/MEK inhibitor resistance in melanoma. Oncogenesis. 2018;7:72. doi: 10.1038/s41389-018-0081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conn CS, Qian SB. mTOR signaling in protein homeostasis: less is more? Cell Cycle. 2011;10:1940–1947. doi: 10.4161/cc.10.12.15858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Basso AD, Solit DB, Chiosis G, Giri B, Tsichlis P, Rosen N. Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J Biol Chem. 2002;277:39858–39866. doi: 10.1074/jbc.M206322200. [DOI] [PubMed] [Google Scholar]
- 62.Broemer M, Krappmann D, Scheidereit C. Requirement of Hsp90 activity for IkappaB kinase (IKK) biosynthesis and for constitutive and inducible IKK and NF-kappaB activation. Oncogene. 2004;23:5378–5386. doi: 10.1038/sj.onc.1207705. [DOI] [PubMed] [Google Scholar]
- 63.Madonna G, Ullman CD, Gentilcore G, Palmieri G, Ascierto PA. NF-κB as potential target in the treatment of melanoma. J Transl Med. 2012;10:53. doi: 10.1186/1479-5876-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith MP, Sanchez-Laorden B, O’Brien K, Brunton H, Ferguson J, Young H, Dhomen N, Flaherty KT, Frederick DT, Cooper ZA, Wargo JA, Marais R, Wellbrock C. The immune microenvironment confers resistance to MAPK pathway inhibitors through macrophage-derived TNFα. Cancer Discov. 2014;4:1214–1229. doi: 10.1158/2159-8290.CD-13-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shao Y, Le K, Cheng H, Aplin AE. NF-κB regulation of c-FLIP promotes TNFα-mediated RAF inhibitor resistance in melanoma. J Invest Dermatol. 2015;135:1839–1848. doi: 10.1038/jid.2015.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lehraiki A, Cerezo M, Rouaud F, Abbe P, Allegra M, Kluza J, Marchetti P, Imbert V, Cheli Y, Bertolotto C, Ballotti R, Rocchi S. Increased CD271 expression by the NF-kB pathway promotes melanoma cell survival and drives acquired resistance to BRAF inhibitor vemurafenib. Cell Discov. 2015;1:15030. doi: 10.1038/celldisc.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xue G, Romano E, Massi D, Mandalà M. Wnt/β-catenin signaling in melanoma: preclinical rationale and novel therapeutic insights. Cancer Treat Rev. 2016;49:1–12. doi: 10.1016/j.ctrv.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 68.Sinnberg T, Levesque MP, Krochmann J, Cheng PF, Ikenberg K, Meraz-Torres F, Niessner H, Garbe C, Busch C. Wnt-signaling enhances neural crest migration of melanoma cells and induces an invasive phenotype. Mol Cancer. 2018;17:59. doi: 10.1186/s12943-018-0773-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson BJ, Saab KR, Ma M, Schatton T, Pütz P, Zhan Q, Murphy GG, Gasser M, Waaga-Gasser AM, Frank NY, Frank MH. ABCB5 maintains melanoma-initiating cells through a pro-inflammatory cytokine signaling circuit. Cancer Res. 2014;74:4196–4207. doi: 10.1158/0008-5472.CAN-14-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hartman ML, Talar B, Noman MZ, Gajos-Michniewicz A, Chouaib S, Czyz M. Gene expression profiling identifies microphthalmia-associated transcription factor (MITF) and Dickkopf-1 (DKK1) as regulators of microenvironment-driven alterations in melanoma phenotype. PLoS ONE. 2014;9:e95157. doi: 10.1371/journal.pone.0095157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu Z, Gholami AM, Kuster B. Systematic identification of the HSP90 candidate regulated proteome. Mol Cell Proteomics. 2012;11(M111):016675. doi: 10.1074/mcp.M111.016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Muller J, Krijgsman O, Tsoi J, Robert L, Hugo W, Song C, Kong X, Possik PA, Cornelissen-Steijger PD, Geukes Foppen MH, Kemper K, Goding CR, McDermott U, Blank C, Haanen J, Graeber TG, Ribas A, Lo RS, Peeper DS. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun. 2014;5:5712. doi: 10.1038/ncomms6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Revach OY, Sandler O, Samuels Y, Geiger B. Cross-talk between receptor tyrosine kinases AXL and ERBB3 regulates invadopodia formation in melanoma cells. Cancer Res. 2019;79:2634–2648. doi: 10.1158/0008-5472.CAN-18-2316. [DOI] [PubMed] [Google Scholar]
