Abstract
Circular RNAs (circRNAs) as a family of non-coding RNAs are increasingly recognized regarding their biogenesis, regulatory roles in gene expression and clinic significance in developmental diseases and cancers. In this study, we aim to identify circRNAs that may be associated with clinicopathological characteristics of patients with bladder cancer. The circRNAs databases CircBase and circ2 Traits were used to seek circRNAs reported to bladder cancer. The expression levels of the circRNA of interest in paired samples of tumor tissue and adjacent normal mucosa from 61 patients with bladder cancer were detected by real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and statistically analyzed. Database search shows that circASXL1 (circBase ID: hsa_circ_0001136) transcribed from the ASXL1 gene locus is among the circRNAs with altered expressions in bladder cancer. Results showed that the expression level of circASXL1 was significantly higher in bladder cancer tissues compared to that in adjacent noncancerous tissues (P<0.001). To be noticed, chi-square tests support that the expression of circASXL1 significantly correlates with tumor grade (P=0.025), tumor stage (P=0.019), lymph node invasion (P=0.011) and distant metastasis (P=0.032). The area under ROC curve (AUC) is 0.770 for circASXL1 in predicting tumor invasion (T2-T4 tumors). Kaplan-Meier survival analysis indicates that tumors of high circASXL1 expression are associated with shorter overall survival compared to tumors of low circASXL1 expression. Further, multivariate analysis reveals that circASXL1 is an independent prognostic factor for overall survival for patients with bladder cancer. Expression of circASXL1 in bladder tumor correlates with TNM classification and may independently predict overall survival for patients with bladder cancer.
Keywords: Circular RNA, circASXL1, bladder cancer, biomarker
Introduction
Bladder cancer is the most common tumor malignancy of the urinary system, with estimated 5-year prevalence of 1, 110, 265 patients worldwide, ranking the 9th in cancer incidence [1]. In China, bladder cancer also has the highest incidence rate across all urinary system tumors and the bladder cancer-related mortality has been increasing overthe past decades. For patients diagnosed with muscle non-invasive bladder cancer which accounts for roughly 75% of bladder cancer, tumor recurrence and progression are significant risk factors linked to patient survival outcomes [2,3]. Therefore, molecular biomarkers predictive of tumorigenesis and development of bladder cancer are greatly needed in clinical course.
Circular RNAs (CircRNAs) are recognized as a naturally occurring family of non-coding RNAs characterized by covalently closed loop structures with neither 5’ to 3’ polarity nor a polyadenylated tail [4-6]. Since the discovery of circRNAs over 20 years ago [7,8], such molecules were typically considered as molecular flukes or products of aberrant RNAs plicing due to their low levels of expression. However, with the advancesin RNA deep sequencing technology and bioinfor matics, recent work has revealed that large numbers of circRNAs are endogenous, abundant, conserved and stable in mammalian cells [9-12]. Studies show that circRNAs can function as microRNA (miRNA) sponges, regulate alternative splicing, and modulate the expression of parental genes [12-16]. Accumulating evidence suggests that circRNAs may be involved in many disorders and cancers [17-19]. Certain circRNAs have been found aberrantly expressed in Hirschsprung’s disease [20] and heart failure [21]. In a series of more recent studies, circRNAs have been proposed as potential biomarkers for gastric cancer [22], colorectal cancer [23-25] and hepato cellular carcinoma [26].
In the latest study, 469 circRNAs were detected to be differentially expressed between bladder carcinoma and matched para-carcinoma tissue [27]. However, currently little is known regarding the clinicopathological implications of circRNAs in bladder cancer. In the present study, we used circRNA databases [28] to explore circRNAs that might be associated with bladder cancer. Database search highlights circASXL1, which is a transcription product from exons 2-3 within the ASXL1 gene locus, as a candidate remaining to be validated. We subsequently analyzed the expression of circASXL1 in tumor tissues and paired normal mucosa from 61 patients with bladder cancer, as well as its correlations with clinical characteristics of the patients.
