Abstract
Most of human malignant melanoma is metastatic and invasive, which has poor therapeutic response and poor prognosis as well as high incidence worldwide. CTHRC1 was overexpressed in metastatic primary melanomas. The relationship between CTHRC1 and miRNA in affecting the melanoma has not been studied. This study was aimed to explore the function of CTHRC1 in human melanoma and finding its regulator miRNA. CTHRC1 and miR155 expression in cancer tissues and paracancer tissues was examined. The cell proliferation, cell cycle, cell migration and cell invasion was evaluated in M21 cell transfected with CTHRC1 siRNA or miR155. The mRNA expression of miR155 in cancer tissues was lower than that in paracancer tissues. The mRNA expression of CTHRC1 in the cancer tissues was higher than that in corresponding paracancer tissues. The mRNA expression of CTHRC1 in the M21 cells was decreased by treated with miR155 mimic. CTHRC1 promoted the cell proliferation, cell cycle process, cell migration and invasion capacity and prevented the cell from apoptosis. miR155 inhibited the cell proliferation, cell migration and cell invasion, arrested the cell cycle process at G1 phase and enhanced the cell apoptosis. The luciferases reporter assay showed that miR155 bound to the 3’-UTR of CTHRC1 and regulated the expression of CTHRC1, further influenced the functions of CTHRC1 in human melanoma development. The study contributed to unearthing a novel potential predictor and therapeutic targets of human melanoma. CTHRC1 was a potential independent predictor and therapeutic target for human melanoma.
Keywords: Human melanoma, M21 cells, collagen triple helix repeat containing 1 (CTHRC1), miR155
Introduction
Human metastatic melanoma is an aggressive skin cancer with high incidence worldwide and poor therapeutic response as well as poor prognosis [1-3]. The incidence of melanoma is about 2.18%. As a metastatic disease and invasive cancer, melanoma evolves rapidly and is resistant to most existing chemotherapies [4]. Surgical excision, systemic chemotherapies, targeted therapy and immunotherapy are common methods to treat melanoma [2]. The long-term survival rate of metastatic melanoma remains very low and the median survival of patients with metastatic melanoma is very short (about 6-12 months) [2,5]. Therefore, uncovering the molecular mechanisms underlying the development and progression of melanomas is very important.
Environmental and individual factors including UV irradiation, cosmetic ingredients, air pollution, smoking, chronic inflammation and higher age could increase the risk of melanoma [5-8]. To response environmental exposure of humans, the expression of microRNA (miR) are markedly various [9]. miRNAs are important posttranscriptional regulators controlling more than 30% of human gene expression via direct binding to the 3’ untranslated region (3’-UTR) of the targeted genes [10]. miRNA signature can obviously differentiate stage III patients into better and worse prognosis groups based on the survival probability [11]. miR155, encoded by the non-coding transcript of the B-cell Integration Cluster (BIC) gene, has been shown to play an oncogenic role in both hematopoietic malignancies and solid cancer including breast, cervical and clear renal cell carcinoma [12,13]. miR155 were significantly elevated in metastatic and primary melanoma samples compared to nevi, which suggested that miR155 may be a useful prognosis marker prior to the development of metastases [11]. Nonetheless, another report has shown that the expression of miR155 is obviously decreased in melanoma cell lines compared with that in melanocytes [14]. Increased expression of miR155 was associated with longer survival of melanoma patients [12]. Anti-proliferative and pro-apoptotic characters of miR155 in melanoma cell lines have been verified, which indicated that miR155 was a direct negative regulator of melanoma cell proliferation and survival [12,14].
A vast amount of mutations, such as those of TERT, BRAF, NRAS, PTEN and CDKN24, have been verified to be associated with the malignant transformation of melanocytes in subgroups of primary melanomas [15-17]. The collagen triple helix repeat containing 1 (CTHRC1), a secreted protein involved in vascular remodeling, bone formation and developmental morphogenesis, has been verified as an oncogene in many types of human cancer, including pancreatic progression, hepatocellular and breast carcinoma [18-20]. CTHRC1 was overexpressed in metastatic primary melanomas, and a high expression of CTHRC1 mRNA was associated with a short survival [21]. CTHRC1 enhanced the melanoma cell adhesion and survival ability [22]. The knockdown of CTHRC1 expression in melanoma cells has been found to decrease melanoma cell migration in vitro [18]. In melanoma, CTHRC1 was expressed in melanoma cells and stromal cells, activated stromal fibroblasts, and blood vessel endothelial cells [21]. The expression of CTHRC1 was induced by the preinvasive and angiogenic transcription factors, such as nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 (NFATC2), transforming growth factor-β (TGF-β) and oncogenic BRAF signaling [21]. All of the results suggested that CTHRC1 might be an attractive target for melanomas therapy. As mentioned, both CTHRC1 and miR155 were closely associated with the development of human melanoma. However, the relationship between CTHRC1 and miR155 in affecting melanoma has not been studied.
