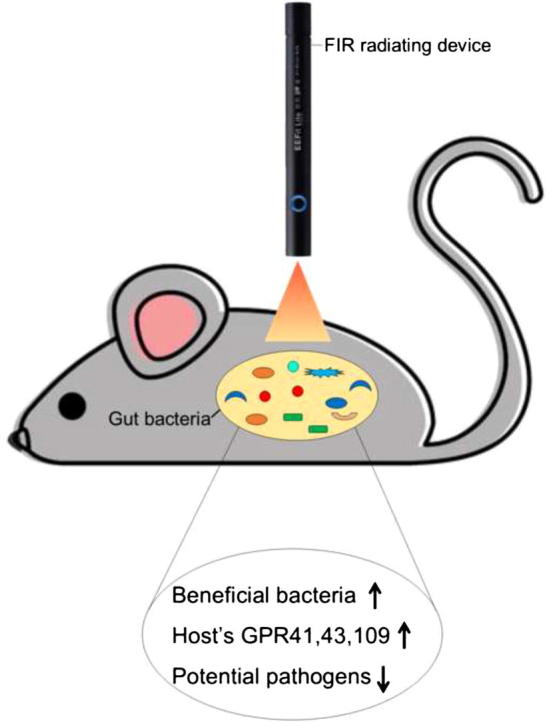
Graphical abstract
Keywords: Far infrared, Gut microbiota, ERIC-PCR, 16S amplicon sequencing, SCFA, GPCR
Highlights
-
•
Transient exposure of FIR induced compositional and temporal changes in gut microbiota of mice.
-
•
FIR exposure stimulated growth of the gut beneficial bacteria.
-
•
FIR exposure promoted growth of the gut SCFAs-producing bacteria.
-
•
FIR treatment upregulated the expressions of the SCFAs-sensing G-protein coupled receptors in the intestinal mucous of the mice.
Abstract
Far infrared radiation (FIR) has been widely used to treat chronic diseases and symptoms; however, the underlying mechanism remains unclear. As gut microbiota (GM) markedly impact the host’s physiology, making GM a potential target for the therapeutic evaluation of FIR. C57BL/6J mice were exposed to five times of 2 min-FIR exposure on the abdomen, with a two-hour interval of each exposure within one day. Fecal samples were collected on day one and day 25 after the FIR/control treatment, and the extracted fecal DNAs were evaluated using ERIC-PCR and 16S amplicon sequencing. Host’s G-protein coupled receptors (GPCR) were analyzed using qRT-PCR. FIR induced immediate changes in the GM composition. A prompt and significant (p < 0.05) reduction in the abundance of phylum Deferribacteres (comprised of several pathogens) was observed in the FIR-irradiated mice compared to the control group. Contrarily, FIR exposure induced beneficial genera such as Alistipes, Barnesiella, and Prevotella. The gut of FIR-irradiated mice was predominated by short-chain fatty acids (SCFAs) producers. Also, FIR stimulated the expression of SCFAs-sensing receptors, GPCR 41, 43, and 109 in the gut epithelial barrier. These findings provide the first-hand evidence in which the beneficial effects of FIR radiation might be partially through the modulation of GM.
Introduction
About 54.3% of the sunlight that arrives the earth is infrared rays [1], [2]. Infrared radiations have been sub-classified, among which, far-infrared (FIR) can transfer energy to other objects in the form of heat [3]. Several studies have reported health-promoting properties of FIR in the murine and cell models. For instance, FIR has shown anti-inflammatory activities by inhibiting IL-6 and TNF-α in a peritonitis mouse model [4]. In another study, Chang et al. reported that FIR could protect spinocerebellar ataxia cells by inhibiting PolyQ protein accumulation and improving mitochondrial function. PolyQ disease is a rare neurodegenerative disease and lacking an effective treatment strategy [5]. Similarly, anti-cancer abilities of FIR have also been observed by the growth arrest of HSC3, A549, and Sa3 cancerous cells through the upregulated expression of the ATF3 gene that led to the activation of tumor suppressor gene p53 [6]. Likewise, FIR has shown to inhibit the growth of spontaneous mammary tumors in a mouse model [7]. However, despite all these beneficial abilities of FIR, the fundamental mechanism is still unknown.
It is well recognized that commensal microbes play an integral role in the host’s digestion and immune systems [8]. Any perturbation in the diversity and composition of gut microbiota (GM) could severely impact the host physiology [9]. Some of the external stimuli that can affect GM composition include ingested foods, dietary supplements, and antibiotic treatments. How would radiation energy, such as FIR, affect the composition of the gut microbiome is unknown. Thus, in this study, we aimed to evaluate the impact of FIR on GM in the C57BL/6J mouse model and to unveil the mystery behind the health benefit of the FIR radiation. In this study, mice were given five consecutive exposures to FIR within 12 h. The fecal GM composition was determined using Enterobacterial Repetitive Intergenic Consensus (ERIC)-PCR and 16S rRNA sequencing. The physiological effect of the host was determined by the expression of SCFA-sensing G-protein coupled receptors in the gut epithelial barrier.