Materials and methods
Clinical sample collection
This study was conducted upon approval of the Ethics Committee of Second Hospital of Tianjin Medical University. Written informed consent was received for all patients. A total of 61 pairs of tumor tissue and adjacent normal mucosa were obtained from bladder cancer patients who underwent radi-cal cystectomy at the Second Hospital of Tianjin Medical University between 2010 and 2011. The patients did not receive any treatment prior to surgery. The clinical specimens were snap-frozen in liquid nitrogen immediately after resection and stored at -80°C until use. All the bladder carcinomas were pathologically confirmed.
Clinical information
The clinical characteristics of patients including gender, age, tumor size, tumor number and vascular tumor embolus were retrieved from patient’s medical reports. Tumor staging was performed according to the World Health Organization classification and the sixth edition of TNM classification of the Union for International Cancer Control (UICC, 2002). Immunohistochemical staining and scoring protocols for Ki67 and p53 were previously described [29-31]. Ki67 level was scored by label index, and Ki67 level was considered high when samples demonstrated 40% or greater positivity. P53 level was semi-quantitated, as 0 for negative, 1+ for mild, 2+ for moderate and 3+ for strong. In our study, patients with bladder cancer were divided into the non-muscle-invasive group (Ta, T1 stage) or the muscle-invasive group (T2, T3, T4 stage) [32]. All patients were followed up retrospectively through hospital records and telephone interviews with either the patients or their close relatives.
Total RNA extraction and cDNA synthesis
Total RNA was isolated from frozen tissues in -80°C using TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacture’s instruction. Concentration and quality of the isolated RNA were analyzed by Nano Drop ND-1000 spectrophotometer (Nano Drop Thermo, Wilmington, DE). After RNA extraction, cDNA was synthesized according to the following method (Transcriptor First Strand cDNA Synthesis Kit, Roche, Germany). First, 2 µg RNA were dilutedinto 10 µl RNase-free water in a 0.2 ml PCR tube, and then 1 µl random primers was added and mixed well. Second, the tubes were put into a PCR thermocycler and incubated at 65°C a for 10 minutes. Third, place the tubes on ice immediately and prepare the reaction mix as following: 4 µL 5× Buffer, 2 µL dNTP (10 mM), 0.5 µL Transcriptor Reverse Transcriptase, 0.5 µL RNase-free water. The reaction mix was incubated at 55°C for 30 minutes, 85°C for 5 minutes and 4°C forever.
Real-timequantitative PCR
qRT-PCR was conducted with 2× lightcycler 480 SYBR Green I Master (Roche). The thermocycler program was set as 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 60 sec, and 72°C for 30 sec. Specific divergent primers for circASXL amplification were designed as below: forward, 5’-TAAACTGCCTGGCCGAATC-3’ and reverse 5’-TCCTTCTGCCTCTATGACCTG-3’, and synthesized by GenePharma Company (Shanghai, China). As internal control, human U6 was amplified using forward primer 5’-CTCGCTTCGGCAGCACA-3’ and reverse primer 5’-AACGCTTCACGAATTTGCGT-3’. The product of qRT-PCR was further confirmed by Sanger sequencing. The relative expression level of circASXL1 wascalculated using 2-ΔΔCt method.
Statistical analysis
All data analyses were performed with SPSS 24.0 software. GraphPad Prism 5.0 was used to plot all graphs. Paired student’s t test was applied to the comparison of circASXL1 expression between bladder cancer tissues and adjacent normal mucosa, while one-way ANOVA was used for the comparison among multiple pathological subgroups. The associations of circASXL1 expression with clinical characteristics of patients were examined by chi-square test.Receiver operating characteristic (ROC) curve was constructed to evaluate the diagnostic accuracy of circASXL1 for tumor invasion. The postoperative survivalrate was analyzed using Kaplan-Meier method, and the log-ranktest was used to assess the level of significance for difference between the survival curves. Multivariate Cox proportional regression analysis was performed to determine the independent contribution of clinicopathological factors for overall survival. P<0.05 was considered statistically significant.
Results
Expression of circASXL1 in bladder cancer
Specific divergent primers were designed for amplification using cDNA as template. Sequencing results show that the RT-PCR product matches circASXL1 in circBase (http://www.circbase.org, ID: has_circ_0001136), indicatinga success forthe detection of circASXL1 in total RNA. Next, we examined the relative expression levels of circASXL1 in paired samples of bladder cancer tissue and adjacent normal mucosa from 61 patients by qRT-PCR. Results show that the relative level of circASXL varies considerably between patients (Figure 1A). And the expression level of lncRNA-n336928 was significantly higher in bladder cancer tissues compared to that in paired normal tissues (Figure 1B, P<0.001).