In this study, we predicted the miR155 was the regulator of CTHRC1. Then, we examined the expression of miR155 and CTHRC1 in melanoma cancer tissues and corresponding paracancer tissues. The cell apoptosis, cell cycle, cell migration and cell invasion of the M21 cells transfected with negative control (NC), miR155 mimic and CTHRC1 overexpression vectors/siRNA were investigated in this study. The results showed that CTHRC1 promoted the cell proliferation, cell cycle process, cell migration and invasion capacity and prevented the cell from apoptosis. Reversly, miR155 inhibited the cell proliferation, cell migration and cell invasion, arrested the cell cycle process at G1 phase and enhanced the cell apoptosis. The luciferase reporter assay showed that miR155 directly bound to the 3’-UTR of CTHRC1 and reduced the expression of CTHRC1, further affected the function of CTHRC1 in human melanoma development.
Materials and methods
Reverse transcription polymerase chain reaction (RT-PCR)
Melanoma tissues and corresponding paracancer tissues of 8 melanoma patients were obtained. The melanoma samples were the tissue taken around the tumor (about 0.5-1.0 cm). The surgery process was as following: disinfection; local infiltration anesthesia; cut the skin and subcutaneous tissue; completely remove the tumor tissue and rapidly send to pathology. No tumor cells were remained in the center and cut edge after surgery. Paracancer tissues were the tissues that were 2 cm away from melanoma tissue, including epidermis, dermis and subcutaneous fat layer. The details of the 8 specimens were listed in Table 1, including the type of the melanoma, the Breslow’s depth, tumor pathology staging.
Table 1.
The detail of the 8 specimens were added in the manuscript, including the type of the melanoma, the Breslow’s depth, tumor pathology staging
| Specimens | Type of the melanoma | Breslow’s depth | Tumor metastasis (Yes or No) | Tumor pathology staging |
|---|---|---|---|---|
| 1 | Primary melanoma | Level 5 | Yes | TanyNM1b |
| 2 | Primary melanoma | Level 4 | Yes | T1~4bN2bM0 |
| 3 | Primary melanoma | Level 3 | No | T3aN0M0 |
| 4 | Primary melanoma | Level 3 | No | T3bN0M0 |
| 5 | Primary melanoma | Level 5 | Yes | TanyNM1c |
| 6 | Primary melanoma | Level 2 | No | T2aN0M0 |
| 7 | Primary melanoma | Level 2 | No | T3aN0M0 |
| 8 | Primary melanoma | Level 3 | Yes | T1~4bN2bM0 |
| 9 | Primary melanoma | Level 5 | Yes | TanyNM1b |
| 10 | Primary melanoma | Level 3 | No | T3aN0M0 |
| 11 | Primary melanoma | Level 2 | No | T2aN0M0 |
| 12 | Primary melanoma | Level 5 | Yes | TanyNM1c |
| 13 | Primary melanoma | Level 4 | Yes | T1~4bN2bM0 |
| 14 | Primary melanoma | Level 2 | No | T2aN0M0 |
The total RNA was extracted from the melanoma tissues and corresponding paracancer tissues. The mRNA expression of miR155 and CTHRC1 in the cancer tissues and paracancer tissues were measured by RT-PCR. cDNA was synthesized by using PrimeScript II 1st Strand cDNA Synthesis Kit (Takara, Japan). The primer sequence used in RT-PCR were shown as follows: CTHRC1 forward primer: 5’ATAATGGAATGTGCTTACAAGG3’, CTHRC1 reverse primer: 5’TTCCCAAGATCTATGCCATAAT3’; 18S forward primer: 5’CCTGGATACCGCAGCTAGGA3’; 18S reverse primer: 5’GCGGCGCAATACGAATGCCCC3’; miR155 RT primer: 5’CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCTGAGA3’; hsa-miR-155-5p forward primer: 5’ACACTCCAGCTGTAAACATCCTACACTCT3’; hsa-miR155-5p reverse primer: 5’CTCAACTGGTGTCGTGGA3’; U6 forward primer: 5’CTCGCTTCGGCAGCACA3’; U6 reverse primer: 5’AACGCTTCACGAATTTGCGT3’. The gene expressions of CTHRC1 and miR-155 were determined by RT-PCR using SYBR Green PC Master Mix (Toyobo, Japan). Reactions were performed on an ABI PRISM® 7500 Sequence Detection System and performed under the following thermocycler conditions: 95°C for 5 min, followed by 40 cycles of 95°C for 15 seconds, 60°C for 15 seconds and 72°C for 30 seconds; then followed by a 60°C for 1 min and 95°C for 15 seconds. Raw Data of all samples were collected and normalized to that of the control (miR155 was normalized to U6 small nuclear RNA and CTHRC1 was normalized to 18S). The gene expression of CTHRC1 and miR155 were calculated using the relative quantification equation (RQ=2-ΔΔCt) [23].
The mRNA expression level of CTHRC1 in M21 cells were also determined by RT-PCR as above-mentioned. The protein expression level of CTHRC1 in M21 cells was evaluated by Western Blotting (WB). Protein concentrations were determined using BCA Protein Assay Kit (Keygen Biotech). Proteins were detected by CTHRC1 monoclonal antibody (ab85739, 1:1000, Abcam, USA), GAPDH polyclonal antibody (KC-5G5, 1:10,000, KangCheng Bio, China) and then visualized by a commercial Immobilon Western HRP Substrate (WBKLS0500, Millipore, USA) under dark conditions.
