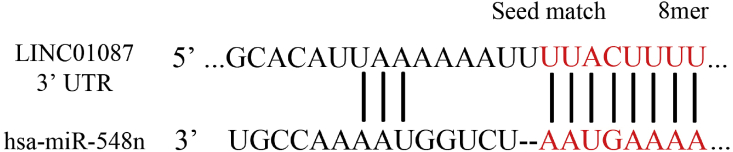Abstract
Apoptosis is a ‘programmed fate’ of all cells participating in diverse physiological and pathological conditions. The role of critical regulators and their involvement in this complex multi-stage process of apoptosis weaved around non-coding RNAs (ncRNAs) is poorly deciphered in breast carcinoma (BC). Aberrant expression patterns of the ncRNAs and their interacting partners, either ncRNAs or coding RNAs or proteins at any point along these pathways, may lead to the malignant transformation of the affected cells, tumour metastasis and resistance to anticancer drugs. Longest non-coding type of ncRNAs (lncRNAs) have been considered as critical factors for the development and progression of breast cancer. The aim of our study was to identify set of novel lncRNAs interacting with microRNAs (miRNAs) or proteins that were significantly dysregulated in breast cancer using RNA-Sequencing (RNA-Seq) technique in different samples acting as oncogenic drivers contributing to cancerous phenotype involved in post-transcriptional processing of RNAs. Four lncRNAs; LINC01087, lnc-CLSTN2-1:1, lnc-c7orf65–3:3 and LINC01559:2 were selected for further analysis. Gene expression analysis of over-expressed LINC01087 in vitro reduced both cell viability and apoptosis. We integrated miRNA and mRNA (hsa-miR-548 and AKT1) expression profiles with curated regulations with lncRNA (LINC01087) which has not been previously associated with any breast cancer type, using different computational tools. The network (lncRNA→ miRNA→ mRNA) is promising for the identification of carcinoma associated genes and apoptosis signaling path highlighting the potential roles of LINC01087, hsa-miR548n, AKT1 gene which may play crucial role in proliferation.
Keywords: Long non-coding RNAs, microRNAs, RNA sequencing, Cancer genomics
1. Introduction
Human genome when analysed shows that it accumulates approximately 20,000 protein coding genes (PCGs), which makes up only ~ 2%, of the whole genome sequence. The majority of the genome is built by ~78% of non-protein-coding transcriptome which undergoes modifications but fails to end up with the process of translation [1]. Apart from the house-keeping non-coding RNAs (ncRNAs) such as ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) our genome also nests small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), micro RNAs (miRNAs), piwi-interacting RNAs (piRNAs) and the longest one, long-non-coding RNAs (lncRNAs) [2,3]. As the name suggests these lncRNAs are longer group members of 200 nucleotides (nts) in length and their expression is controlled by transcriptional as well as miscellaneous epigenetic factors. LncRNAs are governed by RNA polymerase II and undergoes post-transcriptional modifications similar to coding RNAs. Moreover, lncRNAs are less conserved among the different species, as well as lack open reading frames (ORFs) and coding potentials [[4], [5], [6]].
Earlier studies illustrate that the disruptive activity on the histone linked DNA template, like mutation, chromosomal rearrangements, chromosomal deletion/insertion, etc. caused due to chemicals, radiations, stimuli may guide the RNA polymerases to react abnormally [7]. In another process, RNA synthesis gets catalyzed and induces transcription everywhere in the human genome generating the heterogeneous group of lncRNAs randomly [8]. The generated lncRNAs diversely perform activities participating in dosage compensation [9], transcriptional repression [10], regulatory activities [11], growth and differentiation [12]. A widely proposed functional model of lncRNAs serve as guides, decoys or scaffolds thereby regulating different activities of gene transcription, mRNA modification or translation [13]. LncRNAs besides participating in different events form complexes with proteins [14], mRNAs [15] or even miRNAs [16] guiding to several biological processes. However, the mechanisms of action of most lncRNAs remain vague.
Deflecting towards the disease, breast cancer is known to be one of the most prevalent form of female neoplasm across the globe ranking second most among the commonly occurring cancers worldwide (after cardiovascular diseases). This accounts for about 12% of all new cancer cases and 25% of all cancers in women [17,18]. Although considerable amounts of research for understanding the molecular pathology of the disease have been conducted in the past by exploring the proteomic content of the affected organs, the genomic polymorphism and instability due to chromosomal aberrations, epigenetic and genetic alterations have often been overlooked. The focus towards the epigenetic factors can help to characterize the molecular landscape of the disease and contribute in its pathogenesis. Many attempts have been made to predict the involvement of lncRNAs in metastasis of breast cancer mostly by correlating gene expression patterns using high-throughput sequencing (HTS) techniques such as microarray and next generation sequencing (NGS) [19]. Also high profiled machine learning techniques have succeeded in categorizing these lncRNAs on the basis of conserved k-mer patterns [20]. Likewise, with the advent of RNA-Seq technique, the skeptical views of these lncRNAs have changed pushing the study towards its biological significance. Moreover, these lncRNAs are transcribed in single-regulated manner, which categorize the divergent species on the basis of differential expression patterns with the contrasting dataset samples. The dysregulation of lncRNAs have been found to be directly or indirectly associated with the hallmarks of cancers mediated by other interacting partners which can be either non-coding RNAs (miRNAs, siRNAs, and piRNAs) or some proteins, transcription factors (TFs), histone complexes, etc [5,[21], [22], [23]]. However, the less knowledge regarding the interactions shares diminutive and inadequate information about the mechanism of tumorogenesis in cells.
Trending research practice is focused on a greater understanding of the response of different activities, including the role of apoptosis. Cell proliferation and apoptosis governs the normal development of the organ, any loss or gain in the activity may evidence the tumour formation [24,25]. Much of this work is still the subject of research to determine the steadiness between proliferation and apoptosis. Identification of ncRNAs in target driven pathways for apoptosis can lead to several therapeutic benefits [26,27]. With the case of breast cancer, components of several potential signaling pathways can be targeted to improve the survival rate. The aim of the study was to identify one or several of these potential apoptosis regulators associated with ncRNAs in breast cancer cells. Moreover, lncRNAs have been implicated to act as important regulators in tumor-suppressor and oncogenic pathways, and recent studies showed that lncRNAs control the major cancer-driving pathways at epigenetic, transcriptional and post-transcriptional level [28,29]. Several lncRNAs have been found to be dysregulated in breast cancer tissues when compared with normal tissues. LSINCT5, ZFAS1 plays critical role in promoting cell proliferation, LncRNA-Smad7 function as anti-apoptosis gene [30], GAS5 has an established role in inducing apoptosis [31]. Some of the most widely studied lncRNAs in BC are H19, HOTAIR, GAS5, MALAT1, UCA1, CCAT1-2, XIST, and PANDAR [32]. H19 is reported to be up-regulated in cancer cells acting as oncogene and functioning as sponge for miRNA precursors [33]. Similarly MALAT1 [34], UCA1 [35], CCAT1 [36], CCAT2 [37] and HOTAIR [38] acts as oncogenes, functionally regulates different pathways which can detect primary or recurrent BC associating with multiple tumor grades and stages. LncRNAs acting as tumor suppressor genes (TSGs) in BC are even less explored (e.g. GAS5). GAS5 low-expression is shown to predict good prognosis in different cancers including BC where it is downregulated in cancer tissues [39]. Furthermore, MALAT-1 which is reported to be upregulated in most of the cancers is said to be associated with different functional processes such as metastasis, cell proliferation, apoptosis, migration, as well as correlated with clinically unfavourable prognostic parameters. The knockdown experiment, showed the increased proliferation in the cells justifying the dysregulation of the lncRNAs in aberrant conditions [34]. UCA1 induces apoptosis and modulates cell growth by interacting with tumor suppressive miRNA (miR-143) defining a prognostic role in breast cancer [35]. Upregulation of CCAT1 expression is caused due to a cell cycle regulator oncogene that results in the progression of the disease and poor prognosis in breast cancer patients. Similarly, elevated levels of CCAT2 expression leads to conditions indistinguishable from the ones caused by CCAT1 [36,37]. XIST, a widely studied lncRNA for its role in X-chromosome silencing in female cells, is widely expressed by all cells of somatic origin present in females. The expression is said to be lost in female breast-, ovarian-, as well as cervical-cancer cell lines. The dysregulated expression of XIST pin-points its complex and controversial role in breast cancer [40]. A 1.5 kb lengthened lncRNA, PANDAR is reported to be upregulated in breast cancer cell lines and tissues and functions as a tumor-promoting lncRNA by regulating G1/S transition [41]. Therefore, screening based on functional assays could help in identifying novel lncRNAs that are involved in apoptosis regulating breast cancer progression.