- 74.Kamal A, Thao L, Sensintaffar J, Zhang L, Boehm MF, Fritz LC, Burrows FJ. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 75.Senses KM, Ghasemi M, Akbar MW, Isbilen M, Fallacara AL, Frankenburg S, Schenone S, Lotem M, Botta M, Gure AO. Phenotype-based variation as a biomarker of sensitivity to molecularly targeted therapy in melanoma. Medchemcomm. 2017;8:88–95. doi: 10.1039/C6MD00466K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hsieh CC, Shen CH. The potential of targeting P53 and HSP90 overcoming acquired MAPKi-resistant melanoma. Curr Treat Options Oncol. 2019;20:22. doi: 10.1007/s11864-019-0622-9. [DOI] [PubMed] [Google Scholar]
- 77.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Solárová Z, Mojžiš J, Solár P. Hsp90 inhibitor as a sensitizer of cancer cells to different therapies (review) Int J Oncol. 2015;46:907–926. doi: 10.3892/ijo.2014.2791. [DOI] [PubMed] [Google Scholar]
- 79.Terracciano S, Russo A, Chini MG, Vaccaro MC, Potenza M, Vassallo A, Riccio R, Bifulco G, Bruno I. Discovery of new molecular entities able to strongly interfere with Hsp90 C-terminal domain. Sci Rep. 2018;8:1709. doi: 10.1038/s41598-017-14902-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson VA, Singh EK, Nazarova LA, Alexander LD, McAlpine SR. Macrocyclic inhibitors of hsp90. Curr Top Med Chem. 2010;10:1380–1402. doi: 10.2174/156802610792232088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fukuyo Y, Hunt CR, Horikoshi N. Geldanamycin and its anti-cancer activities. Cancer Lett. 2010;290:24–35. doi: 10.1016/j.canlet.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 82.Supko JG, Hickman RL, Grever MR, Malspeis L. Preclinical pharmacologic evaluation of geldanamycin as an antitumor agent. Cancer Chemother Pharmacol. 1995;36:305–315. doi: 10.1007/BF00689048. [DOI] [PubMed] [Google Scholar]
- 83.Li YP, Chen JJ, Shen JJ, Cui J, Wu LZ, Wang Z, Li ZR. Synthesis and biological evaluation of geldanamycin analogs against human cancer cells. Cancer Chemother Pharmacol. 2015;75:773–782. doi: 10.1007/s00280-015-2696-9. [DOI] [PubMed] [Google Scholar]
- 84.Zhang H, Sun GZ, Li X, Pan HY, Zhang YS. A new geldanamycin analogue from Streptomyces hygroscopicus. Molecules. 2010;15:1161–1167. doi: 10.3390/molecules15031161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hao H, Naomoto Y, Bao X, Watanabe N, Sakurama K, Noma K, Motoki T, Tomono Y, Fukazawa T, Shirakawa Y, Yamatsuji T, Matsuoka J, Takaoka M. HSP90 and its inhibitors. Oncol Rep. 2010;23:1483–1492. doi: 10.3892/or_00000787. [DOI] [PubMed] [Google Scholar]
- 86.Babchia N, Calipel A, Mouriaux F, Faussat AM, Mascarelli F. 17-AAG and 17-DMAG-induced inhibition of cell proliferation through B-Raf downregulation in WT B-Raf-expressing uveal melanoma cell lines. Invest Ophthalmol Vis Sci. 2008;49:2348–2356. doi: 10.1167/iovs.07-1305. [DOI] [PubMed] [Google Scholar]
- 87.Beloueche-Babari M, Arunan V, Jackson LE, Perusinghe N, Sharp SY, Workman P, Leach MO. Modulation of melanoma cell phospholipid metabolism in response to heat shock protein 90 inhibition. Oncotarget. 2010;1:185–197. doi: 10.18632/oncotarget.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Calero R, Morchon E, Martinez-Argudo I, Serrano R. Synergistic anti-tumor effect of 17AAG with the PI3 K/mTOR inhibitor NVP-BEZ235 on human melanoma. Cancer Lett. 2017;406:1–11. doi: 10.1016/j.canlet.2017.07.021. [DOI] [PubMed] [Google Scholar]