Cell culture
Human melanoma cells M21 were incubated in the light of the stander process under 37°C supplied with 5% CO2. Cells were treated as follows: negative control mimic (NC group); miR-155 mimics (miR155 group); overexpression of CTHRC1 (CTHRC1 group) and knockdown of CTHRC1 expression by small interfering RNA (si-CTHRC1 group). The transfected processes were performed according to previous research using Lipofectamine 2000 (Invitrogen, USA).
Cell proliferation assay
Cell proliferation was measured using the Cell Counting Kit-8 (CCK-8) assay kit (Dojindo, Kumamato, Japan). The absorbance at 450 nm (OD450) of all samples was measured. The proliferation rates and inhibition rates of cells from day 0 to day 3 were calculated based on OD450 values.
Colony formation assays
The clonogenic assay was performed as previous [24]. M21 cells were plated (clonal density 100 cells/well) in a 96-well dish and incubated for 7 days, then discarded the medium, washed with PBS for 2 times, incubated with crystal violet staining solution of 200 μl/well, washed softly by water and dried at room temperature. The samples were counted, photographed and analyzed by AID EliSpot iSpot Reader System (AID, Germany).
Cell cycle assay
The M21 cells were cultured for 48 h, and then the cells were collected, fixed, then stained on the base of standard process. The cells were stained with a propidium iodide (PI) solution containing 100 μg/ml PI and 50 μg/ml RNase (Sigma, USA) in PBS at 37°C for 30 min in the dark. The samples were analyzed by flow cytometry (BD, USA). Data were collected and analyzed by the CELL Quest and ModFit LT software.
Cell apoptosis analysis
The cells were harvested and washed after 48 h incubation. The cell apoptosis was analyzed by Annexin V-FITC/PI Apoptosis Detection Kit (Catalog no. KGA106; Nanjing Keygen Biotech Co., Ltd) according to the manufacturer’s instruction. After staining, all samples were immediately measured on FACSort flow cytometry (BD, USA). The data were analyzed using the Cell Quest 3.0 software (BD, USA).
Transwell chambers-induced migration and invasion assays
The migration ability of M21 cells was assessed using the Transwell chambers (BD, REF353097) according to the manufacturer’s protocol. Briefly, cells (1×105) were seeded in the upper chamber with serum-free medium. Cells were migrated to lower chamber with 10% fetal bovine serum (FBS) medium. The migratory cells were stained with 0.1% crystal violet for 20 min at room temperature. The stain was eluted with 33% acetic acid and photographed after 48 h incubation. The absorbance at 570 nm (OD570) was measured.
Cells incubated in serum-free medium were added to Transwell chambers consisting of a thin layer of Matrigel basement membrane matrix (8 mm pores), 10% FBS were added to the lower. The invasive cells were stained with 0.1% crystal violet for 20 min at room temperature. The stain was eluted with 33% acetic acid and photographed after 48 h incubation. The absorbance at 570 nm (OD570) was measured.
Luciferase reporter assay
The CTHRC1-3’-UTR fragment containing putative binding sites for miR155 was obtained and ligased (Takara, Japan) into the digested psiCheck-2 plasmid (Promega, USA) between the XhoI and NotI sites. CTHRC1-3’-UTR-targeted site mutations were generated using the KOD-plus mutagenesis kit (Toyobo, Japan) in the light of the manufacturer’s protocol. M21 cells were plated in a 96-well plate and then co-transfected with 200 ng/μl of plasmid and 50 nM miR155, NC, miR155 inhibitor or NC inhibitor. After co-transfection 48 h, luciferase activity was detected by the Dual-Glo luciferase assay kit (Promega, USA). The transfections were performed in duplicate and repeated by three times.
Ethics statement
The experiments were undertaken with the understanding and written consent of each subject, and that the study conforms to The Code of Ethics of the World Medical Association (Declaration of Helsinki), printed in the British Medical Journal (18 July 1964). Ethical approval was obtained from the hospital and fully informed consent were signed by participants before sample collection.
Statistical analysis
All statistical analyses were performed with Statistical Package for the Social Sciences (SPSS) 19.0 software. The data are presented as the mean ± SD from three separate experiments. Statistical significance was determined by paired or unpaired Student’s t-test in cases of standardized expression data.
Results
The mRNA expression of CTHRC1 and miR155 determined by RT-PCR
The mRNA expression of CTHRC1 and miR155 in human melanoma cancer tissues and corresponding paracancer tissues were determined by RT-PCR, which was shown in Figure 1. The mRNA expression of CTHRC1 in human melanoma cancer tissues was higher than that in corresponding paracancer tissues (Figure 1A). The mRNA expression of CTHRC1 between the melanoma group and paracancer tissues group were significantly different (P<0.01). While the miR155 expression level in human melanoma cancer tissues were significantly lower than that in corresponding paracancer tissues (Figure 1B).