Materials and methods
FIR radiating device
Several FIR-emitting devices are commercially available [3]. In this study, we used EEFit® Pen, a FIR emitting device commercially available (Nick Wang Technology Limited). This handheld device emits electromagnetic waves of 4 – 20 µm with 85.61% average FIR emissivity and photon energy level 12.4 MeV–1.7 eV [10].
Animals maintenance and treatment
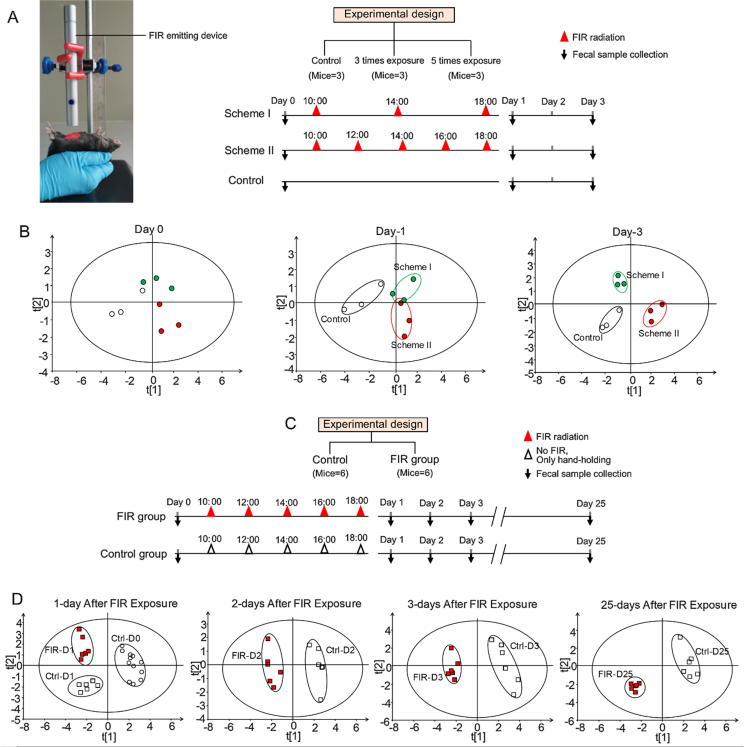
Mice were kept in cages with free access to food (PicoLab®Rodent 20–5053; LabDiet, USA) and water. Mice were housed in a 12 h’ light-dark cycle facility in the Animal Center of the Macau University of Science and Technology. For FIR radiation treatment, the mouse was held by hand with the belly facing up and keeping at a 2 cm distance from the FIR emission device which was mounted on a stand (Fig. 1A). To rule out any stress-induced changes on the GM composition, the control mice were also held by hand for the same time frame as the FIR-treated mice. The treatment schemes are illustrated in Fig. 1. In brief, each FIR radiation lasted for 2 min with either 4 h (Scheme I) or 2 h (Scheme II) interval between each radiation. Fecal samples were collected from individual mice before and after FIR-treatment as indicated in the treatment schemes (Fig. 1). All the fecal samples were stored at −80 °C for later DNA extraction.
Fig. 1.
Graphical presentation of experimental model and ERIC-PCR based analysis of GM in control and FIR-irradiated mice. (A) The setting of the FIR irradiation and the experimental design. FIR-emitting device was mounted on a stand and adjusted to a height of 2 cm against the mouse abdomen. The mouse was held by hand with the belly up for receiving FIR irradiation. Nine mice were housed together in the same cage for 7–10 days before each experiment, then randomly divided into experimental groups in separated cages. (B) PLS-DA plots of ERIC-PCR DNA profiles of the FIR-treated and the control mice (n = 3). Fecal genomic DNAs were subjected to ERIC-PCR and the resulting DNA banding patterns on the gel were digitized by Image Lab 3.0 system (Bio-Rad). Based on the distance and the intensity of each DNA bands, SIMCA-P 14.0 tool (Umetrics, Umea, Sweden) with 95% (p < 0.05) confidence level was applied to obtain the PLS-DA score plots (Chen et al. 2016). Each symbol in the PLS-DA plots (Panels B&D) represents the GM profile of each experimental mouse. All the mice were in same cage before treatment and were marked with green, red or white dots to track down the movement of GM of each mouse over time. (C) FIR-treatment Scheme (n = 6). 12 mice were housed together in the same cage until day-0, then randomly allocated to each experimental group in a separated cage. (D) PLS-DA plots of ERIC-PCR assays for fecal DNAs obtained from the treated and control mice on D1, D2, D3 and D25.