- 89.Mielczarek-Lewandowska A, Sztiller-Sikorska M, Osrodek M, Czyz M, Hartman ML. 17-Aminogeldanamycin selectively diminishes IRE1α-XBP1s pathway activity and cooperatively induces apoptosis with MEK1/2 and BRAF(V600E) inhibitors inmelanoma cells of different genetic subtypes. Apoptosis. 2019;24:596–611. doi: 10.1007/s10495-019-01542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shin MK, Jeong KH, Choi H, Ahn HJ, Lee MH. Heat shock protein 90 inhibitor enhances apoptosis by inhibiting the AKT pathway in thermal-stimulated SK-MEL-2 human melanoma cell line. J Dermatol Sci. 2018;90:357–360. doi: 10.1016/j.jdermsci.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 91.Joshi SS, Jiang S, Unni E, Goding SR, Fan T, Antony PA, Hornyak TJ. 17-AAG inhibits vemurafenib-associated MAP kinase activation and is synergistic with cellular immunotherapy in a murine melanoma model. PLoS ONE. 2018;13:e0191264. doi: 10.1371/journal.pone.0191264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Drysdale MJ, Brough PA, Massey A, Jensen MR, Schoepfer J. Targeting Hsp90 for the treatment of cancer. Curr Opin Drug Discov Dev. 2006;9:483–495. [PubMed] [Google Scholar]
- 93.Sztiller-Sikorska M, Koprowska K, Majchrzak K, Hartman M, Czyz M. Natural compounds’ activity against cancer stem-like or fast-cycling melanoma cells. PLoS ONE. 2014;9:e90783. doi: 10.1371/journal.pone.0090783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang CH, Drechsel DA, Kitson RR, Siegel D, You Q, Backos DS, Ju C, Moody CJ, Ross D. 19-Substituted benzoquinone ansamycin heat shock protein-90 inhibitors: biological activity and decreased off-target toxicity. Mol Pharmacol. 2014;85:849–857. doi: 10.1124/mol.113.090654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ghadban T, Dibbern JL, Reeh M, Miro JT, Tsui TY, Wellner U, Izbicki JR, Güngör C, Vashist YK. HSP90 is a promising target in gemcitabine and 5-fluorouracil resistant pancreatic cancer. Apoptosis. 2017;22:369–380. doi: 10.1007/s10495-016-1332-4. [DOI] [PubMed] [Google Scholar]
- 96.Pacey S, Wilson RH, Walton M, Eatock MM, Hardcastle A, Zetterlund A, Arkenau HT, Moreno-Farre J, Banerji U, Roels B, Peachey H, Aherne W, de Bono JS, Raynaud F, Workman P, Judson I. A phase I study of the heat shock protein 90 inhibitor alvespimycin (17-DMAG) given intravenously to patients with advanced solid tumors. Clin Cancer Res. 2011;17:1561–1570. doi: 10.1158/1078-0432.CCR-10-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kudryavtsev VA, Khokhlova AV, Mosina VA, Selivanova EI, Kabakov AE. Induction of Hsp70 in tumor cells treated with inhibitors of the Hsp90 activity: a predictive marker and promising target for radiosensitization. PLoS ONE. 2017;12:e0173640. doi: 10.1371/journal.pone.0173640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chang YS, Lee LC, Sun FC, Chao CC, Fu HW, Lai YK. Involvement of calcium in the differential induction of heat shock protein 70 by heat shock protein 90 inhibitors, geldanamycin and radicicol, in human non-small cell lung cancer H460 cells. J Cell Biochem. 2006;97:156–165. doi: 10.1002/jcb.20623. [DOI] [PubMed] [Google Scholar]
- 99.Siegel D, Yan C, Ross D. NAD(P)H:quinone oxidoreductase 1 (NQO1) in the sensitivity and resistance to antitumor quinones. Biochem Pharmacol. 2012;83:1033–1040. doi: 10.1016/j.bcp.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Muñoz IG, Morel B, Medina-Carmona E, Pey AL. A mechanism for cancer-associated inactivation of NQO1 due to P187S and its reactivation by the consensus mutation H80R. FEBS Lett. 2017;591:2826–2835. doi: 10.1002/1873-3468.12772. [DOI] [PubMed] [Google Scholar]