Figure 1.

Relative expressions of circASXL1 in bladder cancer tissue and adjacent normal tissue. A. Ratio of circASXL1 expression in BC tissue versus normal bladder mucosa for each patient based on qRT-PCR results. Each column represents the mean of 3 independent experiments. B. Box plot shows the relative expressions of circASXL1 in 61 paired samples of human BC tissues and normal tissues. circASXL1 expression was significantly higher in bladder cancer tissue samples compared to matched normal tissue samples (paired student’s t test, P<0.001). BC = bladder cancer, qRT-PCR = real-time quantitative reverse transcription-polymerase chain reaction.
Correlations between circASXL1 expression and clinicopathological factors in bladder cancer
The circASXL1 expression was categorized as low or high according to the median level of circASXL1 in bladder cancer tissues (n=61) for the convenience for correlation analysis. Chi square test indicates that the level of circASXL1 expressionis not obviously associated with patent’s gender, age, tumor size, tumor number, vasculartumor embolus, Ki67 level and P53 level. However, the level of circASXL1 expression is associated with tumor grade, tumor stage, lymph node invasion and distant metastasis (Table 1). Further, stratification analysis indicates that circASXL1 expression does differ in subgroups based on T classification (Figure 2A), lymph node invasion, (Figure 2B), distant metastasis (Figure 2C) or grade (Figure 2D), showing a tendency that advanced bladder tumors may have higher levels of circASXL1. To evaluate the diagnostic value of circASXL1 on tumor invasion (T2-T4 bladder cancer), ROC curve was plotted. The area under the ROC curve (AUC) is 0.770, and the sensitivity and specificity are 0.686 and 0.769, respectively (Figure 3), suggesting a modest accuracy forcircASXL1 in diagnosis of tumor invasion.
Table 1.
Correlations between circASXL1 expression and clinical parameters in bladder cancer
| Characteristics | No. of patients | CircASXL1 expression | P value | |
|---|---|---|---|---|
|
| ||||
| High | Low | |||
| N (%) | 61 (100.0) | 27 (44.3) | 34 (55.7) | |
| Gender | 0.763 | |||
| Female | 17 (27.9) | 7 (25.9) | 10 (29.4) | |
| Male | 44 (62.1) | 20 (74.1) | 24 (70.6) | |
| Age | 0.842 | |||
| ≤65 | 37 (60.7) | 16 (59.3) | 21 (61.8) | |
| >65 | 24 (39.3) | 11 (40.7) | 13 (38.2) | |
| Tumor size | 0.355 | |||
| ≥3 cm | 21 (34.4) | 11 (40.7) | 10 (29.4) | |
| <3 cm | 40 (65.6) | 16 (59.3) | 24 (70.6) | |
| Tumor number | 0.251 | |||
| ≤2 | 43 (70.5) | 17 (63.0) | 26 (76.5) | |
| >2 | 18 (29.5) | 10 (37.0) | 8 (23.5) | |
| Vascular tumor embolus | 0.158 | |||
| Negative | 46 (75.4) | 18 (66.7) | 28 (82.4) | |
| Positive | 15 (24.6) | 9 (33.3) | 6 (17.6) | |
| Grade | ||||
| G1 | 18 (29.5) | 4 (14.8) | 14 (41.2) | 0.025 |
| G2-G3 | 43 (70.5) | 23 (85.2) | 20 (58.8) | |
| T classification | 0.019 | |||
| Ta-T1 | 26 (42.6) | 7 (25.9) | 19 (55.9) | |
| T2-T4 | 35 (57.4) | 20 (74.1) | 15 (44.1) | |
| LN invasion | 0.011 | |||
| Negative | 42 (68.9) | 14 (51.9) | 28 (82.4) | |
| Positive | 19 (31.1) | 13 (48.1) | 6 (17.6) | |
| Distant metastasis | 0.032 | |||
| Negative | 51 (83.6) | 19 (70.4) | 32 (94.1) | |
| Positive | 10 (16.4) | 8 (29.6) | 2 (5.9) | |
| Ki67 | 0.192 | |||
| ≤40% | 35 (57.4) | 11 (40.7) | 24 (70.6) | |
| >40% | 26 (42.6) | 16 (59.3) | 10 (29.4) | |
| P53 | 0.754 | |||
| 0 | 28 (45.9) | 13 (48.1) | 15 (44.1) | |
| +-+++ | 33 (54.1) | 14 (51.9) | 19 (55.9) | |
Number with percentage is given in brackets. LN = invasion lymph node invasion.