Genomic DNA extraction, ERIC-PCR analysis, and 16S amplicon sequencing
Total genomic DNA was extracted from fecal samples using QIAamp DNA Stool Mini Kit (QIAGEN) following the manufacturer’s protocol. The extracted DNA samples were analyzed for similarity among groups using conserved ERIC regions with a pair of ERIC-1 and ERIC-2 primers and plotted with PLS-DA tool as previously described [11]. DNA samples were sequenced for 16S rRNA genes using Illumina MiSeq (Illumina, San Diego), targeting the V3–V4 region with barcoded 515F and 806R universal primers and processed as previously described [12]. Briefly, dual-index barcodes and Illumina sequencing adapters were used to join the reads using limited PCR cycle. After purification with Agencourt AMPure beads (Agencourt, USA), Nextera XT protocol was used for library normalization. And then, samples were loaded into a single flow cell for sequencing on the MiSeq sequencing platform (Illumina, San Diego) according to the manufacturer’s protocol. Clusters were generated and paired-end sequenced with dual index reads in a single run with a read length of 2 × 300 bp. PANDAseq was used for collecting paired-end sequences, and Raw FASTQ files were obtained. Sequences were trimmed of primers and barcodes. All the reads with ‘N’ and those with sequences <250 bp were removed. The cleaned sequences were then clustered at k = 10 (97% similarity) followed by deletion of chimeras and singleton reads. Finally, operational taxonomic units (OTUs) were classified using BLASTn against a 16S National Center for Biotechnology Information (NCBI)-derived database (http://www.ncbi.nlm.nih.gov, http://rdp.cme.msu.edu). The data has been submitted to NCBI SRA under ID PRJNA514213.
Quantitative real time polymerase chain reaction analysis
At the end of Day 1 and Day 25, mice (n = 5) were euthanized, intestinal mucosal tissues were collected for RNA extraction RNeasy Mini Kit (QIAGEN, Hilden, German) according to the manufacturer's protocol. The mRNA expressions of GPRs 41, 43 and 109 were conducted using qRT-PCR analysis as previously described [11]. The q-PCR analysis was performed on fecal DNA samples to identify the relative abundance of SCFA- producing bacteria and the specific gene sequences encoded the bacterial enzymes (butyrate transferase, butyrate kinase and mm-CoA decarboxylase) involved in the synthesis of the SCFAs. Specific primer sequences used for the PCR analysis were listed in Table 1. β-actin was employed as the internal control for qRT-PCR. All qPCR assays were normalized with a universal primer set for the 16S rRNA gene of total bacteria. The 2–ΔΔCt method was applied to calculate the fold change of relative gene expression. ΔΔCt = (Ct treatment_target gene − Ct treatment_reference gene) − (Ct control_target gene − Ct control_reference gene).
Table 1.
List of primers used for Clostridial cluster and GPR amplification.
| Target gene | Nucleotide sequence of primer (5′–3′) |
Reference | |
|---|---|---|---|
| Forward | Reverse | ||
| C. Cluster IV | GCACAAGCAGTGGAG T | CTTCCTCCGTTTTGTCAA | [1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40] |
| C. Cluster XIV | TGACCGGCCACATTGGGACTG | TCATCCCCACCTTCCTCCAG | [34] |
| C. Cluster XIVa | CTTTGAGTTTCATTCTTGCGAA | GCAGTGGGG AATATTGCA | [35] |
| But Transferase | ggWatWggMgsYatgcc | aaRtcaaSctgKccDc | [36] |
| But Kinase | tgctgtWgttggWagaggYgga | gcaacIgcYttttgatttaatgcatgg | [36] |
| mm-CoA decarboxylase | AATGACTCGGGIGGIGCIMGNATHCARGA | GATTGTTACYTTIGGIACNGTNGCYTC | [37] |
| GPR41 | GGGGTCGATACAAGAGT | CTGGCGGAGCTACGTGCT | [38] |
| GPR43 | TTCTTACTGGGCTCCCTGCC | TACCAGCGGAAGTTGGATGC | [39] |
| GPR109 | TCAGATCTGACTCGTCCACC | CCATTGCCCAGGAGTCCGAAC | [40] |
Statistical analysis
R packages phyloseq (1.22.3) was used for alpha and beta diversity analysis. Linear discriminant analysis effect size (LEfSe) was performed with bioBakery (version 17). Alpha values were set to 0.05 whereas the threshold on the logarithmic score of linear discriminant analysis was ≥2.0. SPSS (version 22) for statistical analysis. Data normality was ascertained with Kolmogorov–SmirnovD test. Statistical significance was ascertained with Mann-Whitney U and Wilcoxon signed-rank tests.
Results
Optimization of the FIR exposure scheme
Since there was no prior reference to the effect of FIR radiation on gut microbes, our first task was to check whether GM would respond to FIR radiation and what would be the FIR exposure scheme to detect significant changes in GM. For this purpose, nine mice from the same cage were randomly divided into three groups: namely, the control group, the 1st group received three exposures of FIR radiation and designated as Scheme I, and the 2rd group received five exposures of FIR and designated as Scheme II (Fig. 1A). Based on the ERIC-PCR results, we found that the Scheme II yielded greater separation between the FIR-treated and the control groups (R2 = 0.63) compared to the group separation (R2 = 0.37) in Scheme I based on the PERMANOVA test (Fig. 1B).
Transient exposure to FIR induced compositional and temporal changes in GM profile assessed by ERIC-PCR based analysis
Henceforth, the Scheme II was adopted for the subsequent experiments (Fig. 1C). Under this scheme, we evaluated both the transient and the longitudinal effects of FIR using ERIC-PCR analysis on fecal samples collected on Day1 (D1), Day 2 (D2), Day3 (D3) and Day 25 (D25) from both treated and the control groups. To our surprise, the FIR-induced changes in GM composition sustained up to 25 days after the initial exposure on D0 (Fig. 1D).