- 101.Gaspar N, Sharp SY, Pacey S, Jones C, Walton M, Vassal G, Eccles S, Pearson A, Workman P. Acquired resistance to 17-allylamino-17-demethoxygeldanamycin (17-AAG, tanespimycin) in glioblastoma cells. Cancer Res. 2009;69:1966–1975. doi: 10.1158/0008-5472.CAN-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kasai S, Arakawa N, Okubo A, Shigeeda W, Yasuhira S, Masuda T, Akasaka T, Shibazaki M, Maesawa C. NAD(P)H: quinone oxidoreductase-1 expression sensitizes malignant melanoma cells to the HSP90 inhibitor 17-AAG. PLoS ONE. 2016;11:e0153181. doi: 10.1371/journal.pone.0153181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee J, Grenier L, Holson E, Slocum K, Coco J, Ge J, Normant E, Hoyt J, Lim A, Cushing J, Sydor J, Wright J (2008) IPI-493, a potent, orally bioavailable HSP90 inhibitor of the ansamycin class. In: Proceedings of the EORTC-NCI-AACR symposium on “molecular targets and cancer therapeutics”
- 104.Floris G, Sciot R, Wozniak A, Van Looy T, Wellens J, Faa G, Normant E, Debiec-Rychter M, Schöffski P. The Novel HSP90 inhibitor, IPI-493, is highly effective in human gastrostrointestinal stromal tumor xenografts carrying heterogeneous KIT mutations. Clin Cancer Res. 2011;17:5604–5614. doi: 10.1158/1078-0432.CCR-11-0562. [DOI] [PubMed] [Google Scholar]
- 105.Solit DB, Osman I, Polsky D, Panageas KS, Daud A, Goydos JS, Teitcher J, Wolchok JD, Germino FJ, Krown SE, Coit D, Rosen N, Chapman PB. Phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with metastatic melanoma. Clin Cancer Res. 2008;14:8302–8307. doi: 10.1158/1078-0432.CCR-08-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bentke A, Małecki J, Ostrowska B, Krzykowska-Petitjean K, Laidler P. Tanespimycin and tipifarnib exhibit synergism in inducing apoptosis in melanoma cell lines from later stages of tumor progression. Cancer Invest. 2013;31:545–549. doi: 10.3109/07357907.2013.830736. [DOI] [PubMed] [Google Scholar]
- 107.Huang SW, Kao JK, Wu CY, Wang ST, Lee HC, Liang SM, Chen YJ, Shieh JJ. Targeting aerobic glycolysis and HIF-1alpha expression enhance imiquimod-induced apoptosis in cancer cells. Oncotarget. 2014;5:1363–1381. doi: 10.18632/oncotarget.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ito A, Saito H, Mitobe K, Minamiya Y, Takahashi N, Maruyama K, Motoyama S, Katayose Y, Ogawa J. Inhibition of heat shock protein 90 sensitizes melanoma cells to thermosensitive ferromagnetic particle-mediated hyperthermia with low Curie temperature. Cancer Sci. 2009;100:558–564. doi: 10.1111/j.1349-7006.2008.01072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chai RC, Vieusseux JL, Lang BJ, Nguyen CH, Kouspou MM, Britt KL, Price JT. Histone deacetylase activity mediates acquired resistance towards structurally diverse HSP90 inhibitors. Mol Oncol. 2017;11:567–583. doi: 10.1002/1878-0261.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cheung KM, Matthews TP, James K, Rowlands MG, Boxall KJ, Sharp SY, Maloney A, Roe SM, Prodromou C, Pearl LH, Aherne GW, McDonald E, Workman P. The identification, synthesis, protein crystal structure and in vitro biochemical evaluation of a new 3,4-diarylpyrazole class of Hsp90 inhibitors. Bioorg Med Chem Lett. 2005;15:3338–3343. doi: 10.1016/j.bmcl.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 111.Sharp SY, Boxall K, Rowlands M, Prodromou C, Roe SM, Maloney A, Powers M, Clarke PA, Box G, Sanderson S, Patterson L, Matthews TP, Cheung KM, Ball K, Hayes A, Raynaud F, Marais R, Pearl L, Eccles S, Aherne W, McDonald E, Workman P. In vitro biological characterization of a novel, synthetic diaryl pyrazole resorcinol class of heat shock protein 90 inhibitors. Cancer Res. 2007;67:2206–2216. doi: 10.1158/0008-5472.CAN-06-3473. [DOI] [PubMed] [Google Scholar]