Figure 1.

The mRNA expression of CTHRC1 and miR155 in human melanoma cancer tissues and corresponding paracancer tissues were determined by RT-PCR. A. The mRNA expression of CTHRC1. B. The mRNA expression of miR155. *indicates P<0.05.
The miR155 inhibited the expression of CTHRC1 in M21 cells
The mRNA expression of CTHRC1 in M21 cells transfected with NC and miR155 vectors or treated with miR155 inhibitor were determined by RT-PCR (Figure 2A). The result showed that the mRNA expression of CTHRC1 in miR155 group was significantly lower than that in NC group and miR155 inhibitor group. The expression of CTHRC1 in miR155 group was about 3.2 times of that in NC group. The results suggested that miR155 inhibited the expression of CTHRC1, while miR155 inhibitor promoted the expression of CTHRC1. The protein expression of CTHRC1 in M21 cells transfected with NC and miR155 mimics were detected by WB (Figure 2B). The protein expression of CTHRC1 was significantly decreased in miR155 group compared with the M21 cell group and the NC group. There was no significantly difference of the expression of CTHRC1 between NC group and M21 cell group. The result suggested that the miR155 indeed inhibited the expression of CTHRC1.
Figure 2.
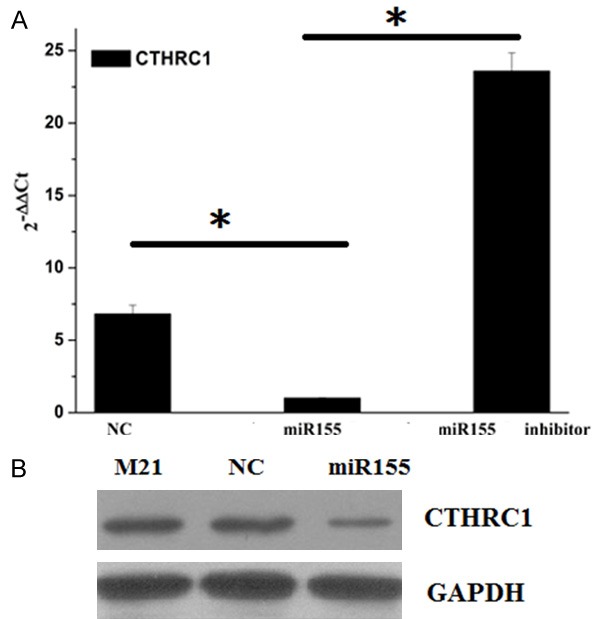
The expression of CTHRC1 in M21 cells. A. The mRNA expression of CTHRC1 in M21 cells transfected with negative control (NC) and miR155 vectors or treated with miR155 inhibitor were determined by RT-PCR. B. The protein expression of CTHRC1 in M21 cells transfected with NC and miR155 vectors by WB. *indicates P<0.05.
Evaluating the effect of CTHRC1 on cells growth
To investigate the functions of miR155 and CTHRC1 in melanoma cell M21, M21 cells were treated with miR155 mimics, overexpressing CTHRC1 and siRNA used to knockdown CTHRC1 as well as NC. Then, proliferation rate and inhibition rate of the M21 cells were determined and calculated (Figure 3A-C). Overexpression of CTHRC1 enhanced the OD450 values, promoted the cells proliferation rate and inhibited the cells inhibition rate, while overexpression of miR155 or downregulation of CTHRC1 lowered the OD450 values, inhibited the cells proliferation and promoted the cells inhibition rate. All these results suggested that CTHRC1 promoted the cells proliferation. Knockdown of CTHRC1 by siRNA could inhibit the cells proliferation.
Figure 3.
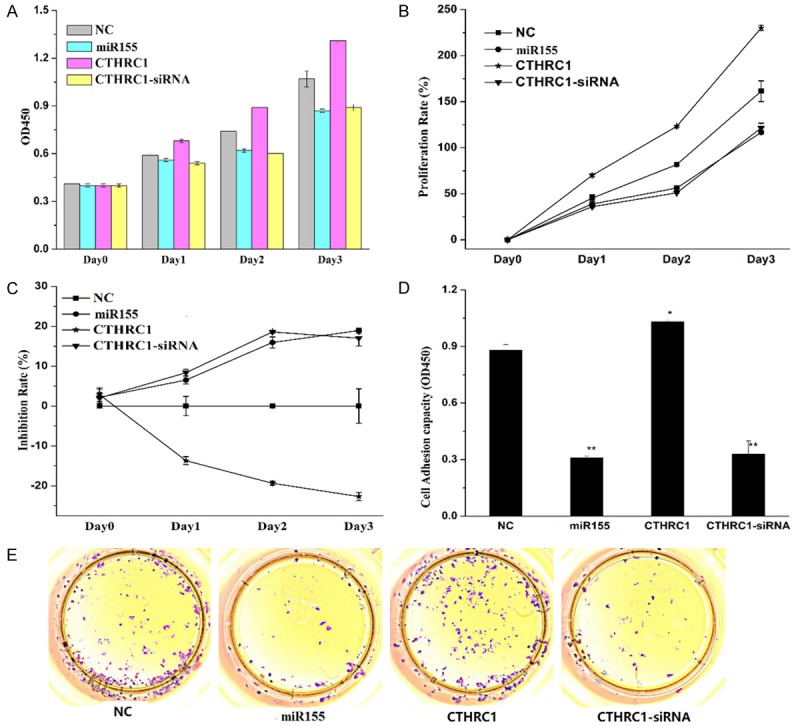
The effects of miR155 and CTHRC1 on M21 cell proliferation. A-C. Then OD450, proliferation rate and inhibition rate of the M21 cells treated with NC, miR155, CTHRC1 and si-CTHRC1 were determined and calculated. D and E. The cell adhesion of M21 cells affected by miR155 and CTHRC1. Data shows the representative of the three independent experiments, *P<0.05, **P<0.01.