16S sequencing of the mouse fecal DNAs revealed the FIR treatment is in favor of the growth of beneficial bacteria
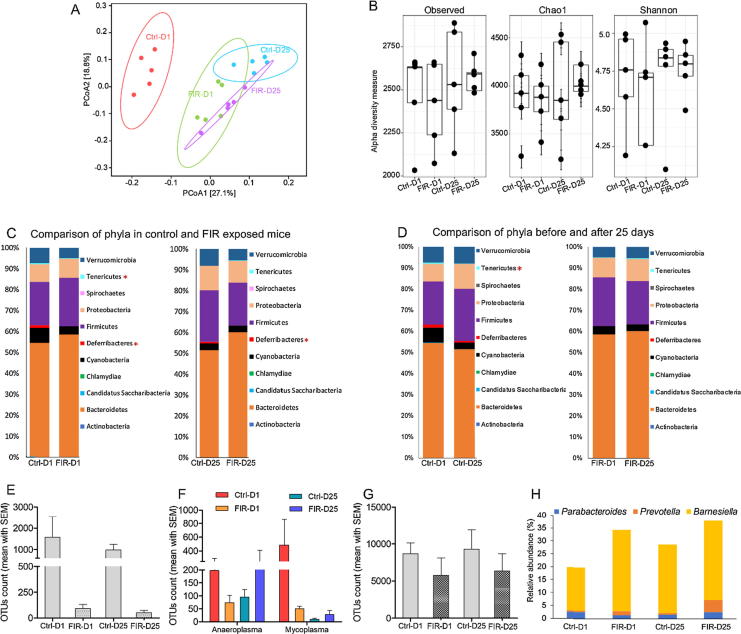
For in-depth GM analysis, 16S sequencing experiment was carried out (Fig. 2A). Like ERIC-PCR analysis, UniFrac analysis separated 16S sequences of the FIR-treated mice from that of control mice (Fig. 2A). There was a slight decrease in the OTUs diversity in the FIR-D1 (Chao1 = 3844.97, Shannon = 4.6) compared to the Ctrl-D1 mice (Chao1 = 3875.83, Shannon = 4.6); however, enrichment of OTUs diversity was observed later at D25 (Chao1 = 4057.67, Shannon = 4.7) (Fig. 2B).
Fig. 2.
16S rRNA gene sequencing analysis of the fecal DNAs collected from the FIR-treated and the control mice. The mice were exposed to FIR radiation according to the Scheme II described in Fig. 1C. The control mice were held by hand without FIR treatment. N = 5/group. (A) Weighted UniFrac distance analysis of the top 200 most abundant OTUs in the fecal samples collected from animal model shown in Fig. 1C. X and Y axis are showing GM separation among the groups based on distance analysis. Each dot represents the top 200 OTUs of the individual mouse. (B) Alpha diversity analysis of GM in the samples collected from the mouse model shown in Fig. 1C. Every dot is representing total OTUs of the individual mouse. Data presented in Fig. 2B and C was analyzed and visualized with R package phyloseq. (C) Comparison of the relative abundance of the detected phyla in the control and FIR-irradiated mice. Statistical significance was done with Mann-Whitney U test. (D) Longitudinal comparison of the relative abundance of the phyla detected in the control and FIR exposed mice. Statistical significance was done with Wilcoxon signed-rank test. (E) Number of OTUs assigned to genus Mucispirillum. (F) Number of OTUs assigned to genera Anaeroplasma and Mycoplasma. (G) Number of OTUs assigned to Akkermansia muciniphila. (H) Relative percentile abundance of the three genera that are contributing to the enrichment of phylum Bacteroidetes in FIR treated mice.
Particularly, the abundance of phyla Tenericutes and Deferribacteres was significantly (p = 0.032 and p = 0.049, respectively) reduced in the gut of FIR-D1 compared to Ctrl-D1 mice (Fig. 2C). These two phyla are comprised several pathogenic bacteria that dwell in the gut and the buccal mucosa of the host [13], [14]. Lower abundance of Deferribacteres in the FIR-irradiated mice (Fig. 2C) was mainly related to the reduced abundance of Mucispirillum (Fig. 2E). Similarly, the decrease of Tenericutes (Fig. 2C and D) was mainly related to the decreased abundance of genus Mycoplasma (Fig. 2F). Moreover, the suppressed abundance of Verrucomicrobia with FIR exposure (Fig. 2C) was contributed to the lower OTUs count of Akkermansia muciniphila (Fig. 2G), a commensal bacterium found to be negatively correlated with inflammation and obesity [15]. Furthermore, an increasing trend in the prevalence of Bacteroidetes was observed in FIR-exposed mice (Fig. 2C), which is mainly caused by the enhanced growth of Barnesiella and Prevotella species (Fig. 2H). Complete lists of FIR-enhanced and -suppressed bacteria are shown in Fig. S1 and Fig. S2, respectively.