The dysregulated or defective apoptosis regulation forms the base of standard therapies like chemotherapy and radiotherapy [42]. Apoptosis induced tumor progression is a fundamental aspect of the tumor biology. Non-surgically the cancer cells can be removed by inducing the mechanism of apoptosis which may activate the cell death of malignant cells or may repair the mechanism in the defective ones [43,44]. Hence, deeper perceptive of the molecular mechanisms of apoptosis targeted via epigenetic regulators transcriptionally or post-transcriptionally opens the new opportunities for researchers to develop drugs which could eradicate the tumor formation.
In this study, we identified significantly dysregulated lncRNAs in BC profiling expression analysis using RNA-Seq technique. RNA-Seq and microarray technology both are determined as a means of generating transcriptome information. However, RNA-Seq has a number of advantages over the microarray technique regarding the experimental design, data acquisition, and data analysis. RNA-Seq technology detects transcripts that may or may not correspond to existing genomic sequencing, compared to microarray technology where the prior information about the sequence is required. This quality of the RNA-Sequencing method aids in investigating both known transcripts as well as exploring new ones. RNA-Seq delivers low background noise unambiguously mapping to the unique regions compared to the microarray where the noise is generated due to cross-hybridization. RNA-Seq has the ability to quantify a large dynamic range of expression levels, with absolute values determined by means of differential expression analysis. RNA-Seq analysis is cost effective and can be performed with high levels of reproducibility between large numbers of replicates. However, RNA-Seq costs more per sample than microarray and requires high power computing facilities [[45], [46], [47]]. The samples gathered from the curated data available on NCBI SRA database was used to identify putative lncRNAs active in different samples of breast cells viz. normal and tumorous conditions. Furthermore, a standalone application freely accessible web interface called ‘DeepLNC’ for distinguishing coding and non-coding RNAs from the sequence specific dataset was used, available at our website https://bioserver.iiita.ac.in/deeplnc/. The curated regulations considered in our study identified significantly up-regulated lncRNAs (LINC01087 and lnc-CLSTN2-1:1) and down-regulated lncRNAs (lnc-c7orf65–3:3 and LINC01559:2) which were further characterized in vitro using RT-PCR techniques. We contemplated that the dysregulated lncRNAs exhibit oncogenic functions in breast cancer cells by regulating associated miRNAs with pathways related to cell growth/proliferation/apoptosis. Furthermore, we generated a network from the components of the genome such as lncRNA-miRNA-mRNA and identified the novel regulatory mechanisms involved in apoptosis signaling pathway which might participate in the oncogenic activity in breast cancer. The network created is first of its type which is an attempt to link the genomic macromolecules, challenging the age old phenomenon of central dogma.
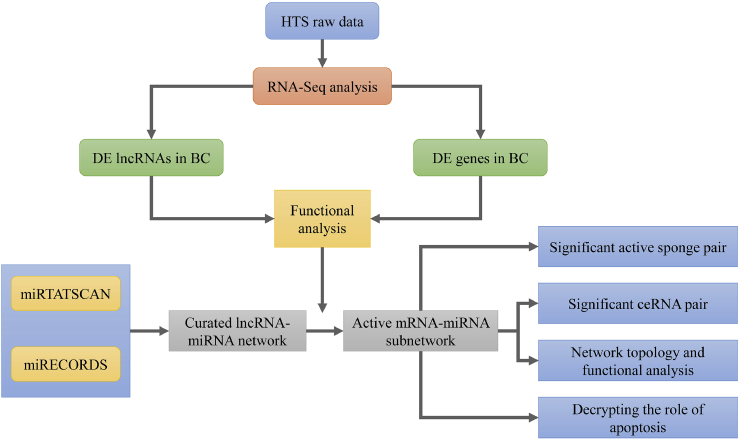
In addition, our candidate lncRNA was found to interact with some regulatory proteins involved in functions significant to different cellular processes which can also be of oncogenic purpose. This sponge-interaction of ncRNAs in association with mRNA mediated genes is more promising for investigation and diagnosis of the disease [[48], [49], [50]]. The outline of the steps followed in the analysis is outlined in a flowgram provided in Fig. 1.
Fig. 1.
A basic flowchart to show the steps involved in processing of the data.
Thus, the work done in this paper displays group of lncRNAs and interacting miRNAs possessing novel potential of being critical regulators during the disease occurrence and progression. This regulatory network and pathway analysis involving the ncRNA components is first of its kind specifically in terms of interacting partners. The pair will provide a skeleton for implementing the research work in the area targeting crucial steps in disease progression strapping up these for therapeutic benefits.
2. Materials and methods
2.1. Gene and lncRNA expression profile of breast cancer
Breast cancer known to be one of the most prevalent form of female neoplasm across the globe accounts for about 25% of all death. To explore the molecular pathology of the disease in terms of non-protein coding content we carried the RNA-Seq analysis for differential expression testing of novel lncRNAs. The primary NGS datasets (GSE71862) generated using Illumina's HiSeq 1500 platform and related RNA-Seq library clinical information was obtained from Gene expression omnibus (GEO) database. The NGS dataset comprised of 6 samples including 3 normal breast samples as control and 3 samples of breast cancer. For gene expression analysis, pre-processing of data was performed using different set of tools. The fastq data was filtered using FASTQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) tool, which helped with the graphical images provided during the adapter trimming and quality check. The mammalian genome data was mapped using the algorithm developed by the tool Bowtie 2 (http://bowtiebio.sourceforge.net/bowtie2/index.shtml) vastly supporting the gap alignment. The aligned data was mapped to the hg19 genome extracted from the UCSC genome browser (https://genome.ucsc.edu/). The number of the reads mapping to the particular area, which helps with the quantification purpose for the expression analysis, was done using HT-seq tool (http://wwwhuber.embl.de/HTSeq/doc/count.html). The count based algorithm was followed by the differential expression analysis using edgeR (http://bioconductor.org/packages/release/bioc/html/edgeR.html), an R based tool. The differentially expressed genes were subjected to the specific regulation process by calculating the logFC value. The lincRNAs name was according to LNCipedia 4.0 nomenclature rules decided as per the HGN Committee [51].
2.2. Functional analysis of DE lncRNAs
The differentially expressed (DE) genes were subdivided according to the biological, molecular and cellular processes using Panther database (http://pantherdb.org/), BiomaRt (http://www.biomart.org/), and DAVID (https://david.ncifcrf.gov/) tool search. The chromosomal location of the several bp long transcribed lncRNA was analysed using IGV tool (http://software.broadinstitute.org/software/igv/), whereas the coding potential was declared using ENSEMBL (http://www.ensembl.org/index.html) genome browser, PRIDE reprocessing tool (v 2.0), PhyloCSF score, CPAT, Randfold tool, respectively. The presence of miRNAs target region and locus conservation was searched using MirTarget2 in another species, as assessed by the Emsembl Compara API, respectively. The manually curated and experimentally validated dysregulated lncRNAs with an established role in the cancer were screened out for further analysis. We additionally mined miRNA databases: miRTarget (http://www.mirdb.org/), LncBAse analysis (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=site/help&topic=lncBase), TargetScan (www.targetscan.org/), miRTarBase (http://mirtar.mbc.nctu.edu.tw/human/) to obtain miRNAs associated with lncRNAs targeting mRNA genes which may be involved in BC or some other cancer for our study.