To further investigate the effect of miR155 and CTHRC1 on cell growth and proliferation, the clonogenic capacity of M21 cell was evaluated by colony formation assay (Figure 3D and 3E). Overexpression of CTHRC1 significantly enhanced M21 colony establishment. Downregulation of CTHRC1 (si-CTHRC1 group) with siRNA or overexpression of miR155 obviously inhibited the cells clonogenic capacities. CTHRC1 improved M21 cell holoclone formation and clonogenic capacity, which was consistent with that CTHRC1 could promote the cells proliferation.
Investigating the effects of CTHRC1 on cell apoptosis and cell cycle
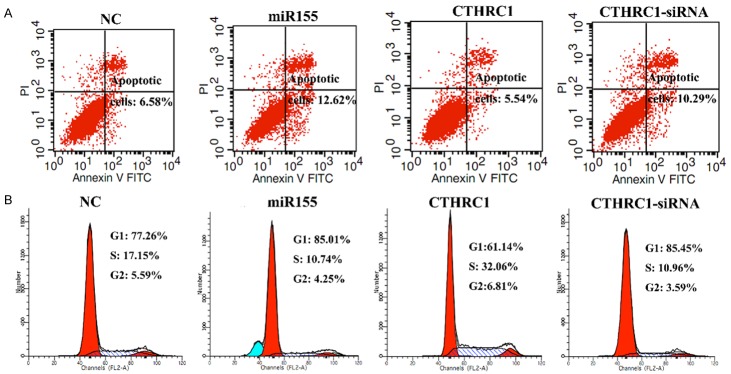
To determine the effect of miR155 and CTHRC1 on cell death, the apoptosis of M21 cells transfected with NC, miR155, CTHRC1 and si-CTHRC1 vectors were determined by flow cytometry (Figure 4A). The shifts in cell population with the different treatments clearly indicated that apoptotic rate of the si-CTHRC1-treated or miR155 group was higher than that of the CTHRC1 group and the NC group. The apoptotic rate of the si-CTHRC1-treated group and miR155 group was 1.9 and 1.6 times higher than that in NC group, respectively. CTHRC1 overexpression decreased the apoptotic rate to 84% of NC group. All these results suggested that downregulating the expression of CTHRC1 or overexpressing miR155 promoted the cells apoptosis, while overexpression of CTHRC1 prevented the cells from apoptosis.
Figure 4.
The effects of miR155 and CTHRC1 on M21 apoptosis and cell cycle determined by flow cytometry. A. The apoptosis of M21 cells transfected with NC, miR155, CTHRC1 and si-CTHRC1 vectors. B. The cell cycle of M21 cells transfected with NC, miR155, CTHRC1 and si-CTHRC1 vectors.
We further examined the effect of miR155 and CTHRC1 on the cell cycle of M21 cells by flow cytometry (Figure 4B). The percentage of G1 in CTHRC1 group was lower than that in NC group miR155, and si-CTHRC1 group. The percentage of S and G2 phases in CTHRC1 group were higher than that in NC group, miR155, and si-CTHRC1 group. The percentages of G1 in miR155 and si-CTHRC1 group were higher than that in NC group. All of the results suggested that overexpression of CTHRC1 promoted the cell cycle process, while miR155 or downregulation of CTHRC1 arrested the cell cycle in G1 phase. The result was further verified that CTHRC1 promoted the cells proliferation, while miR155 inhibited the cell proliferation.
Discovering effects of CTHRC1 on the cell migration, invasion and adhesion
The migration and invasion assays were performed to study the effects of miR155 and CTHRC1 on migration and invasion of M21 cells. The migration capacity of M21 cells was shown in Figure 5A and 5C. The invasion capacity of M21 was showed in Figure 5B and 5D. The migrated and invaded cell number in CTHRC1 group were obviously higher than that in control group and si-CTHRC1 group, which suggested that overexpression of CTHRC1 enhanced the migration and invasion of cells (Figure 5). The migrated and invaded cell number in si-CTHRC1 and miR155 group were lower than that in NC group, which indicated that downregulation of CTHRC1 or overexpression of miR155 reduced the migration and invasion capacity of cells.
Figure 5.