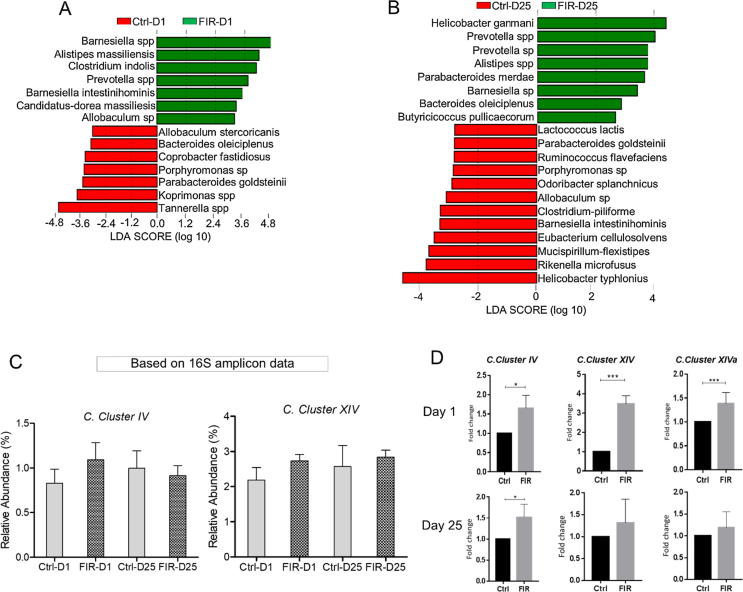
Based on the linear discriminant analysis (LEfSe), on D1, FIR radiation mainly induced the growth of Barnesiella spp., Alistipes massiliensis, Clostridium indolis, Prevotella spp., and Barnesiella intestinihominis (Fig. 3A). On D25, the FIR-treated mice displayed enrichment of Bacteroides oleiciplenus, Butyricicoccus pullicaecorum, Parabacteroides merdae, and Helicobacter ganmani (Fig. 3B).
Fig. 3.
Comparison of the differentially abundant species in the fecal DNAs from the FIR-treated and the control mice using linear discriminant analysis (LEfSe), and qPCR assay. The analysis was performed on OTUs comprising ≥97% of the total abundance in each group. LEfSe analysis of the bacterial species between control and FIR-exposed mice on D1 (A) and D25 (B) as described in Fig. 3A. Data analysis was carried out with bioBakery. Threshold parameters were set as: p = 0.05 Kruskal-Wallis; LDA score was set to 3, and multi-class analysis = all against all. (C) Comparison of the relative abundance of Clostridium cluster IV and XIV. Species that belong to these clusters were subset and comparatively analyzed. (D) qPCR analysis of the main SCFAs-producers in fecal DNAs of the FIR-treated and control mice on D1 and D25. Panels 3C-E were generated using GraphPad Prism version 5.01.
FIR enhanced the relative abundance of short-chain fatty acids (SCFAs) producing bacteria
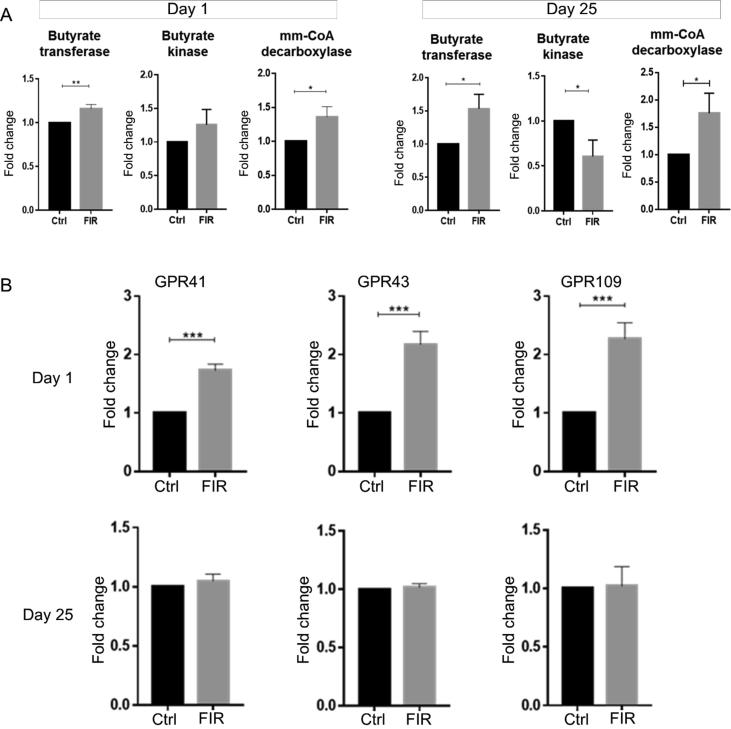
Another interesting finding was the striking enhancement of the relative abundance of Clostridium clusters in the gut of FIR-irradiated mice (Fig. 3C). These clusters are known to produce SCFAs, especially for butyrate [16], [17], [18]. The enrichment of Clostridia clusters was further confirmed through the qPCR assays with the species-specific primers (Table 1) (Fig. 3D). Remarkably, the FIR-induced effects sustained 25 days from the initial FIR exposure (Fig. 3C and D). We also examined and compared the relative levels of SCFAs-producing bacteria based on the presence of the gene sequences that are involved in the SCFAs synthesis pathways. Using sets of specific degenerate PCR primers, we conducted qPCR on three relevant gene sequences in the terminal steps of SCFAs synthesis, i.e., butyrate transferase (But), butyrate kinase (BuK) and malonyl‐CoA decarboxylase (MatA). The results showed that the relative levels of all three gene sequences were elevated in FIR-treated mice compared to the control, and the elevated level was detectible at least until D25 (Fig. 4A).