2.3. Clinical samples, RNA extraction, cDNA synthesis and RNA-Seq
For the cell culture and morphological studies, human breast adenocarcinoma cell line (MCF-7) was used. The methods in the study were carried out in accordance with the approved guidelines and regulations. Detailed information regarding the cell lines, RNA extraction, cDNA synthesis is as follows: the frozen vials of cells (MCF-7) stored in liquid Nitrogen (N2) were thawed, pelleted down and were re-suspended in culture medium (DMEM), supplemented with 15% foetal bovine serum (FBS), 1% penicillin/streptomycin. Cells were grown in T25 cell culture flask, kept in CO2 incubator at 37 °C until 70–80% of the flask surface was covered with the cells, in between changing the media after every alternate day. When the cells became confluent, media was aspirated out and cells were washed with 1X PBS followed by trypsinization with ~1 ml trypsin/EDTA solution at 37 °C for 1–2 minutes. When around 50% cells start floating, reaction was terminated by adding 2–3 ml of complete DMEM medium and the suspended cells were transferred to 15 ml falcon. After centrifugation at 1000–1500 rpm for 2 min at Room temperature (RT), the cell pellets were re-suspended in 1 ml complete DMEM medium, counted and seeded at the concentration of 0.1 million cells/ml in T25 cell culture flask and were grown overnight in cell culture incubator at 37 °C and 5% CO2 around at 70–80% of the flask surface was covered by the cells, in between changing the media after every alternate day. After 24 h the medium was replaced, the morphological changes in the cells or changes in their cell growth were observed in between 12 and 48 h and photographs were taken under phase contrast microscope (Nikon TiS). When the cells became confluent, media was aspirated out and cells were washed, trypsinized. Total RNA from the MCF-7 cell lines were extracted using Trizol reagent (Sigma Aldrich) as per the manufacturers' instructions. To prevent RNA degradation the RNAs were treated with RNase-free DNAse I enzyme (Promega). cDNAs were synthesized from 1 μg of DNAse-I Treated total RNA and 0.5 μg of oligo(dT) 12–18 primers (Promega) using 100 U of M-MLV Reverse Transcriptase and 20 U of RNAsin (Promega). The above mixture was suitable for 25 μl reaction mixture. For strand specific PCR, 1 μM of sense/antisense strand-specific LINC01087 (Fwd 5′ CAAAGAGCAACCAGCCA 3’; Rev 5′ AACCAGTACCAGCCACTA 3’) primers was used, generated using Primer3D tool (primer3d.com/). 1 mM dNTPs were used for generating cDNAs at 50 °C, 2.5 μl of cDNAs were further used for the PCR amplification using positive reference control GAPDH. The amplified product was run on 1.5% agarose-TBE gel electrophoresis. 1 μl of the specific primer along with 2.5 μl of cDNA template (of MCF-7 cell total RNA) was used in a reaction mixture and the amplified product along with the control was loaded. The 3516 bp (LINC01087) and 452 bp (GAPDH) amplicons were quantified and the result was plotted using IDV values.
2.4. Sponge-pair interactions
A curated regulatory relationship was established among lncRNAs, miRNAs and target genes obtained from different database using network topology. In our analysis, we tried to build a new association between coding RNAs and non-coding RNAs giving equal weight age to RNA-RNA interactions. LncRNAs have been least explored and their association with miRNAs or any other ncRNAs may help to throw light on the mechanism of their action specifically in breast cancer system. We generated a curated miRNA-mRNA regulatory network using Genemania (https://genemania.org/). The network targeted genes were involved in tumor suppressive or oncogenic role which are likely to play important regulatory roles in the pathogenesis of the disease.
2.5. Statistical analysis
Co-expression analysis between lncRNA and miRNA was studied. The correlation coefficient is used as a measure of gene co-expression, where negative r-value indicates negative correlation and positive value indicates positive co-relation. Kaplan-Meier plot was used to demonstrate the relationship between hsa-miR548n, AKT1 and PAWR gene expression and survival of patients with breast cancer.
3. Results and discussion
3.1. Differentially expressed lncRNAs in breast cancer
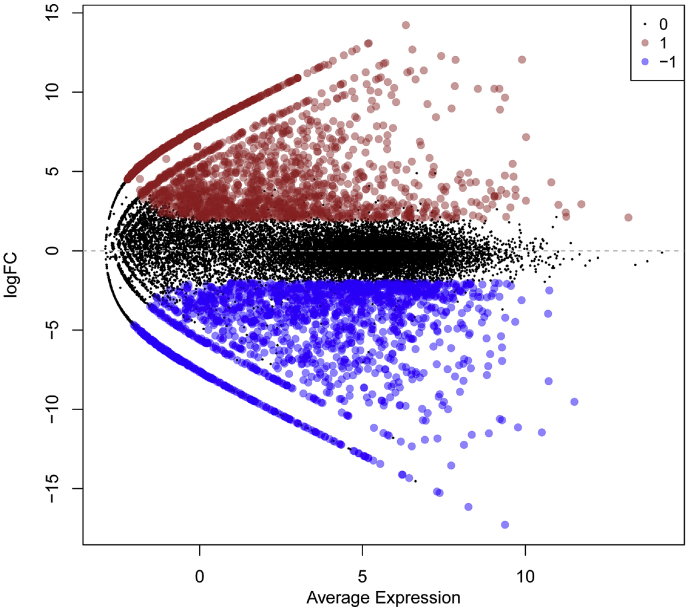
RNA-Seq analysis of the two data sets, consisting of 3 normal and 3 cancerous samples, using HTS tools (Tuxedo pipeline) identified 19435 non-protein coding transcripts viz. long intergenic non-coding RNAs (lincRNAs), antisense (as) RNAs, miRNAs, snoRNAs, snRNAs and miscellaneous RNAs (miscRNAs). The most recurrently occurring category among non-coding transcripts was lincRNAs followed by as lncRNA. Using edgeR, significantly dysregulated lincRNAs were identified in breast cancer cells when compared to normal breast cells (p < 0.05 and fold change ≥ 2.0). (Fig. 2). The identified lncRNAs were further demonstrated to be linked with 33 miRNAs and 44347 mRNA genes, with p < 0.05 and fold change ≥2.0. Functional analysis of the dysregulated genes using DAVID [52] was used to spot the ones involved in apoptosis of cancer cells.
Fig. 2.
The significantly dysregulated lncRNAs in breast cancer cells when compared to normal breast cells (p < 0.05 and fold change ≥ 2.0). Distribution of significantly differentially expressed non-protein coding transcripts (lincRNAs, antisense RNAs, miRNAs, snoRNAs, snRNAs and miscRNAs) obtained from SRA dataset, identified using edgeR (in colour) and annotated according to hg19 fasta file. Mean centered genes were displayed in black colour.