- 112.Graham B, Curry J, Smyth T, Fazal L, Feltell R, Harada I, Coyle J, Williams B, Reule M, Angove H, Cross DM, Lyons J, Wallis NG, Thompson NT. The heat shock protein 90 inhibitor, AT13387, displays a long duration of action in vitro and in vivo in non-small cell lung cancer. Cancer Sci. 2012;103:522–527. doi: 10.1111/j.1349-7006.2011.02191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smyth T, Paraiso KHT, Hearn K, Rodriguez-Lopez AM, Munck JM, Haarberg HE, Sondak VK, Thompson NT, Azab M, Lyons JF, Smalley KSM, Wallis NG. Inhibition of HSP90 by AT13387 delays the emergence of resistance to BRAF inhibitors and overcomes resistance to dual BRAF and MEK inhibition in melanoma models. Mol Cancer Ther. 2014;13:2793–2804. doi: 10.1158/1535-7163.MCT-14-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shapiro GI, Kwak E, Dezube BJ, Yule M, Ayrton J, Lyons J, Mahadevan D. First-in-human phase I dose escalation study of a second-generation non-ansamycin HSP90 inhibitor, AT13387, in patients with advanced solid tumors. Clin Cancer Res. 2015;21:87–97. doi: 10.1158/1078-0432.CCR-14-0979. [DOI] [PubMed] [Google Scholar]
- 115.Canella A, Welker AM, Yoo JY, Xu J, Abas FS, Kesanakurti D, Nagarajan P, Beattie CE, Sulman EP, Liu J, Gumin J, Lang FF, Gurcan MN, Kaur B, Sampath D, Puduvalli VK. Efficacy of onalespib, a long-acting second-generation HSP90 inhibitor, as a single agent and in combination with temozolomide against malignant gliomas. Clin Cancer Res. 2017;23:6215–6226. doi: 10.1158/1078-0432.CCR-16-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Eccles SA, Massey A, Raynaud FI, Sharp SY, Box G, Valenti M, Patterson L, deHaven Brandon A, Gowan S, Boxall F, Aherne W, Rowlands M, Hayes A, Martins V, Urban F, Boxall K, Prodromou C, Pearl L, James K, Matthews TP, Cheung KM, Kalus A, Jones K, McDonald E, Barril X, Brough PA, Cansfield JE, Dymock B, Drysdale MJ, Finch H, Howes R, Hubbard RE, Surgenor A, Webb P, Wood M, Wright L, Workman P. NVP-AUY922: a novel heat shock protein 90 inhibitor active against xenograft tumor growth, angiogenesis, and metastasis. Cancer Res. 2008;68:2850–2860. doi: 10.1158/0008-5472.CAN-07-5256. [DOI] [PubMed] [Google Scholar]
- 117.Yeramian A, Vea A, Benítez S, Ribera J, Domingo M, Santacana M, Martinez M, Maiques O, Valls J, Dolcet X, Vilella R, Cabiscol E, Matias-Guiu X, Marti RM. 2-phenylethynesulphonamide (PFT-μ) enhances the anticancer effect of the novel hsp90 inhibitor NVP-AUY922 in melanoma by reducing GSH levels. Pigment Cell Melanoma Res. 2016;29:352–371. doi: 10.1111/pcmr.12472. [DOI] [PubMed] [Google Scholar]
- 118.London CA, Bear MD, McCleese J, Foley KP, Paalangara R, Inoue T, Ying W, Barsoum J. Phase I evaluation of STA-1474, a prodrug of the novel HSP90 inhibitor ganetespib, in dogs with spontaneous cancer. PLoS ONE. 2011;6:e27018. doi: 10.1371/journal.pone.0027018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nagaraju GP, Long TE, Park W, Landry JC, Taliaferro-Smith L, Farris AB, Diaz R, El-Rayes BF. Heat shock protein 90 promotes epithelial to mesenchymal transition, invasion, and migration in colorectal cancer. Mol Carcinog. 2015;54:1147–1158. doi: 10.1002/mc.22185. [DOI] [PubMed] [Google Scholar]
- 120.Gomez-Casal R, Bhattacharya C, Epperly MW, Basse PH, Wang H, Wang X, Proia DA, Greenberger JS, Socinski MA, Levina V. The HSP90 inhibitor ganetespib radiosensitizes human lung adenocarcinoma cells. Cancers (Basel) 2015;7:876–907. doi: 10.3390/cancers7020814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wu X, Marmarelis ME, Hodi FS. Activity of the heat shock protein 90 inhibitor ganetespib in melanoma. PLoS ONE. 2013;8:e56134. doi: 10.1371/journal.pone.0056134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Acquaviva J, Smith DL, Jimenez JP, Zhang C, Sequeira M, He S, Sang J, Bates RC, Proia DA. Overcoming acquired BRAF inhibitor resistance in melanoma via targeted inhibition of Hsp90 with ganetespib. Mol Cancer Ther. 2014;13:353–363. doi: 10.1158/1535-7163.MCT-13-0481. [DOI] [PubMed] [Google Scholar]