Fig. 4.
(A) Quantitative analysis of the three main key enzymes in the SCFA synthetic pathways using qPCR technique. Specific primer used in each PCR reactions are listed in Table 1. Significance was generated with t-test. (B) mRNA expressions of SCFAs-sensing receptors. qRT-PCR was performed for the expressions of GPR41, 43, and 109 in the intestinal mucosa of the FIR-treated and the control mice. The GPRs specific primers used for the qRT-PCR were listed in Table 1. Figures were plotted using GraphPad Prism version 5.01.
FIR upregulated key GPCR genes in mice
One of the important biological effects of SCFAs is the activation of various host’s GPCRs and influence an array of cellular responses to the benefits of the host’s health [19]. GPCRs are expressed at the surface of the gut epithelial cells, play essential roles in promoting gut homeostasis, and regulating inflammatory responses [20]. Bacterial metabolites, such as SCFAs, bind to the GPCRs as ligands and mimic the host signaling molecules [21]. We found upregulation of three main GPCRs genes, i.e., GPRs 41, 43, and 109a in the mice 24 h after the FIR exposure (Fig. 4B).
Discussion
FIR application is growing in various biomedical fields and health providers, yet there is no measurable parameter to assess its beneficial effects. GM is known to play an essential role in host metabolism and immunity; and, any event-induced perturbation in this community imposes direct implication on the host’s health [22]. Therefore, make GM a potential biological system to assess the unknown healing abilities of FIR. In this study, we evaluated FIR effects on GM diversity and composition in C57BL/6J mice. Also, the observed microbial changes were associated with the expression of GPCR encoding genes. It is worth mentioning that the EFFit Pen emitting radiation can be gaged and delivered to a confined area of the mouse abdomen in our study.
In this study, we showed that several potential pathogenic bacteria were significantly reduced in mice upon FIR radiation. The genus Mycoplasma under Tenericutes constitutes several mucosal pathogenic bacteria that can cause acute infection in the host [14]. On the other hand, another genus Anaeroplasma from Tenericutes remained at high abundance in the FIR-exposed mice on D25. Anaeroplasma is considered a potential new probiotic genus against chronic inflammatory diseases. This group of bacteria possesses anti-inflammatory properties by inducing cytokine TGF-β and improves the intestinal barrier by enhancing mucosal IgA in mice [23]. Similarly, the FIR radiation inhibited the growth of family Deferribacteraceae and genus Mucispirillum. Mucispirillum sp. is a mucus degrading bacterium and can trigger spontaneous colitis in mice [24].
Among the FIR-induced beneficial bacteria, we observed instantaneous growth of Barnesiella spp, Alistipes massiliensis, Clostridium indolis, Prevotella spp., and Barnesiella intestinihominis in FIR-D1 (Fig. 3A). Apart from Barnesiella, the growth of these groups of bacteria sustained or even further propagated on D25 (Fig. 3B). Prevotella can degrade the undigested polysaccharide, improve the host’s glucose metabolism, and plays an important role in energy homeostasis [25]. Species belonged to the genus Barnesiella are common dwellers in the human gut and protect the host against vancomycin-resistant Enterococcus faecalis [26]. In addition, Barnesiella species could help host against obesity as these species are found abundantly in the gut of normal weight compared to the obese people [27]. In another experiment, Everard et al., (2011) found that prebiotics-induced Barnesiella spp. improved leptin sensitivity and metabolic parameters in ob/ob mice [28]. Furthermore, Weiss et al. (2014) reported enhancement of Barnesiella spp. in mice fed with oligosaccharides 2-fucosyllactose-derivatives and 3-fucosyllactose-derivatives [29].
Alistipes bacteria are essential for the efficacy of dietary therapy against Crohn’s disease. The prevalence of Alistipes sp., Barnesiella spp., and Prevotella spp. have been correlated with the production of monosaccharides and short-chain fatty acids [30]. Another bacterium Butyricicoccus pullicaecorum remained elevated in FIR-treated mice on D25. This bacterium is a butyrate-producing bacterium with anti-colitis properties by strengthening the epithelium function [31]. We noticed the prevalence of Helicobacter ganmani in FIR treated mice. This bacterium, although belongs to a pathogenic genus, is unable to induce typhlitis in laboratory mice [32]. In addition to the data obtained from 16S sequencing, we verified the enhancement of SCFAs-producing bacteria by qPCR analysis of three main Clostridia clusters IV, XIV, and XIVa and the genes encoded enzymes in the SCFAs synthetic pathways, especially butyrate (Figs. 3E and 4A). Butyrate is an energy source for colonocytes and has been reported for anti-inflammatory and anti-cancer properties [21].