This research was centered towards the topmost up- & down-regulated lncRNAs from which four of the most differentially expressed lncRNAs were selected for detailed functional characterization and in vitro studies. All four selected candidates displayed nill/low predicted protein-coding scores using different tools. The coding potential was substantiated using ENSEMBL (http://www.ensembl.org/index.html) genome browser, PRIDE reprocessing tool (v 2.0) [53], PhyloCSF score [54], CPAT [55], and Randfold tool [56]. The presence of miRNAs target region was scanned using LncBase analysis tool [57], whereas the locus conservation across vertebrate species (Mouse and Zebrafish) was searched using MirTarget2 (as assessed by the Emsembl Compara API) [58], exclusively for four dysregulated lncRNAs LINC01087, lnc-CLSTN2-1:1, lnc-c7orf65–3:3, LINC01559:2. This led to the conclusion of a possible conserved function passed on through evolutionarily means (Table 1). The differential expression of the lncRNAs was validated using data entry from Genecard, NONCODE, LNCipedia databases.
Table 1.
Predicted protein-coding scores for the four candidates using different tools.
| Sets | C vs. T (U) | C vs. T (U) | C vs. T (D) | C vs. T (D) |
|---|---|---|---|---|
| Differentially expressed lncRNAs | LINC01087 | lnc-CLSTN2-1:1 | lnc-c7orf65–3:3 | LINC01559:2 |
| Length (bp) | 3516 | 5692 | 609 | 1003 |
| PhyloCSF score | 25.7357 | −56.324 | −44.4845 | – |
| CPAT coding probability | 0.48% | 2.19% | 5.28% | 0.63% |
| Bazzini small ORFs | 0 | 0 | 0 | 0 |
| Locus conservation | No | Yes | No | No |
| No | No | No | No | |
| R and fold minimum free energy | −917.46 | −188 | −177.8 | −789.43 |
| R and fold P-value | 0.016 | 0.073 | 0.078 | 0.045 |
| Targeting miRNAs | hsa-miR-548n, hsa-miR-548as-5p, hsa-miR-548as-5p | – | – | – |
C=Control, T = Treated, U=Up-regulated, D = Down-regulated.
3.2. In-vitro approach for functional characterization of lncRNAs
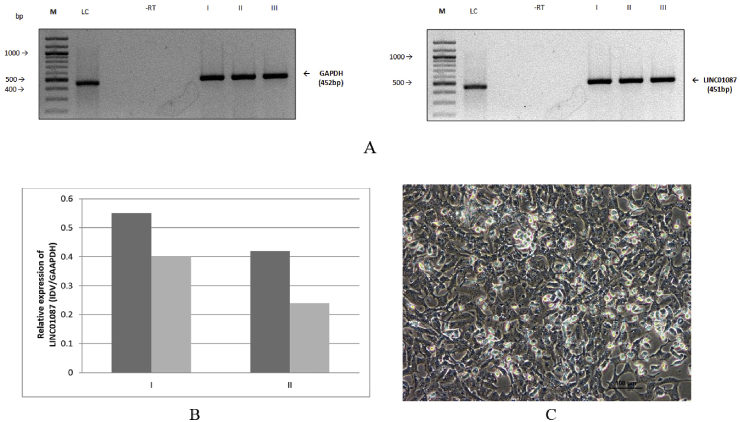
RNA-Seq analysis for a panel of tumorigenic lncRNAs was classified with high/low expression. MCF-7 cell line, which belongs to a metastases producing human breast cancer cell line, was used to investigate the functional impact of LINC01087. All selected cell lines were derived from human breast adenocarcinoma cells and were utilized for the cell culture and morphological studies obtained from National Centre for Cell Science (NCCS), Pune, India. Total RNA extracted from the cell lines was used to determine the expression of our candidate lncRNAs. The RNA was treated with RNase-free DNase I (Promega) to prevent amplification which may be caused due to DNA contamination. cDNAs were synthesized using 1 μg of DNase I treated total RNA extracted in RNase free environment. For the amplification of LINC01087 and GAPDH, which was taken as a positive internal control, RT-PCR was carried out using 5.0 and 2.5 μl of cDNA, respectively in a 25 μl PCR reaction mixture containing 1X Taq buffer, 0.2 mM dNTPs, 1U Taq polymerase and 25pm of LINC01087 and GAPDH specific primer pairs. The amplification conditions used were: initial denaturation at 95 °C for 4 min; followed by 35 cycles of: denaturation at 95 °C for 45 s, annealing at 54 °C for 45 s and extension at 72 °C for 45 s; and a final extension at 72 °C for 10 min. The amplified product (1/5th for 5.5 and LINC01087 and 1/10th for GAPDH) was checked on 1.5% agarose-TBE gel, run at 70 V or 100 mA and their respective concentrations were quantified by measuring integrated density value (IDV) of the amplified band using AlphaEase FCTM software and Alpha Imager 3400 (Aplhainnotech corporation). RT-PCR analysis revealed a higher expression of LINC01087 in MCF-7 cell lines (Fig. 3a). Successful isolation of pure cytoplasmic fractions was confirmed by a marker RNA molecule with known cytoplasmic (GAPDH) enrichment (Fig. 3b).
Fig. 3.
(a) Expression of LINC010187 in MCF-7 cells. RT-PCR of LINC01087 by using RNA isolated from MCF-7 cells. GAPDH was taken as internal control. M: 100bp DNA ladder; LC: loading control (500 bp). (b) Relative expression of LINC01087 in MCF-7 determined by RT-PCR (c) Microscopic morphological analysis of MCF-7 cells cultured in complete media captured at 24 h at 20X resolution.
To explore various pathways related to programmed cell death, cellular morphology studies were performed. Cellular morphology can be used as the basis for defining different programmed cell death modes. We assessed the effects of LINC01087 on proliferation, a crucial step in cancer progression and metastatic colonization. MCF-7 cells grown in complete media displayed cell rounding and shrinkage, plasma membrane blebbing followed by subsequent detachment of the cells reflecting slow apoptosis (Fig. 3c). Overall, the results conclude the specific functional role of LINC01087 in the regulation of cancerous cells growth and proliferation. The product can act as regulating factor in slowing down the process of overgrowth in cancerous cell and putting a check on the procedure of cell death/apoptosis.