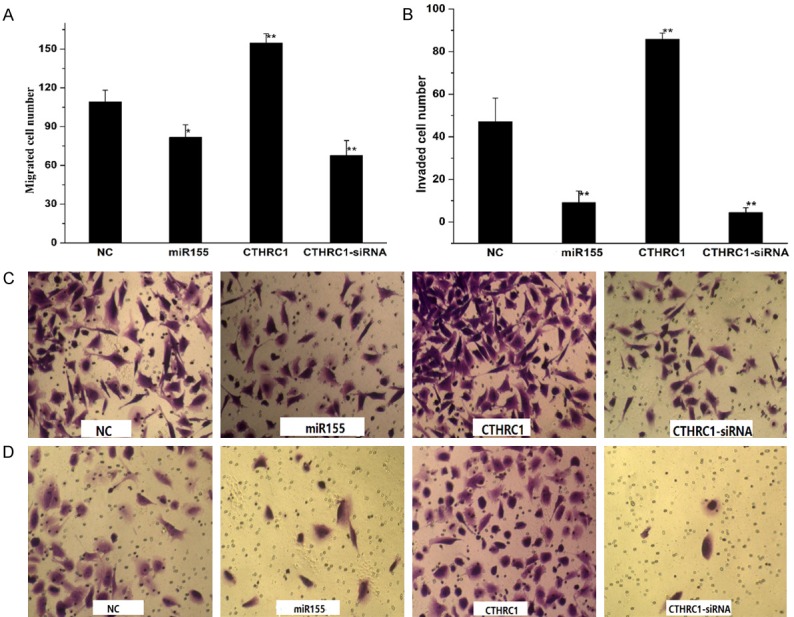
The effects of miR155 and CTHRC1 on cell migration and invasion of M21 treated with NC, miR155, CTHRC1 and si-CTHRC1 vectors. A and C. The migration assay of M21 cells. B and D. The invasion assays of M21 cells. Data shows the representative of the three independent experiments, *P<0.05, **P<0.005.
CTHRC1 was a direct target of miR155
To determine the molecular mechanism by which CTHRC1 promoted cells growth, cell migration, cell invasion and cell survival. We used open-target prediction programs, such as picTar, TargetScan and miRanda, to predict that CTHRC1 was a target gene of miR-155. To confirm our previous hypothesis, we compared the sequence of 3’-UTR of CTHRC1 with miR155. The 3’-UTR of CTHRC1 mRNA contained a complementary site for the seed region of miR155 (Figure 6A). CTHRC1 was a notably attractive candidate because it plays important roles in the development and progression of melanoma.
Figure 6.
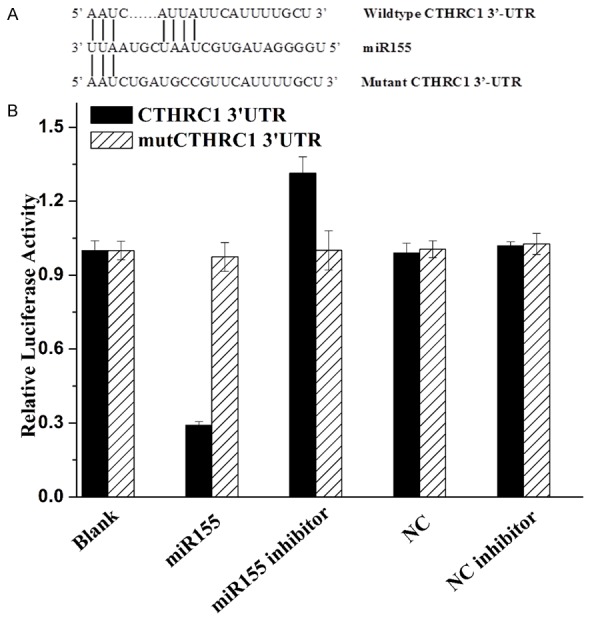
Luciferase reporter assay. A. Sequence alignment between miR155 and the 3’-UTR of CTHRC1 mRNA seed-matching region or seed-mutated region. B. Relative luciferase activity was calculated as follows: (Rluc miRNA/Luc miRNA)/(Rluc no-target/Luc nontarget). The data are presented as the average of triplicate values.
To determine whether CTHRC1 was the direct target gene for miR155, a dual-luciferase reporter system was employed, the results showed in Figure 6B. The luciferase reporter assay indicated that the luciferase activity of reporter containing the CTHRC1 gene’s wide-type 3’-UTR decreased by 71% following treatment with miR155 mimics. By contrast, the inhibitory effect of miR155 mimics was abolished in the mutated construct. Moreover, miR155 inhibitor treated in CTHRC1 group induced the increase of luciferase activity (increased by 31% compared with the blank). The luciferase activity between mutCTHRC1-3’UTR groups had no difference. The results indicated that miR155 most likely suppresses gene expression of CTHRC1 through miR155-binding sequence at the 3’-UTR of CTHRC1. Therefore, the data indicated that miR155 reduced CTHRC1 expression by inhibiting translation and/or causing mRNA instability.