Interestingly, some of the FIR-induced changes in the Clostridia clusters were long-lasting. In addition, we also evaluated SCFAs sensing receptors (GPCRs) to evaluate whether FIR-modulated GM could impact mice physiology. Therefore, three genes encoding GPR41, GPR43, and GPR109 were evaluated and found upregulated in the mice exposed to FIR radiations.
Conclusion
Our findings showed that transient FIR-radiation could induce long-lasting alterations of gut microbial composition. The study also revealed, for the first time, that the health benefit of FIR treatment might be in part through the modulation of GM and the responses of host’s signaling mediators such as SCFA-sensing GPCRs.
Compliance with Ethics Requirements
All Institutional and National Guidelines for the care and use of animals (mice) were followed.
Acknowledgments
Acknowledgements
This research is partially by FDCT 103/2016/A3. Our gratitude also goes to the Nick Wang Technology Limited who provides partial support and the FIR emitting device (EEFit Pen®).
Conflict of interest
The authors have declared no conflict of interest
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2019.12.003.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Kochevar I.E., Pathak M., Parrish J. McGraw-Hill; New York, NY: 1999. Photophysics, photochemistry and photobiology. Fitzpatrick’s Dermatology in General Medicine. [Google Scholar]
- 2.Tanaka Y. Impact of near-infrared radiation in dermatology. World J Dermatol. 2012 [Google Scholar]
- 3.Vatansever F., Hamblin M.R. Far infrared radiation (FIR): its biological effects and medical applications. Photonics Lasers Med. 2012;4:255–266. doi: 10.1515/plm-2012-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang Y. The effect of far infrared radiation therapy on inflammation regulation in lipopolysaccharide-induced peritonitis in mice. SAGE Open Med. 2018;6 doi: 10.1177/2050312118798941. 2050312118798941–2050312118798941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang J.-C., Wu S.-L., Hoel F., Cheng Y.-S., Liu K.-H., Hsieh M. Far-infrared radiation protects viability in a cell model of Spinocerebellar Ataxia by preventing polyQ protein accumulation and improving mitochondrial function. Sci Rep. 2016;6:30436. doi: 10.1038/srep30436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kikuji Yamashita, Dalkhsuren Shine-Od, Ishikawa Tatsuo, Sumida Kaori, Ishibashi Jun, Hosokawa Hiroyoshi. Far infrared ray radiation inhibits the proliferation of A549, HSC3 and Sa3 cancer cells through enhancing the expression of ATF3 gene. J Electromagn Anal Appl. 2010;2:13. [Google Scholar]
- 7.Udagawa Y., Nagasawa H., Kiyokawa S. Inhibition by whole-body hyperthermia with far-infrared rays of the growth of spontaneous mammary tumours in mice. Anticancer Res. 1999;19:4125–4130. [PubMed] [Google Scholar]
- 8.Maslowski K.M., Mackay C.R. Diet, gut microbiota and immune responses. Nat Immunol. 2011;12:5–9. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 9.Carding S., Verbeke K., Vipond D.T., Corfe B.M., Owen L. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. 2015;26. doi: 10.3402/mehd.v26.26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.https://www.nwtech.com.hk/en/.
- 11.Chen L., Brar M.S., Leung F.C.C., Hsiao W.L.W. Triterpenoid herbal saponins enhance beneficial bacteria, decrease sulfate-reducing bacteria, modulate inflammatory intestinal microenvironment and exert cancer preventive effects in ApcMin/+ mice. Oncotarget. 2016;7(31226–42) doi: 10.18632/oncotarget.8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowd S.E., Sun Y., Wolcott R.D., Domingo A., Carroll J.A. Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathog Dis. 2008;5:459–472. doi: 10.1089/fpd.2008.0107. [DOI] [PubMed] [Google Scholar]
- 13.Robertson B.R., O’Rourke J.L., Neilan B.A., Vandamme P., On S.L.W., Fox J.G. Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol. 2005 doi: 10.1099/ijs.0.63472-0. [DOI] [PubMed] [Google Scholar]
- 14.Embree J.E., Embil J.A. Mycoplasmas in diseases of humans. Can Med Assoc J. 1980;123:105–111. [PMC free article] [PubMed] [Google Scholar]
- 15.Schneeberger M., Everard A., Gomez-Valades A.G., Matamoros S., Ramirez S., Delzenne N.M. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci Rep. 2015;5:16643. doi: 10.1038/srep16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pryde S.E., Duncan S.H., Hold G.L., Stewart C.S., Flint H.J. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217 doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 17.Lopetuso L.R., Scaldaferri F., Petito V., Gasbarrini A. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5:23. doi: 10.1186/1757-4749-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van den Abbeele P., Belzer C., Goossens M., Kleerebezem M., De Vos W.M., Thas O. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013;7:949–961. doi: 10.1038/ismej.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flint H.J., Duncan S.H., Scott K.P., Louis P. Links between diet, gut microbiota composition and gut metabolism. Proc Nutr Soc. 2015;74:13–22. doi: 10.1017/S0029665114001463. [DOI] [PubMed] [Google Scholar]