3.3. Identification of active lncRNA-miRNA-mRNA sub-network
Overall the most promising result of the in vitro analysis in accordance with in silico analysis was shown by LINC01087. Therefore, we set out to identify lncRNA-miRNA interaction partners to further elucidate the mechanisms of action of the dysregulated lincRNA. We scrutinized the ability of coding and ncRNAs to act as competing endogenous RNAs (ceRNAs) in human carcinoma and identified a new-fangled network of sponge interactions. This interaction can further act as mediator of crosstalk between different regulatory pathways. A curated gene-miRNA regulatory network was constructed. The generated network included 33 miRNAs and 44347 target genes. From 44347 genes and miRNAs of active sub-networks, 116 genes were found to be enriched in 8 apoptotic signaling pathways as obtained. As finding the combination of all possible RNA/miRNA/mRNA triplets is computationally taxing, only RNA/RNA pairs showing binding category of more than 7-mer were considered, thus using a priori information on putative or validated seeds to complementing expression data via LncBase analysis. The curative role of LINC01087 as possible sponge regulators of miRNAs activity on target mRNAs was established by Chromatin Immunoprecipitation sequencing (ChIP-seq) analysis. LncBase analysis showed the association of LINC01087 to several miRNAs. Among these miRNAs, 33 miRNAs whose binding energy >82.0 were only selected for the further analysis (Fig. 4, Supplementary Table 1). We further, explored miRNA decoy mechanism within gene regulatory circuitry using expression data from tumor and matched normal samples of breast carcinoma, provided by NCBI SRA and TCGA databases. In this study, two networks of lncRNA-miRNA interactions and miRNA-mRNA interactions were constructed. The reduced dimensionality of this configuration space, the data obtained from lncRNA-centered approach reduced the configuration space dimension, making the computational burden manageable, with the additional advantage of using a purely data-driven approach. Our study revealed the existence of a complex regulatory network of miRNA-mediated interactions that is step ahead of all biological significant studies. As a result, tumour-suppressive and oncogenic activity of some specific lncRNAs, exploiting a decoy mechanism, is speculated therein. Furthermore, the established lncRNA-miRNA-mRNA network participating in apoptotic activity triggered the more functional probability of sponge pair interactions in cancer tissues. A specific potential genes-miRNA regulatory sub-network mapped total 116 apoptotically active genes with 3 miRNAs having role as active seed nodes (Supplementary Table 2). The miRNAs (hsa-miR-4778–3p, hsa-miR-4482–3p, hsa-miR-335–3p) were found to be associated with 1 apoptotic pathway whereas 2 miRNAs (hsa-miR-3613–3p, hsa-miR-548n) were associated with >1 apoptotic pathway. Interestingly, hsa-miR-3613–3p, hsa-miR-548n, and hsa-miR-335–3p were identified to be associated with mRNA genes which have established function in tumor-suppression and oncogenic activity using target pair identification tool TargetScan (Supplementary Table 3). We obtained 3 miRNA hubs (hsa-miR-3548n, hsa-miR-3613–3p and hsa-miR-335–3p) associated with 13 TSGs (NF1, PAWR, PHLAD3, SIAH1, TP53INP1, TSC22D1, BCL2L11, DFNA5, HTATIP, PEG3, SMAD3, EGR1, YAP1) and 7 PGs (AKT1, NOTCH2, SOX4, PIM3, RAF1, CDKN1B, MDM2) (Table 2). These regulatory hubs/molecules might play important role in apoptotic signaling associated with carcinoma (Supplementary Fig. 1). Many of these miRNAs were involved in the regulation and initiation of translation and in post-transcriptional modifications of RNAs. Interestingly, miR-193a-5p is known to be less expressed in IBC patients and plays a suppressive role in breast cancer [59], miR-548w member has been found to be downregulated in pancreatic cancer [60], miR-185 is said to be downregulated in breast cancer and inhibits the proliferation of breast cancer cells [61,62]. Using TargetScan (http://www.targetscan.org/), physical association between LINC01087 and miR-548n was confirmed in data samples. Finally, the related miRNA resulted in interaction probabilities ranging between 0.04 and 0.0007 indicating that miR-548n and LINC01087 are likely to act together with each other (Supplementary Table 4). This interaction is novel in its kind and has not yet been reported in breast cancer. Network topology showed that the set of 33 miRNAs are clustered in two hubs which might be regulated in a similar way, since group of miRNAs which have similar pattern of expression can be considered to be in the same cluster (Supplementary Fig. 2). Furthermore, computationally predicted significant lncRNA-miRNAs interacting pair was found to be involved in different apoptotic regulatory pathways viz. apoptotic signaling pathway (Supplementary Fig. 3), intrinsic apoptotic signaling pathway (Supplementary Fig. 4), cellular component disassembly involved in execution phase of apoptosis (Supplementary Fig. 5), and positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway (Supplementary Fig. 6).
Fig. 4.
The binding site of hsa-miR548n has canonical 8-mer binding site in 3′UTR of LINC01087 obtained using TARGETSCAN.
Table 2.
List of miRNA genes interacting with mRNA genes classified functionally as tumor-suppressive genes (TSGs) and oncogenes (OGs).
| miRNAs | Genes | TSG | OG |
|---|---|---|---|
| has-miR-3613–3p | DFNA5 | ✓ | |
| NOTCH2 | ✓ | ||
| EGR1 | ✓ | ||
| MDM2 | ✓ | ||
| PIM3 | ✓ | ||
| TP53INP1 | ✓ | ||
| has-miR-335–3p | NF1 | ✓ | |
| PHLDA3 | ✓ | ||
| RAF1 | ✓ | ||
| HTATIP2 | ✓ | ||
| SMAD3 | ✓ | ||
| CDKN1B | ✓ | ||
| TP53INP1 | ✓ | ||
| has-miR-548n | SOX4 | ✓ | |
| SMADS | ✓ | ||
| PAWR | ✓ | ||
| SIAH1 | ✓ | ||
| BCL2L11 | ✓ | ||
| AKT1 | ✓ | ||
| TSC22D1 | ✓ |
3.4. Co-expression analysis
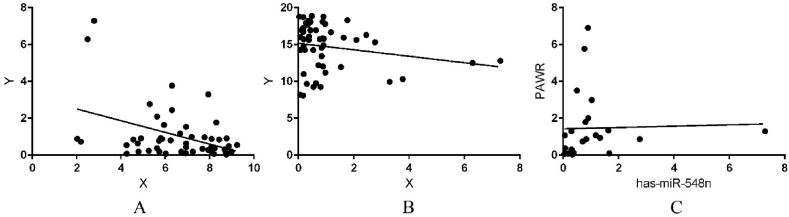
The significantly dysregulated lncRNA in the predicted network associated with breast cancer is likely to show significant co-expression relationship of biological interest with miRNA and mRNA genes. The analysis adds meaningful insight to our study since the co-expressed genes are said to be governed by the same factors which may be coherently related [[63], [64], [65]]. Hence, co-expression analysis of some of the path components between the critical upstream regulator LINC01087, and its modulated oncogenic partners-hsa-miR548n, AKT1 & PAWR gene were performed to identify whether these are significantly co-expressed in breast cancer. Analysis suggests that these dysregulated cancer-associated lncRNAs ignites the function of paired up miRNAs which further push the cascade of events strengthening the whole lncRNA-miRNA-mRNA network in breast cancer samples. We analysed three crucial components of the lncRNA-miRNA-mRNA gene network, using statistical analysis, as obtained from the NGS data and found LINC01087 to be highly upregulated and hsa-miR548n to be downregulated in breast cancer subtypes. Co-expression analysis was used to infer lncRNA–miRNA association from genome-wide expression (FPKM/TPM from SRAdb and TCGA, respectively) counts to interrogate and analyse the interaction. LINC01087 showed a significant negative correlation with miRNA-548n in cancer samples with correlation coefficient R2 = 0.5375 and p-value < 0.001 (Fig. 5a). This co-expression analysis inferred the higher expression of LINC01087 associated with low-expression of miR-548n. We hypothesized that LINC01087 upregulation in the diseased samples might be responsible for the downregulation of hsa-miR-548n in breast cancer cells inhibiting the normal functioning of the cell by promoting the proliferation. Similarly the co-expression analysis of miR-548n and AKT1, showed that miRNA-548n is significantly negatively correlated with AKT1 genes with correlation coefficient R2 = 0.04093 and p-value < 0.001 (Fig. 5b). AKT1 leads to inhibition of kinase activity and phosphorylation of GSK3 isoform to antiapoptotic activities (impairs proliferation) [66]. The upregulation of LINC01087 from our NGS and RT-PCR analysis was in accordance with co-expression analysis, where high expression was in accordance with the low-expression of miRNA-548n. Interestingly, another member of family miR-548 has been reported to have low expression rate in some cancer types (except the Breast cancer) [67,68]. Therefore, we hypothesized that miR-548n in low expressed potential can inhibit the normal activity of AKT1 gene of proliferation and can act as potential candidate for prognostic indicator of chemotherapeutic benefits in breast cancer. Equivalently, miRNA-548n is found to be positively correlated with PAWR genes from the expression data of cancer samples indicating miRNA inhibiting the pro-apoptotic activity of the gene (R2 = 0.0009489, p-value = 0.8864) (Fig. 5c). PAWR normally functions to sensitize the cells to diverse apoptotic stimuli and cause regression of tumors [69]. We also hypothesized miR-548n and PAWR as potential candidates for prognostic indicator of chemotherapeutic benefits in breast cancer in deregulated conditions. Therefore, the three nodes can act as potential candidate markers for prognostic indicator of chemotherapeutic benefits in breast cancer diagnosis.