Discussion
Melanoma, which is known as a heterogeneous and invasive malignancy, remains need more attention to uncover more target genes and understand its molecular mechanism [3]. In recent years, more and more reports have shown that miRNA play important roles in cancer tumorigenesis and development via mediating its target genes [25,26]. miR21, miR155, miR211 and so on have been verified to be associated with the human melanoma [10,14,27]. CTHRC1 has been demonstrated to express aberrantly and be involved in the cancer cell migration and invasion [18]. The CTHRC1 is upregulated in melanoma and further promotes the invasion of melanoma [22]. The relationship between miR155 and CHTRC1 in melanoma has not been studied.
In our research, we aimed to discover the relationship between miR155 and CTHRC1 in melanoma and further understand the functions of miR155 and CTHRC1 in melanoma. We first predicted that miR155 probably regulate the CTHCR1, then we studied the effects of miR155 and CTHRC1 on melanoma cell proliferation, apoptosis, migration and invasion. The CTHCR1 was upregulated in the human melanoma cancer tissues compared with that in corresponding paracancer tissues, which was consistent with the previous results [18,21,22]. CTHRC1 promoted the M21 cell cycle process, cell migration and cell invasion and cell survival, which was also demonstrated in previous results [22]. The miR155 was downregulated in human melanoma cancer tissues, which was contradictory with the previous result [11]. MiR155 inhibited the M21 cell migration, cell invasion and promoted the cell apoptosis, which was consistent with the previous results [14]. The luciferase reporter assay showed that miR155 bound to the 3’-UTR of CTHRC1 and affected the luciferase intensity. We concluded that CTHRC1 promoted the development of melanoma, which was mediated by miR155.
In melanoma, the expression of CTHRC1 was upregulated by TGF-β [21]. The activin/transforming growth factor-β (TGF-β) pathway played an important role in tumorigenesis by its tumor suppressor or tumor promoting effect [28,29]. TGF-β signaling facilitated metastasis in advanced malignancy, and CTHRC1 also participated in the invasion of cancer [29]. Therefore, the TGF-β and CTHRC1 synergistically affect the invasion and metastasis of melanoma. Previous reports also have shown that miR155 is upregulated by TGF-β in breast cancer and miR155 plays a key role in breast cancer metastasis and cell migration and invasion induced by TGF-β [30]. Another report has shown that TGF-β increases the expression of miR155 in intestinal T cells [31]. In this paper, we found that the miR155 decreased the expression of CTHRC1 via direct binding to the 3’-UTR of CTHRC1 in melanoma. The expression of miR155 and CTHRC1 in melanoma tissues and cells are converse, being consistent with the results that miR155 is downregulated in melanoma while CTHRC1 is upregulated in melanoma. However, it is contradictory with the previous results that TGF-β positively regulated the expression of CTHRC1 or miR155. The relationship between CTHRC1, miR155 and TGF-β and their functions in melanoma needs further investigation.
Conclusion
Melanoma remains one of the refractory cancers to be treated. Investigating the molecular mechanism and discovering more target genes of melanoma is still urgent. Both CTHRC1 and miR155 are associated with the melanoma. Herein, we further investigated the relationship between CTHRC1 and miR155 in melanoma, then found that CTHRC1 facilitated the development of melanoma and the expression of CTHRC1 was regulated by miR155.
Acknowledgements
This work was financially supported by Shandong Provincial Key Research and Development Program (No. 2016GSF201110).
Disclosure of conflict of interest
None.
References
- 1.Demirsoy S, Martin S, Maes H, Agostinis P. Adapt, recycle, and move on: proteostasis and trafficking mechanisms in melanoma. Front Oncol. 2016;6:240. doi: 10.3389/fonc.2016.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olszanski AJ. Current and future roles of targeted therapy and immunotherapy in advanced melanoma. J Manag Care Pharm. 2014;20:346–356. doi: 10.18553/jmcp.2014.20.4.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melanoma and other skin cancers.pdf. U.S. Department of health and human services national institutes of health 10-76252010 [Google Scholar]
- 4.Holzel M, Bovier A, Tuting T. Plasticity of tumour and immune cells: a source of heterogeneity and a cause for therapy resistance? Nat Rev Cancer. 2013;13:365–376. doi: 10.1038/nrc3498. [DOI] [PubMed] [Google Scholar]
- 5.Dengel LT, Slingluff CL Jr. Short length of stay and rapid recovery to normal function after surgery for metastatic melanoma to abdominal and retroperitoneal viscera. J Surg Oncol. 2009;100:481–483. doi: 10.1002/jso.21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaminska-Winciorek G, Wydmanski J, Gajda M, Tukiendorf A. Melanoma awareness and prevalence of dermoscopic examination among internet users: a cross-sectional survey. Postepy Dermatol Alergol. 2016;33:421–428. doi: 10.5114/pdia.2016.63297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang AJ, Rambhatla PV, Eide MJ. Socioeconomic and lifestyle factors and melanoma: a systematic review. Br J Dermatol. 2015;174:885–915. doi: 10.1111/bjd.13500. [DOI] [PubMed] [Google Scholar]