- 20.Reimann F., Tolhurst G., Gribble F.M. G-protein-coupled receptors in intestinal chemosensation. Cell Metab. 2012;15:421–431. doi: 10.1016/j.cmet.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 21.Husted A.S., Trauelsen M., Rudenko O., Hjorth S.A., Schwartz T.W. GPCR-mediated signaling of metabolites. Cell Metab. 2017;25:777–796. doi: 10.1016/j.cmet.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt T.S.B., Raes J., Bork P. The human gut microbiome: from association to modulation. Cell. 2018;172:1198–1215. doi: 10.1016/j.cell.2018.02.044. [DOI] [PubMed] [Google Scholar]
- 23.Beller A., Kruglov A., Durek P., von Goetze V., Hoffmann U., Maier R. P104 Anaeroplasma, a potential anti-inflammatory probiotic for the treatment of chronic intestinal inflammation. Ann Rheum Dis. 2019;78:A45–A46. [Google Scholar]
- 24.Bordon Y. A microbial trigger for colitis. Nat Rev Immunol. 2019;19:350–351. doi: 10.1038/s41577-019-0175-y. [DOI] [PubMed] [Google Scholar]
- 25.Precup G., Vodnar D.C. Gut Prevotella as a possible biomarker of diet and its eubiotic versus dysbiotic roles-A comprehensive literature review. Br J Nutr. 2019 doi: 10.1017/S0007114519000680. [DOI] [PubMed] [Google Scholar]
- 26.Ubeda C., Bucci V., Caballero S., Djukovic A., Toussaint N.C., Equinda M. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun. 2013;81:965–973. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Del Chierico F., Abbatini F., Russo A., Quagliariello A., Reddel S., Capoccia D. Gut microbiota markers in obese adolescent and adult patients: age-dependent differential patterns. Front Microbiol. 2018;9:1210. doi: 10.3389/fmicb.2018.01210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Everard A., Lazarevic V., Derrien M., Girard M., Muccioli G.G., Neyrinck A.M. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes. 2011;60:2775–2786. doi: 10.2337/db11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss G.A., Chassard C., Hennet T. Selective proliferation of intestinal Barnesiella under fucosyllactose supplementation in mice. Br J Nutr. 2014;111:1602–1610. doi: 10.1017/S0007114513004200. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y., Wu J., Li J.V., Zhou N.-Y., Tang H., Wang Y. Gut microbiota composition modifies fecal metabolic profiles in mice. J Proteome Res. 2013;12:2987–2999. doi: 10.1021/pr400263n. [DOI] [PubMed] [Google Scholar]
- 31.Geirnaert A., Steyaert A., Eeckhaut V., Debruyne B., Arends J.B.A., Van Immerseel F. Butyricicoccus pullicaecorum, a butyrate producer with probiotic potential, is intrinsically tolerant to stomach and small intestine conditions. Anaerobe. 2014;30:70–74. doi: 10.1016/j.anaerobe.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Alvarado C.G., Kocsis A.G., Hart M.L., Crim M.J., Myles M.H., Franklin C.L. Pathogenicity of Helicobacter ganmani in mice susceptible and resistant to infection with H. hepaticus. Comp Med. 2015;65:15–22. [PMC free article] [PubMed] [Google Scholar]
- 33.Dostal A., Lacroix C., Bircher L., Pham V.T., Follador R., Zimmermann M.B. Iron modulates butyrate production by a child gut microbiota in vitro. MBio. 2015;6:e01453–e1515. doi: 10.1128/mBio.01453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boets E., Deroover L., Houben E., Vermeulen K., Gomand S.V., Delcour J.A. Quantification of in vivo colonic short chain fatty acid production from inulin. Nutrients. 2015;7:8916–8929. doi: 10.3390/nu7115440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwiertz A., Taras D., Schäfer K., Beijer S., Bos N.A., Donus C. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 36.Vital M., Penton C.R., Wang Q., Young V.B., Antonopoulos D.A., Sogin M.L. A gene-targeted approach to investigate the intestinal butyrate-producing bacterial community. Microbiome. 2013;1:8. doi: 10.1186/2049-2618-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reichardt N., Duncan S.H., Young P., Belenguer A., McWilliam Leitch C., Scott K.P. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veprik A., Laufer D., Weiss S., Rubins N., Walker M.D. GPR41 modulates insulin secretion and gene expression in pancreatic beta-cells and modifies metabolic homeostasis in fed and fasting states. FASEB J Off Publ Fed Am Soc Exp Biol. 2016;30:3860–3869. doi: 10.1096/fj.201500030R. [DOI] [PubMed] [Google Scholar]
- 39.Dewulf E.M., Cani P.D., Neyrinck A.M., Possemiers S., Van Holle A., Muccioli G.G. Inulin-type fructans with prebiotic properties counteract GPR43 overexpression and PPARgamma-related adipogenesis in the white adipose tissue of high-fat diet-fed mice. J Nutr Biochem. 2011;22:712–722. doi: 10.1016/j.jnutbio.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Singh N., Thangaraju M., Prasad P.D., Martin P.M., Lambert N.A., Boettger T. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J Biol Chem. 2010;285:27601–27608. doi: 10.1074/jbc.M110.102947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.