Fig. 5.
Co-expression analysis (a) between lncRNA and its regulated miRNA showing a negative correlation between them indicating upregulation of LINC01087 and downregulation of hsa-mir548n (b) negative correlation between miRNA and AKT1 gene showing downregulation of hsa-mir548n and upregulation of AKT1 gene (c) positive correlation analysis between hsa-mir548n and PAWR gene both being downregulated in breast cancer associated with inhibition of apoptosis.
3.5. Survival analysis
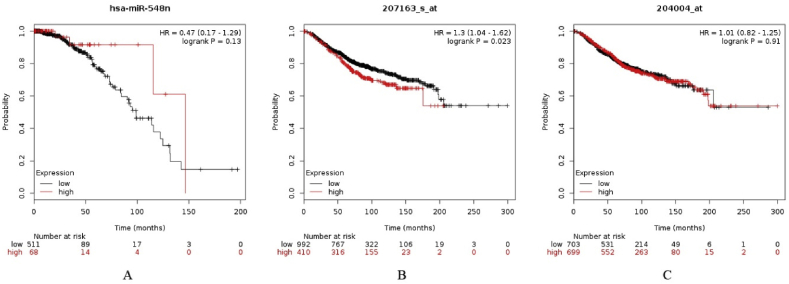
Kaplan-Meier (KM) survival analysis curve [70] was plotted to associate the expression data with the survival rate of breast cancer patients. We checked the expression of novel putative lncRNA an initial component of the path as obtained from the NGS data and found it to be highly up-regulated in breast cancer subtypes using RNA-Seq technique. The subsequent analysis steps wherein the data obtained from TCGA database were subjected to Survival analysis using KM plot. The interacting miR-548n low expression and AKT1 protein coding genes high expression, was seen to be associated with decreased survival rate in breast cancer patients (HR = 0.47, logrank P = 0.13 and HR = 1.3, logrank P = 0.023, respectively). Survival analysis for PAWR gene showed that the low-expression is associated with lower survival rate of breast cancer patients (HR = 1.01, logrank P = 0.91). Therefore, it can be concluded from the statistical analysis that AKT1 in up- and miR-548n in down-regulated condition might play oncogenic and tumor suppressive role, respectively (Fig. 6a and b). PAWR and miR-548n both are down-regulated in abnormal conditions indicating tumor suppressive role for both. The dysregulated expression of hsa-miR-548n has not yet been studied in the breast cancer system and the association with the lncRNA is first of its kind (Fig. 6c).
Fig. 6.
(a) Kaplan-Meier plot demonstrating the relationship between hsa-miR548n expression and survival of patients with breast cancer obtained from analysis of TCGA data. KM plot indicating the association of low expression of hsa-miR548n in breast cancer samples with low survival of patients (b) Kaplan-Meier plot demonstrating the relationship between AKT1 gene expression and survival of patients with breast cancer. KM plot showing the association of high expression of hsa-miR548n in breast cancer samples with low survival of patients (c) Kaplan-Meier plot demonstrating the relationship between PAWR gene expression and survival of patients with breast cancer. KM plot showing the association of low expression of PAWR in breast cancer sample with low survival of patients.
Moreover, starBase v2.0 database [71] was used to establish the relationship of in vitro assayed dysregulated lncRNA of mammalian cells (infected with breast cancer) and proteins. An additional study was performed using Cross-linking Immunoprecipitation sequencing (CLIP-seq) analysis where LINC01087 was found to target three ribosomal proteins RBPs- FUS, U2AF65 and hnRNPC. However, the novel cross-talks among the diverse functioning will help gain understanding of their role in development of breast cancer and other cancers. Further validation of associated proteins in clinical carcinoma tissue samples and in vivo studies can be done to establish the findings at the clinical level as potential candidate of prognostic indicator for chemotherapeutic benefits in cancer metastasis.
4. Conclusion
Dysregulation of the non-coding RNAs at the transcriptional and post-transcriptional level plays a major role in the oncogenesis of breast cells. In this study, an lncRNA-miRNA-mRNA regulatory network was established that could help unravel the mechanism behind the multi-stage process of apoptosis of breast cancer at both coding and non-coding level. There is vast literature dedicated to the study of coding RNAs, however non-coding regulators and their effects on the apoptotic pathway in breast cancer are rarely studied [39,72]. Understanding the role of regulators and how they influences apoptosis is an essential step in improving the outcome of the patient and developing novel therapeutics. Therefore, an lncRNA-miRNA-mRNA regulatory network in breast cancer was constructed with the help of curated regulations and deduced critical regulators, novel apoptotic signaling components and significant active paths which have illuminated the most significant regulatory nodes in metastatic cascades operating in breast cancer. This work represents the first lncRNA-miRNA-mRNA regulatory network in breast cancer.
We obtained top differentially expressed lncRNAs from the RNA-Seq analysis, validated four novel lncRNAs and checked non-coding potential of the candidates. The top lncRNAs were subjected to RT-PCR analysis followed by functional analysis to determine the pathways associated with the linked ncRNAs. From the active sub-network analysis novel sponge-pair interacting partners were shown to participate in the events of apoptosis associated with several genes which did not have any known significant functions in breast cancer. Among them, number of pathways have role reported in apoptosis of cancers, such as CyclinA1 [73], c-Myc [74], DUSP6 [75], WNT-16 [76], Bcl-XL [77], CD-40 [78], p53 [79], p21 [80], BAIAP3 [81], IL6 [82] signaling pathways and few more which pave tumor proliferation. Our statistical analysis also confirmed p53 signaling pathway to be critical in the apoptosis of carcinoma cells as well and ideally placed to regulate other signaling pathways involved in tumorigenesis. These events transcriptional misregulation in cancer could be linked to direct/indirect regulation of protein coding mRNAs involved in cell apoptosis and proliferation. Very few lncRNAs have been characterized and experimentally validated thus far. Among them, the ceRNA in humans and their important sponge interactions can act as icing on cake benefitting the RNA world [61,62]. We speculated the LINC01087/hsa-miR548n pair might be involved in regulating the translation of mRNAs, a hypothesis that is supported by the binding of other proteins related to translational regulation in the LINC01087 ChIP-Seq analysis. Alternatively, LINC010187 may also be involved in oncogenic activity by controlling the AKT1and PAWR gene. Nevertheless further analyses are required to elucidate the mechanisms of action and functional role of the LINC01087-hsa-miR548n-AKT1 (as well as the other associations identified in the RNA-Seq analysis). In conclusion, we identified a significantly dysregulated lncRNAs in breast cancer compared to normal mammary cells.
Declaration of competing interest
The authors confirm that this article content has no conflict of interest.
Acknowledgements
The authors acknowledge the department of Bioinformatics & Applied Sciences, Indian Institute of Information Technology, Allahabad for providing computing facility.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2019.12.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
figs5.
figs6.