- 8.Candido S, Rapisarda V, Marconi A, Malaponte G, Bevelacqua V, Gangemi P, Scalisi A, McCubrey JA, Maestro R, Spandidos DA, Fenga C, Libra M. Analysis of the B-RafV600E mutation in cutaneous melanoma patients with occupational sun exposure. Oncol Rep. 2014;31:1079–1082. doi: 10.3892/or.2014.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vrijens K, Bollati V, Nawrot TS. MicroRNAs as potential signatures of environmental exposure or effect: a systematic review. Environ Health Perspect. 2015;123:399–411. doi: 10.1289/ehp.1408459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melnik BC. MiR-21: an environmental driver of malignant melanoma? J Transl Med. 2015;13:202. doi: 10.1186/s12967-015-0570-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philippidou D, Schmitt M, Moser D, Margue C, Nazarov PV, Muller A, Vallar L, Nashan D, Behrmann I, Kreis S. Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer Res. 2010;70:4163–4173. doi: 10.1158/0008-5472.CAN-09-4512. [DOI] [PubMed] [Google Scholar]
- 12.Segura MF, Belitskaya-Levy I, Rose AE, Zakrzewski J, Gaziel A, Hanniford D, Darvishian F, Berman RS, Shapiro RL, Pavlick AC, Osman I, Hernando E. Melanoma MicroRNA signature predicts post-recurrence survival. Clin Cancer Res. 2010;16:1577–1586. doi: 10.1158/1078-0432.CCR-09-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levati L, Alvino E, Pagani E, Arcelli D, Caporaso P, Bondanza S, Di Leva G, Ferracin M, Volinia S, Bonmassar E, Croce CM, D’Atri S. Altered expression of selected microRNAs in melanoma: antiproliferative and proapoptotic activity of miRNA-155. Int J Oncol. 2009;35:393–400. [PubMed] [Google Scholar]
- 15.Flaherty KT, Hodi FS, Fisher DE. From genes to drugs: targeted strategies for melanoma. Nat Rev Cancer. 2012;12:349–361. doi: 10.1038/nrc3218. [DOI] [PubMed] [Google Scholar]
- 16.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, Schadendorf D, Kumar R. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 17.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang L, Dai DL, Su M, Martinka M, Li G, Zhou Y. Aberrant expression of collagen triple helix repeat containing 1 in human solid cancers. Clin Cancer Res. 2006;12:3716–3722. doi: 10.1158/1078-0432.CCR-06-0030. [DOI] [PubMed] [Google Scholar]
- 19.Jiang N, Cui Y, Liu J, Zhu X, Wu H, Yang Z, Ke Z. Multidimensional roles of collagen triple helix repeat containing 1 (CTHRC1) in malignant cancers. J Cancer. 2016;7:2213–2220. doi: 10.7150/jca.16539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park EH, Kim S, Jo JY, Kim SJ, Hwang Y, Kim JM, Song SY, Lee DK, Koh SS. Collagen triple helix repeat containing-1 promotes pancreatic cancer progression by regulating migration and adhesion of tumor cells. Carcinogenesis. 2013;34:694–702. doi: 10.1093/carcin/bgs378. [DOI] [PubMed] [Google Scholar]
- 21.Eriksson J, Le Joncour V, Nummela P, Jahkola T, Virolainen S, Laakkonen P, Saksela O, Hölttä E. Gene expression analyses of primary melanomas reveal CTHRC1 as an important player in melanoma progression. Oncotarget. 2016;7:15065–15092. doi: 10.18632/oncotarget.7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ip W, Wellman-Labadie O, Tang L, Su M, Yu R, Dutz J, Wang Y, Huang S, Zhang X, Huang C, Zhou Y. Collagen triple helix repeat containing 1 promotes melanoma cell adhesion and survival. J Cutan Med Surg. 2011;15:103–110. doi: 10.2310/7750.2011.10014. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 25.Price C, Chen J. MicroRNAs in cancer biology and therapy: current status and perspectives. Genes Dis. 2014;1:53–63. doi: 10.1016/j.gendis.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macfarlane LA, Murphy PR. MicroRNA biogenesis, function and role in cancer. Curr Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller DW, Rehli M, Bosserhoff AK. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J Invest Dermatol. 2009;129:1740–1751. doi: 10.1038/jid.2008.452. [DOI] [PubMed] [Google Scholar]
- 28.Butz H, Rácz K, Hunyady L, Patócs A. Crosstalk between TGF-β signaling and the microRNA machinery. Trends Pharmacol Sci. 2012;33:382–393. doi: 10.1016/j.tips.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Wendt MK, Tian M, Schiemann WP. Deconstructing the mechanisms and consequences of TGF-beta-induced EMT during cancer progression. Cell Tissue Res. 2012;347:85–101. doi: 10.1007/s00441-011-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, Cheng JQ. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das LM, Torres-Castillo MD, Gill T, Levine AD. TGF-beta conditions intestinal T cells to express increased levels of miR-155, associated with down-regulation of IL-2 and itk mRNA. Mucosal Immunol. 2013;6:167–176. doi: 10.1038/mi.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]