- 123.Vermeulen K, Naus E, Ahamed M, Attili B, Siemons M, Luyten K, Celen S, Schymkowitz J, Rousseau F, Bormans G. Evaluation of [(11)C]NMS-E973 as a PET tracer for in vivo visualisation of HSP90. Theranostics. 2019;9:554–572. doi: 10.7150/thno.27213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mbofung RM, McKenzie JA, Malu S, Zhang M, Peng W, Liu C, Kuiatse I, Tieu T, Williams L, Devi S, Ashkin E, Xu C, Huang L, Zhang M, Talukder AH, Tripathi SC, Khong H, Satani N, Muller FL, Roszik J, Heffernan T, Allison JP, Lizee G, Hanash SM, Proia D, Amaria R, Davis RE, Hwu P. HSP90 inhibition enhances cancer immunotherapy by upregulating interferon response genes. Nat Commun. 2017;8:451. doi: 10.1038/s41467-017-00449-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shah S, Luke JJ, Jacene HA, Chen T, Giobbie-Hurder A, Ibrahim N, Buchbinder EL, McDermott DF, Flaherty KT, Sullivan RJ, Lawrence DP, Ott PA, Hodi FS. Results from phase II trial of HSP90 inhibitor, STA-9090 (ganetespib), in metastatic uveal melanoma. Melanoma Res. 2018;28:605–610. doi: 10.1097/CMR.0000000000000509. [DOI] [PubMed] [Google Scholar]
- 126.Beebe K, Mollapour M, Scroggins B, Prodromou C, Xu W, Tokita M, Taldone T, Pullen L, Zierer BK, Lee MJ, Trepel J, Buchner J, Bolon D, Chiosis G, Neckers L. Posttranslational modification and conformational state of heat shock protein 90 differentially affect binding of chemically diverse small molecule inhibitors. Oncotarget. 2013;4:1065–1074. doi: 10.18632/oncotarget.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Moulick K, Ahn JH, Zong H, Rodina A, Cerchietti L, Gomes DaGama EM, Caldas-Lopes E, Beebe K, Perna F, Hatzi K, Vu LP, Zhao X, Zatorska D, Taldone T, Smith-Jones P, Alpaugh M, Gross SS, Pillarsetty N, Ku T, Lewis JS, Larson SM, Levine R, Erdjument-Bromage H, Guzman ML, Nimer SD, Melnick A, Neckers L, Chiosis G. Affinity-based roteomics reveal cancer-specific networks coordinated by Hsp90. Nat Chem Biol. 2011;7:818–826. doi: 10.1038/nchembio.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gallerne C, Prola A, Lemaire C. Hsp90 inhibition by PU-H71 induces apoptosis through endoplasmic reticulum stress and mitochondrial pathway in cancer cells and overcomes the resistance conferred by Bcl-2. Biochim Biophys Acta. 2013;1833:1356–1366. doi: 10.1016/j.bbamcr.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 129.Cerchietti LC, Lopes EC, Yang SN, Hatzi K, Bunting KL, Tsikitas LA, Mallik A, Robles AI, Walling J, Varticovski L, Shaknovich R, Bhalla KN, Chiosis G, Melnick A. A purine scaffold Hsp90 inhibitor destabilizes BCL-6 and has specific antitumor activity in BCL-6-dependent B cell lymphomas. Nat Med. 2009;15:1369–1376. doi: 10.1038/nm.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li HK, Matsumoto Y, Furusawa Y, Kamada T. PU-H71, a novel Hsp90 inhibitor, as a potential cancer-specific sensitizer to carbon-ion beam therapy. J Radiat Res. 2016;57:572–575. doi: 10.1093/jrr/rrw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Speranza G, Anderson L, Chen AP, Do K, Eugeni M, Weil M, Rubinstein L, Majerova E, Collins J, Horneffer Y, Juwara L, Zlott J, Bishop R, Conley BA, Streicher H, Tomaszewski J, Doroshow JH, Kummar S. First-in-human study of the epichaperome inhibitor PU-H71: clinical results and metabolic profile. Invest New Drugs. 2018;36:230–239. doi: 10.1007/s10637-017-0495-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang X, Wang S, Liu Y, Huangm D, Zheng K, Zhang Y, Wang X, Liu Q, Yang D, Wang Y. Comparative effects of SNX-7081 and SNX-2112 on cell cycle, apoptosis and Hsp90 client proteins in human cancer cells. Oncol Rep. 2015;33:230–238. doi: 10.3892/or.2014.3552. [DOI] [PubMed] [Google Scholar]