References
- 1.Zhang K., Shi Z.M., Chang Y.N., Hu Z.M., Qi H.X., Hong W. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene. 2014;547:1–9. doi: 10.1016/j.gene.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 2.Wright M.W., Bruford E.A. Naming 'junk': human non-protein coding RNA (ncRNA) gene nomenclature. Hum. Genom. 2011;5:90–98. doi: 10.1186/1479-7364-5-2-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinkovics J.G. 2016. RNA/DNA and Cancer: Springer. [Google Scholar]
- 4.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat. Rev. Drug Discov. 2013;12:433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 5.Peschansky V.J., Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9:3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarroux J., Morillon A., Pinskaya M. History, discovery, and classification of lncRNAs. Adv. Exp. Med. Biol. 2017;1008:1–46. doi: 10.1007/978-981-10-5203-3_1. [DOI] [PubMed] [Google Scholar]
- 7.Lin R., Maeda S., Liu C., Karin M., Edgington T.S. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 8.Mann R.S. How genomes generate animals: a view from Pasadena genomic control process: development and evolution. FASEB J. 2015;29:4764–4765. doi: 10.1096/fj.15-1203ufm. [DOI] [PubMed] [Google Scholar]
- 9.Duret L., Chureau C., Samain S., Weissenbach J., Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- 10.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J.T. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 12.Arun G., Diermeier S., Akerman M., Chang K.C., Wilkinson J.E., Hearn S., Kim Y., MacLeod A.R., Krainer A.R., Norton L., Brogi E., Egeblad M., Spector D.L. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisler S., Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon J.H., Abdelmohsen K., Kim J., Yang X., Martindale J.L., Tominaga-Yamanaka K., White E.J., Orjalo A.V., Rinn J.L., Kreft S.G., Wilson G.M., Gorospe M. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan Y., Zhang L., Jiang Y., Xu T., Mei Q., Wang H., Qin R., Zou Y., Hu G., Chen J., Lu Y. LncRNA and mRNA interaction study based on transcriptome profiles reveals potential core genes in the pathogenesis of human glioblastoma multiforme. J. Cancer Res. Clin. Oncol. 2015;141:827–838. doi: 10.1007/s00432-014-1861-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jalali S., Bhartiya D., Lalwani M.K., Sivasubbu S., Scaria V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trichopoulos D., Li F.P., Hunter D.J. What causes cancer? Sci. Am. 1996;275:80–87. doi: 10.1038/scientificamerican0996-80. [DOI] [PubMed] [Google Scholar]
- 18.Hammond M.E., Hayes D.F., Dowsett M., Allred D.C., Hagerty K.L., Badve S., Fitzgibbons P.L., Francis G., Goldstein N.S., Hayes M., Hicks D.G., Lester S., Love R., Mangu P.B., McShane L., Miller K., Osborne C.K., Paik S., Perlmutter J., Rhodes A., Sasano H., Schwartz J.N., Sweep F.C., Taube S., Torlakovic E.E., Valenstein P., Viale G., Visscher D., Wheeler T., Williams R.B., Wittliff J.L., Wolff A.C., American Society of Clinical O., College of American P. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version) Arch. Pathol. Lab Med. 2010;134:e48–72. doi: 10.5858/134.7.e48. [DOI] [PubMed] [Google Scholar]
- 19.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 20.Tripathi R., Patel S., Kumari V., Chakraborty P., Varadwaj P.K. DeepLNC, a long non-coding RNA prediction tool using deep neural network. Netw. Model. Anal. Health Inform. Bioinform. 2016;5:21. [Google Scholar]
- 21.Sana J., Faltejskova P., Svoboda M., Slaby O. Novel classes of non-coding RNAs and cancer. J. Transl. Med. 2012;10:103. doi: 10.1186/1479-5876-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng G., Sui G. Noncoding RNA in oncogenesis: a new era of identifying key players. Int. J. Mol. Sci. 2013;14:18319–18349. doi: 10.3390/ijms140918319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang G., Lu X., Yuan L. LncRNA: a link between RNA and cancer. Biochim. Biophys. Acta. 2014;1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Biswas D.K., Shi Q., Baily S., Strickland I., Ghosh S., Pardee A.B., Iglehart J.D. NF-kappa B activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10137–10142. doi: 10.1073/pnas.0403621101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mingo-Sion A.M., Marietta P.M., Koller E., Wolf D.M., Van Den Berg C.L. Inhibition of JNK reduces G2/M transit independent of p53, leading to endoreduplication, decreased proliferation, and apoptosis in breast cancer cells. Oncogene. 2004;23:596–604. doi: 10.1038/sj.onc.1207147. [DOI] [PubMed] [Google Scholar]
- 26.Tuo Y.L., Li X.M., Luo J. Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. Eur. Rev. Med. Pharmacol. Sci. 2015;19:3403–3411. [PubMed] [Google Scholar]
- 27.Yang F., Bi J., Xue X., Zheng L., Zhi K., Hua J., Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 28.Pickard M.R., Williams G.T. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: implications for chemotherapy. Breast Canc. Res. Treat. 2014;145:359–370. doi: 10.1007/s10549-014-2974-y. [DOI] [PubMed] [Google Scholar]
- 29.Shi Y., Lu J., Zhou J., Tan X., He Y., Ding J., Tian Y., Wang L., Wang K. Long non-coding RNA Loc554202 regulates proliferation and migration in breast cancer cells. Biochem. Biophys. Res. Commun. 2014;446:448–453. doi: 10.1016/j.bbrc.2014.02.144. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y., Sharma S., Watabe K. Roles of lncRNA in breast cancer. Front. Biosci. 2015;7:94–108. doi: 10.2741/s427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Malih S., Saidijam M., Malih N. A brief review on long noncoding RNAs: a new paradigm in breast cancer pathogenesis, diagnosis and therapy. Tumour Biol. 2016;37:1479–1485. doi: 10.1007/s13277-015-4572-y. [DOI] [PubMed] [Google Scholar]
- 33.Vennin C., Spruyt N., Dahmani F., Julien S., Bertucci F., Finetti P., Chassat T., Bourette R.P., Le Bourhis X., Adriaenssens E. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget. 2015;6:29209–29223. doi: 10.18632/oncotarget.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Z., Chen C., Liu Y., Wu C. 17beta-Estradiol treatment inhibits breast cell proliferation, migration and invasion by decreasing MALAT-1 RNA level. Biochem. Biophys. Res. Commun. 2014;445:388–393. doi: 10.1016/j.bbrc.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Huang J., Zhou N., Watabe K., Lu Z., Wu F., Xu M., Mo Y.Y. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) Cell Death Dis. 2014;5 doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X.F., Liu T., Li Y., Li S. Overexpression of long non-coding RNA CCAT1 is a novel biomarker of poor prognosis in patients with breast cancer. Int. J. Clin. Exp. Pathol. 2015;8:9440–9445. [PMC free article] [PubMed] [Google Scholar]
- 37.Redis R.S., Sieuwerts A.M., Look M.P., Tudoran O., Ivan C., Spizzo R., Zhang X., de Weerd V., Shimizu M., Ling H., Buiga R., Pop V., Irimie A., Fodde R., Bedrosian I., Martens J.W., Foekens J.A., Berindan-Neagoe I., Calin G.A. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations. Oncotarget. 2013;4:1748–1762. doi: 10.18632/oncotarget.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorensen K.P., Thomassen M., Tan Q., Bak M., Cold S., Burton M., Larsen M.J., Kruse T.A. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Canc. Res. Treat. 2013;142:529–536. doi: 10.1007/s10549-013-2776-7. [DOI] [PubMed] [Google Scholar]