Figure 2.
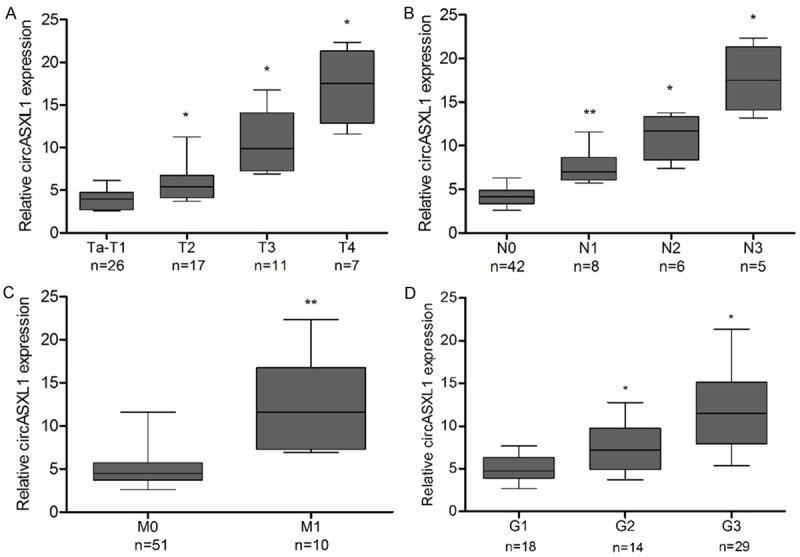
Box plots show the relative expressions of circASXL1 in bladder tumors in terms of T classification (A), N classification (B), distant metastasis (C), or histopathologic grade (D). *P<0.05, **P<0.01.
Figure 3.
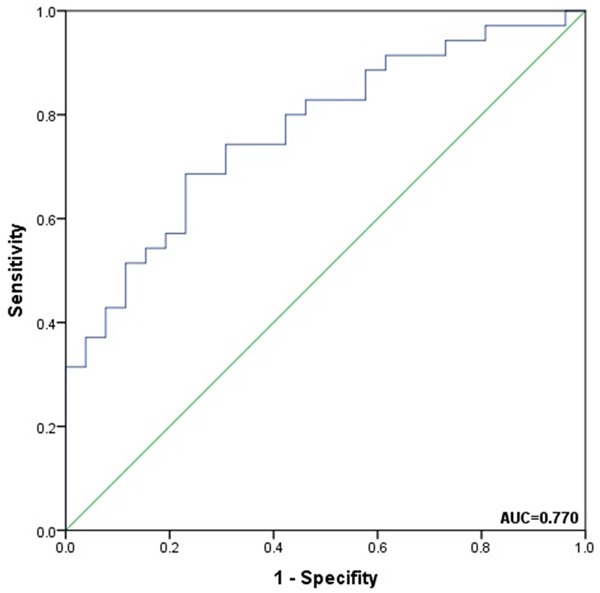
ROC curves for CircASXL1 in TNM (Ta-T1, T2-T4) diagnosis in bladder cancer. The area under curves (AUC) is 0.770; the sensitivity and specificity are 0.686 and 0.769, respectively. ROC = receiver operating characteristic, AUC = area under the curve.