- 133.Liu KS, Ding WC, Wang SX, Liu Z, Xing GW, Wang Y, Wang YF. The heat shock protein 90 inhibitor SNX-2112 inhibits B16 melanoma cell growth in vitro and in vivo. Oncol Rep. 2012;27:1904–1910. doi: 10.3892/or.2012.1738. [DOI] [PubMed] [Google Scholar]
- 134.Liu KS, Liu H, Qi JH, Liu QY, Liu Z, Xia M, Xing GW, Wang SX, Wang YF. SNX-2112, an Hsp90 inhibitor, induces apoptosis and autophagy via degradation of Hsp90 client proteins in human melanoma A-375 cells. Cancer Lett. 2012;318:180–188. doi: 10.1016/j.canlet.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 135.Rajan A, Kelly RJ, Trepel JB, Kim YS, Alarcon SV, Kummar S, Gutierrez M, Crandon S, Zein WM, Jain L, Mannargudi B, Figg WD, Houk BE, Shnaidman M, Brega N, Giaccone G. A phase I study of PF-04929113 (SNX-5422), an orally bioavailable heat shock protein 90 inhibitor, in patients with refractory solid tumor malignancies and lymphomas. Clin Cancer Res. 2011;17:6831–6839. doi: 10.1158/1078-0432.CCR-11-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Infante JR, Weiss GJ, Jones S, Tibes R, Bauer TM, Bendell JC, Hinson JM, Jr, Von Hoff DD, Burris HA, 3rd, Orlemans EO, Ramanathan RK. Phase I dose-escalation studies of SNX-5422, an orally bioavailable heat shock protein 90 inhibitor, in patients with refractory solid tumours. Eur J Cancer. 2014;50:2897–2904. doi: 10.1016/j.ejca.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 137.Massey AJ, Schoepfer J, Brough PA, Brueggen J, Chène P, Drysdale MJ, Pfaar U, Radimerski T, Ruetz S, Schweitzer A, Wood M, Garcia-Echeverria C, Jensen MR. Preclinical antitumor activity of the orally available heat shock protein 90 inhibitor NVP-BEP800. Mol Cancer Ther. 2010;9:906–919. doi: 10.1158/1535-7163.MCT-10-0055. [DOI] [PubMed] [Google Scholar]
- 138.Haarberg HE, Paraiso KH, Wood E, Rebecca VW, Sondak VK, Koomen JM, Smalley KS. Inhibition of Wee1, AKT, and CDK4 underlies the efficacy of the HSP90 inhibitor XL888 in an in vivo model of NRAS-mutant melanoma. Mol Cancer Ther. 2013;12:901–912. doi: 10.1158/1535-7163.MCT-12-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lyman SK, Crawley SC, Gong R, Adamkewicz JI, McGrath G, Chew JY, Choi J, Holst CR, Goon LH, Detmer SA, Vaclavikova J, Gerritsen ME, Blake RA. High-content, high-throughput analysis of cell cycle perturbations induced by the HSP90 inhibitor XL888. PLoS ONE. 2011;6:e17692. doi: 10.1371/journal.pone.0017692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rebecca VW, Wood E, Fedorenko IV, Paraiso KH, Haarberg HE, Chen Y, Xiang Y, Sarnaik A, Gibney GT, Sondak VK, Koomen JM, Smalley KS. Evaluating melanoma drug response and therapeutic escape with quantitative proteomics. Mol Cell Proteom. 2014;13:1844–1854. doi: 10.1074/mcp.M113.037424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Paraiso KH, Haarberg HE, Wood E, Rebecca VW, Chen YA, Xiang Y, Ribas A, Lo RS, Weber JS, Sondak VK, John JK, Sarnaik AA, Koomen JM, Smalley KS. The HSP90 inhibitor XL888 overcomes BRAF inhibitor resistance mediated through diverse mechanisms. Clin Cancer Res. 2012;18:2502–2514. doi: 10.1158/1078-0432.CCR-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Phadke M, Gibney GT, Rich CJ, Fedorenko IV, Chen YA, Kudchadkar RR, Sondak VK, Weber J, Messina JL, Smalley KSM. XL888 limits vemurafenib-induced proliferative skin events by suppressing paradoxical MAPK activation. J Invest Dermatol. 2015;135:2542–2544. doi: 10.1038/jid.2015.