- 39.Mourtada-Maarabouni M., Pickard M.R., Hedge V.L., Farzaneh F., Williams G.T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 40.Weakley S.M., Wang H., Yao Q., Chen C. Expression and function of a large non-coding RNA gene XIST in human cancer. World J. Surg. 2011;35:1751–1756. doi: 10.1007/s00268-010-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sang Y., Tang J., Li S., Li L., Tang X., Cheng C., Luo Y., Qian X., Deng L.M., Liu L., Lv X.B. LncRNA PANDAR regulates the G1/S transition of breast cancer cells by suppressing p16(INK4A) expression. Sci. Rep. 2016;6:22366. doi: 10.1038/srep22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghobrial I.M., Witzig T.E., Adjei A.A. Targeting apoptosis pathways in cancer therapy. CA Cancer J Clin. 2005;55:178–194. doi: 10.3322/canjclin.55.3.178. [DOI] [PubMed] [Google Scholar]
- 43.Williams G.T. Programmed cell death: apoptosis and oncogenesis. Cell. 1991;65:1097–1098. doi: 10.1016/0092-8674(91)90002-g. [DOI] [PubMed] [Google Scholar]
- 44.Shintani T., Klionsky D.J. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 47.Tripathi R., Sharma P., Chakraborty P., Varadwaj P.K. Next-generation sequencing revolution through big data analytics. Front. Life Sci. 2016;9:119–149. [Google Scholar]
- 48.Paci P., Colombo T., Farina L. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst. Biol. 2014;8:83. doi: 10.1186/1752-0509-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou M., Wang X., Shi H., Cheng L., Wang Z., Zhao H., Yang L., Sun J. Characterization of long non-coding RNA-associated ceRNA network to reveal potential prognostic lncRNA biomarkers in human ovarian cancer. Oncotarget. 2016;7:12598–12611. doi: 10.18632/oncotarget.7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Volders P.J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J., Mestdagh P. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:D174–D180. doi: 10.1093/nar/gku1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vizcaino J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q.W., Wang R., Hermjakob H. Update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin M.F., Jungreis I., Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W. CPAT: coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yotsukura S., duVerle D., Hancock T., Natsume-Kitatani Y., Mamitsuka H. Computational recognition for long non-coding RNA (lncRNA): software and databases. Briefings Bioinf. 2017;18:9–27. doi: 10.1093/bib/bbv114. [DOI] [PubMed] [Google Scholar]
- 57.Paraskevopoulou M.D., Georgakilas G., Kostoulas N., Reczko M., Maragkakis M., Dalamagas T.M., Hatzigeorgiou A.G. DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 2013;41:D239–D245. doi: 10.1093/nar/gks1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X. Computational prediction of microRNA targets. Methods Mol. Biol. 2010;667:283–295. doi: 10.1007/978-1-60761-811-9_19. [DOI] [PubMed] [Google Scholar]
- 59.Shi Y., Qiu M., Wu Y., Hai L. MiR-548-3p functions as an anti-oncogenic regulator in breast cancer. Biomed. Pharmacother. 2015;75:111–116. doi: 10.1016/j.biopha.2015.07.027. [DOI] [PubMed] [Google Scholar]
- 60.Taucher V., Mangge H., Haybaeck J. Non-coding RNAs in pancreatic cancer: challenges and opportunities for clinical application. Cell. Oncol. 2016;39:295–318. doi: 10.1007/s13402-016-0275-7. [DOI] [PubMed] [Google Scholar]
- 61.Ferracin M., Querzoli P., Calin G.A., Negrini M. MicroRNAs: toward the clinic for breast cancer patients. Semin. Oncol. 2011;38:764–775. doi: 10.1053/j.seminoncol.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 62.Su H., Jin X., Zhang X., Xue S., Deng X., Shen L., Fang Y., Xie C. Identification of microRNAs involved in the radioresistance of esophageal cancer cells. Cell Biol. Int. 2014;38:318–325. doi: 10.1002/cbin.10202. [DOI] [PubMed] [Google Scholar]
- 63.Kapranov P., Willingham A.T., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 64.Gennarino V.A., D'Angelo G., Dharmalingam G., Fernandez S., Russolillo G., Sanges R., Mutarelli M., Belcastro V., Ballabio A., Verde P., Sardiello M., Banfi S. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res. 2012;22:1163–1172. doi: 10.1101/gr.130435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kartha R.V., Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front. Genet. 2014;5:8. doi: 10.3389/fgene.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hennessy B.T., Smith D.L., Ram P.T., Lu Y., Mills G.B. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat. Rev. Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 67.Fang L., Zhang H.B., Li H., Fu Y., Yang G.S. miR-548c-5p inhibits proliferation and migration and promotes apoptosis in CD90(+) HepG2 cells. Radiol. Oncol. 2012;46:233–241. doi: 10.2478/v10019-012-0025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang T., Guo L., Liu C. Genome-wide analysis of mir-548 gene family reveals evolutionary and functional implications. J. Biomed. Biotechnol. 2012:679563. doi: 10.1155/2012/679563. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnstone R.W., Ruefli A.A., Lowe S.W. Apoptosis: a link between cancer genetics and chemotherapy. Cell. 2002;108:153–164. doi: 10.1016/s0092-8674(02)00625-6. [DOI] [PubMed] [Google Scholar]
- 70.Goel M.K., Khanna P., Kishore J. Understanding survival analysis: Kaplan-Meier estimate. Int. J. Ayurveda Res. 2010;1:274–278. doi: 10.4103/0974-7788.76794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huarte M., Rinn J.L. Large non-coding RNAs: missing links in cancer? Hum. Mol. Genet. 2010;19:R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hsia D.A., Tepper C.G., Pochampalli M.R., Hsia E.Y., Izumiya C., Huerta S.B., Wright M.E., Chen H.W., Kung H.J., Izumiya Y. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9671–9676. doi: 10.1073/pnas.1000401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liao D.J., Dickson R.B. c-Myc in breast cancer. Endocr. Relat. Cancer. 2000;7:143–164. doi: 10.1677/erc.0.0070143. [DOI] [PubMed] [Google Scholar]
- 75.Nunes-Xavier C.E., Tarrega C., Cejudo-Marin R., Frijhoff J., Sandin A., Ostman A., Pulido R. Differential up-regulation of MAP kinase phosphatases MKP3/DUSP6 and DUSP5 by Ets2 and c-Jun converge in the control of the growth arrest versus proliferation response of MCF-7 breast cancer cells to phorbol ester. J. Biol. Chem. 2010;285:26417–26430. doi: 10.1074/jbc.M110.121830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Katoh M. WNT and FGF gene clusters (review) Int. J. Oncol. 2002;21:1269–1273. [PubMed] [Google Scholar]
- 77.Moore M.R., Conover J.L., Franks K.M. Progestin effects on long-term growth, death, and Bcl-xL in breast cancer cells. Biochem. Biophys. Res. Commun. 2000;277:650–654. doi: 10.1006/bbrc.2000.3728. [DOI] [PubMed] [Google Scholar]
- 78.Hislop T.G., Waxler N.E., Coldman A.J., Elwood J.M., Kan L. The prognostic significance of psychosocial factors in women with breast cancer. J. Chronic Dis. 1987;40:729–735. doi: 10.1016/0021-9681(87)90110-x. [DOI] [PubMed] [Google Scholar]
- 79.Gasco M., Shami S., Crook T. The p53 pathway in breast cancer. Breast Cancer Res. 2002;4:70–76. doi: 10.1186/bcr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mira J.P., Benard V., Groffen J., Sanders L.C., Knaus U.G. Endogenous, hyperactive Rac3 controls proliferation of breast cancer cells by a p21-activated kinase-dependent pathway. Proc. Natl. Acad. Sci. U. S. A. 2000;97:185–189. doi: 10.1073/pnas.97.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Palmer R.E., Lee S.B., Wong J.C., Reynolds P.A., Zhang H., Truong V., Oliner J.D., Gerald W.L., Haber D.A. Induction of BAIAP3 by the EWS-WT1 chimeric fusion implicates regulated exocytosis in tumorigenesis. Cancer Cell. 2002;2:497–505. doi: 10.1016/s1535-6108(02)00205-2. [DOI] [PubMed] [Google Scholar]
- 82.Korkaya H., Kim G.I., Davis A., Malik F., Henry N.L., Ithimakin S., Quraishi A.A., Tawakkol N., D'Angelo R., Paulson A.K., Chung S., Luther T., Paholak H.J., Liu S., Hassan K.A., Zen Q., Clouthier S.G., Wicha M.S. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol. Cell. 2012;47:570–584. doi: 10.1016/j.molcel.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.