Correlation of circASXL1 expression and patient survival
We explored the correlation of circASXL1 expression level (high vs. low) with patient overall survival in bladder cancer. Kaplan-Meier curves show that tumors of high circASXL1 expression are associated with shorter overall survival compared to tumors of low circASXL1 expression. In this study, the median follow-up was 34 months (range, 1 to 60 months). The median overall survival time was 44.6 months in the high expression group versus 54.3 months in the low expression group (Figure 4, P=0.003). To determine the prognostic significance of circASXL1 expression, univariate analysis was conducted first and led to the finding that tumor grade, TNM classification and circASXL1 expression are factors linked with overall survival. Multivariate Cox proportional hazards regression analyses with adjustment for tumor grade, stage, lymph node invasion and metastasis indicate that circASXL1 expression level is an independent prognostic factor for 5-year overall survival of patients with bladder cancer in the cohort (Table 2).
Figure 4.
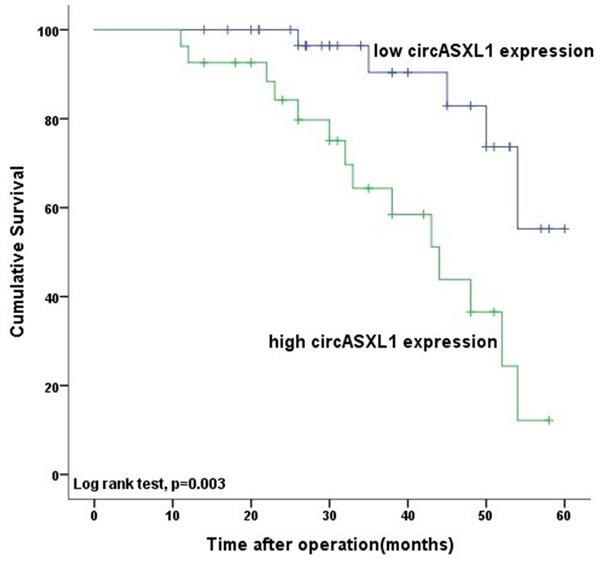
Kaplan-Meier curves show the overall survival rates of patient groups classified by the circASXL1 expression level in tumor tissue. Patients with high circASXL1 expression show significantly shorter overall survival than those with low circASXL1 expression (P=0.003, log-rank test).
Table 2.
Univariate and multivariate analysis of prognostic factors with respect to 5-year overall survival of patients with bladder cancer
| Covariant | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Exp (B) | 95% CI | P value | Exp (B) | 95% CI | P value | |
| Gender | 0.686 | 0.361-0.976 | 0.716 | |||
| Age | 0.964 | 0.379-2.455 | 0.939 | |||
| Tumor size | 1.898 | 0.749-4.809 | 0.177 | |||
| Tumor number | 1.702 | 0.678-4.273 | 0.257 | |||
| Vascular tumor embolus | 1.603 | 0.556-4.626 | 0.382 | |||
| Grade | 3.539 | 0.892-11.238 | 0.028 | 2.813 | 0.642-9.281 | 0.072 |
| T classification | 4.082 | 1.333-12.500 | 0.016 | 1.723 | 1.013-6.353 | 0.038 |
| LN invasion | 6.312 | 2.084-19.115 | 0.008 | 2.828 | 1.124-6.193 | 0.028 |
| Distant metastasis | 3.904 | 1.561-9.767 | 0.011 | 2.138 | 1.043-5.849 | 0.026 |
| Ki67 | 1.135 | 0.460-2.800 | 0.784 | |||
| P53 | 1.102 | 0.433-2.804 | 0.838 | |||
| circASXL1 expression | 4.831 | 1.718-13.514 | 0.007 | 2.214 | 1.002-5.647 | 0.046 |
CI confidence interval, LN = invasion lymph node invasion.
Discussion
The emerging circRNAs are increasingly ac knowledged as a kind of non-coding RNAs that play roles in gene regulation and disease pathogenesis. The diagnostic and prognostic values of circRNAs in cancers are gaining more interest as exemplified in studies of gastric cancer [22], colorectal cancer [23-25] and hepatocellular carcinoma [26].
More recently, a number of circRNAs werereportedto be differentially expressed in bladder cancer tissue compared with adjacent non-cancerous tissuein terms of microarray data [27]. Among the differentially regulated circRNAs, circASXL1, a circular RNA transcribed from the ASXL1 gene locus draw our attention. In the present study, we detected the expression of circASXL1 in paired samples of tumor tissue and normal mucosa from 61 patients with bladder cancer by using qRT-PCR. Based on our data, the expression level of circASXL1 varies considerably between individuals, and we observed that circASXL1 expression was significantly higher in bladder tumor tissues compared to that in matched adjacent normal mucosa.