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Eroglu Z, Chen YA, Gibney GT, Weber JS, Kudchadkar RR, Khushalani NI, Markowitz J, Brohl AS, Tetteh LF, Ramadan H, Arnone G, Li J, Zhao X, Sharma R, Darville LNF, Fang B, Smalley I, Messina JL, Koomen JM, Sondak VK, Smalley SM. Combined BRAF and HSP90 inhibition in patients with unresectable BRAF (V600E)-mutant melanoma. Clin Cancer Res. 2018;24:5516–5524. doi: 10.1158/1078-0432.CCR-18-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Azimi A, Caramuta S, Seashore-Ludlow B, Boström J, Robinson JL, Edfors F, Tuominen R, Kemper K, Krijgsman O, Peeper DS, Nielsen J, Hansson J, Egyhazi Brage S, Altun M, Uhlen M, Maddalo G. Targeting CDK2 overcomes melanoma resistance against BRAF and Hsp90 inhibitors. Mol Syst Biol. 2018;14:e7858. doi: 10.15252/msb.20177858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Belofsky GN, Jensen PR, Fenical W. Sansalvamide: a new cytotoxic cyclic depsipeptide produced by a marine fungus of the genus Fusarium. Tetrahedron Lett. 1999;40:2913–2916. doi: 10.1016/S0040-4039(99)00393-7. [DOI] [Google Scholar]
- 146.Liu Y, Zhang G, Wang H, Liu S, Chen J, Zhao L, Li J, Shan B. Novel cyclic pentapeptide H-15 induces differentiation and inhibits proliferation in murine melanoma B16 cells. Oncol Lett. 2016;11:1251–1255. doi: 10.3892/ol.2015.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang G, Liu S, Liu Y, Wang F, Ren J, Gu J, Zhou K, Shan B. A novel cyclic pentapeptide, H-10, inhibits B16 cancer cell growth and induces cell apoptosis. Oncol Lett. 2014;8:248–252. doi: 10.3892/ol.2014.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang X, Yao X, Fan S, Xiang C, Liu R, Feng J, Huang J, Liu S. A LY-15, a novel cyclic pentapeptide that inhibits B16 cell proliferation and migration and induces cell apoptosis. Oncol Lett. 2018;15:5887–5892. doi: 10.3892/ol.2018.8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nordenberg J, Albukrek D, Hadar T, Fux A, Wasserman L, Novogrodsky A, Sidi Y. Novobiocin-induced anti-proliferative and differentiating effects in melanoma B16. Br J Cancer. 1992;65:183–188. doi: 10.1038/bjc.1992.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nordenberg J, Kornfeld J, Wasserman L, Shafran M, Halabe E, Beery E, Landau O, Novogrodsky A, Sidi Y. Novobiocin modulates colchicine sensitivity in parental and multidrug-resistant B16 melanoma cells. J Cancer Res Clin Oncol. 1994;120:599–604. doi: 10.1007/BF01212814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Marcu MG, Chadli A, Bouhouche I, Catelli M, Neckers LM. The heat shock protein 90 antagonist novobiocin interacts with a previously unrecognized ATP-binding domain in the carboxyl terminus of the chaperone. J Biol Chem. 2000;275:37181–37186. doi: 10.1074/jbc.M003701200. [DOI] [PubMed] [Google Scholar]
- 152.Marcu MG, Schulte TW, Neckers L. Novobiocin and related coumarins and depletion of heat shock protein 90-dependent signaling proteins. J Natl Cancer Inst. 2000;92:242–248. doi: 10.1093/jnci/92.3.242. [DOI] [PubMed] [Google Scholar]
- 153.Donnelly AC, Zhao H, Kusuma BR, Blagg BSJ. Cytotoxic sugar analogues of an optimized novobiocin scaffold. Med Chem Commun. 2010;1:165–170. doi: 10.1039/C0MD00063A. [DOI] [Google Scholar]
- 154.Samadi AK, Zhang X, Mukerji R, Donnelly AC, Blagg BS, Cohen MS. A novel C-terminal HSP90 inhibitor KU135 induces apoptosis and cell cycle arrest in melanoma cells. Cancer Lett. 2011;312:158–167. doi: 10.1016/j.canlet.2011.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Prince TL, Kijima T, Tatokoro M, Lee S, Tsutsumi S, Yim K, Rivas C, Alarcon S, Schwartz H, Khamit-Kush K, Scroggins BT, Beebe K, Trepel JB, Neckers L. Client proteins and small molecule inhibitors display distinct binding preferences for constitutive and stress-induced HSP90 isoforms and their conformationally restricted mutants. PLoS ONE. 2015;10:e0141786. doi: 10.1371/journal.pone.0141786. [DOI] [PMC free article] [PubMed] [Google Scholar]