Importantly, our study shows that the expression of circASXL1 is significantly associated with tumor grade and TNM classification, and that more aggressive tumors tend to have higher expression of circASXL1, implicating a diagnostic role of circASXL1 in bladder cancer. Thus, we employed ROC curves to evaluate the performance of circASXL1 in tumor invasion prediction. Although specificity and sensitivity results simply support a modest efficacy of circASXL1 for tumor invasion diagnosis, it is expected to improve diagnostic accuracy by using circASXL1 combined with other circRNAs in future studies. Here, our study also highlights a prognostic role of circASXL1 in bladder cancer. Multivariate analysis suggests that circASXL1 can independently predict the patient overall survival. However, considering the potential bias that may result from relatively small patient number and high ratio of patient with high-grade T1 tumors in our study, the prognostic significance should be validated by multiple independent studies with larger cohorts before it is taken generally.
Although circRNAs have been associated with clinic characteristics of patients with bladder cancer, the mechanisms by which circRNAs play pathological roles in bladder cancer are just begun to be revealed. In a recent study by Zhong et al, circTCF25 was shown to down-regulate miR-103a-3p and miR-107, increase CDK6 expression, and promote proliferation and migration in vitro and vivo in bladder carcinoma [27]. Additionally, quite a few studies have demonstrated the regulatory role of circRNAs in gene expression by functioning as microRNA sponges [17]. Therefore, it will be interesting to explore microRNAs that are targeted by circASXL1 in bladder cancer using cell line models in future studies.
In summary, our study for the first time shows circASXL1 is associated with tumor TNM classification and may predict overall survival in bladder cancer. Further studies are expected to reveal the mechanisms underlying the pathological roles of circASXL1 in bladder cancer.
Acknowledgements
This study was supported in part by grants from the Natural Science Foundation of Tianjin (No. 15JCYBJC24600).
Disclosure of conflict of interest
None.
References
- 1.Ploeg M, Aben KK, Kiemeney LA. The present and future burden of urinary bladder cancer in the world. World J Urol. 2009;27:289–293. doi: 10.1007/s00345-009-0383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao F, Lin T, He W, Han J, Zhu D, Hu K, Li W, Zheng Z, Huang J, Xie W. Knockdown of a novel lincRNA AATBC suppresses proliferation and induces apoptosis in bladder cancer. Oncotarget. 2015;6:1064–1078. doi: 10.18632/oncotarget.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhuang J, Lu Q, Shen B, Huang X, Shen L, Zheng X, Huang R, Yan J, Guo H. TGFbeta1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci Rep. 2015;5:11924. doi: 10.1038/srep11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 8.Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 9.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 15.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 16.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 17.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 18.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng L, Chen G, Zhu Z, Shen Z, Du C, Zang R, Su Y, Xie H, Li H, Xu X, Xia Y, Tang W. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung’s disease. Oncotarget. 2017;8:808–818. doi: 10.18632/oncotarget.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 22.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 23.Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG, Zhang B. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982. doi: 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, Liang L. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, Wang C. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular rna biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltimore) 2016;95:e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng C, Wang L, Ding G, Ding Q, Zhou Z, Jiang H, Wu Z. Predictive value of clinicopathological markers for the metachronous bladder cancer and prognosis of upper tract urothelial carcinoma. Sci Rep. 2014;4:4015. doi: 10.1038/srep04015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Feng C, Ding G, Ding Q, Zhou Z, Jiang H, Wu Z. Ki67 and TP53 expressions predict recurrence of non-muscle-invasive bladder cancer. Tumour Biol. 2014;35:2989–2995. doi: 10.1007/s13277-013-1384-9. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Feng C, Ding G, Zhou Z, Jiang H, Wu Z. Relationship of TP53 and Ki67 expression in bladder cancer under WHO 2004 classification. J BUON. 2013;18:420–424. [PubMed] [Google Scholar]
- 32.Sobin LH, Wittekind CH. International union against cancer (UICC) 2002 [Google Scholar]


