Abstract
Biosynthesis for the preparation of antimicrobial silver nanoparticles (Ag NPs) is a green method without the use of cytotoxic reducing and surfactant agents. Herein, shape-controlled and well-dispersed Ag NPs were biosynthesized using yeast extract as reducing and capping agents. The synthesized Ag NPs exhibited a uniform spherical shape and fine size, with an average size of 13.8 nm. The biomolecules of reductive amino acids, alpha-linolenic acid, and carbohydrates in yeast extract have a significant role in the formation of Ag NPs, which was proved by the Fourier transform infrared spectroscopy analysis. In addition, amino acids on the surface of Ag NPs carry net negative charges which maximize the electrostatic repulsion interactions in alkaline solution, providing favorable stability for more than a year without precipitation. The Ag NPs in combination treatment with ampicillin reversed the resistance in ampicillin-resistant E. coli cells. These monodispersed Ag NPs could be a promising alternative for the disinfection of multidrug-resistant bacterial strains, and they showed negligible cytotoxicity and good biocompatibility toward Cos-7 cells.
Keywords: Silver nanoparticles, Biosynthesis, Yeast Extract, Stability, Antibacterial activity
Introduction
Drug-resistant infections are a major cause of death and have resulted in a serious risk to public health. Additionally, increasing resistance to antimicrobial drugs is emerging as an urgent problem in medicine [1]. A number of strains of Staphylococcus aureus are resistant to methicillin and are the major cause of acquired infections in hospitals. Furthermore, other antibiotic-resistant bacteria include penicillin-resistant Neisseria gonorrhoeae and multidrug-resistant Escherichia coli (E. coli) [2, 3]. The major mechanisms of resistance are increased efflux and the reduced absorption of antibiotics [4]. Another mechanism of drug resistance is the expression of enzymes that modify the molecular structure of antibiotics [5]. Although much effort has been focused on developing the next generation of antimicrobial agents, there is an increased need for superior disinfection methods.
Silver nanoparticles (Ag NPs) have been used in many applications, such as protein carriers, radiosensitizers, solar fuel cell efficiency improvement, and antibacterial agents [6–8]. Nanoparticles, including metal-containing nanoparticles, Ag NPs are the most widely used as antimicrobial agents [9]. In reality, silver nanoparticles have shown significant antimicrobial activity against bacterial strains but negligible cytotoxicity to animal cells [10, 11]. Moreover, Ag NPs have exhibited antimicrobial activity against fungi, certain viruses, and antibiotic-resistant bacterial strains. With regard to their mechanism of action, suppression of DNA replication, blockage of the electrical potential difference needed in cytoplasmic membranes, and suppression of the respiratory chain are the main mechanisms of action of Ag NPs. Thus, the size, surface structure, and controlled shapes of Ag NPs play crucial roles in their antimicrobial activity and other applications. The general method for the preparation of Ag NPs involves the reduction of silver ions in the presence of an appropriate surfactant to achieve the controlled growth of Ag NPs [12]. The majority of reducing and surfactant agents show cytotoxicity to human tissue cells and potentially cause environmental contamination. Therefore, more effort in developing green methods for the preparation of shape-controlled Ag NPs is essential.
In this work, we present a novel route for the biosynthesis of Ag NPs by utilizing yeast extract. During the process, yeast extract supplies reducing and capping agents, including amino acids, vitamins, and carbohydrates, whereas silver ions serve as an electron acceptor. As a result, the favorable stability provided by the organic capping agents on the surface, the monodispersed Ag NPs, can be preserved for more than a year without precipitation. It was found that Ag NPs displayed a superior antibacterial activity compared to ampicillin against ampicillin-resistant E. coli cells. Compared to conventional synthetic methods, the biosynthesis approach presented herein is biocompatible, cost-effective, and environmentally benign. Furthermore, the shape-controlled and well-dispersed Ag NPs displayed good antibacterial effects toward E. coli.
Methods
Materials
Silver nitrate (AgNO3), sucrose (C12H22O11), sodium chloride (NaCl), and sodium hydroxide (NaOH) were purchased from Sinopharm Co., Ltd. Dry baker’s yeast was obtained from AB/MAURI Co., Ltd. E. coli was purchased from TransGen Biotech Co., Ltd. The CellTiter 96® Aqueous One Solution Cell Proliferation Assay kit (MTS) was purchased from Promega Biotech Co., Ltd. pcDNA3.4 plasmid, 1 × NuPAGE® LDS sample buffer, Dulbecco’s modified Eagle medium (DMEM) and fetal bovine serum (FBS) were purchased from Thermo Fisher Scientific Inc. Ampicillin and Luria-Bertani (LB) medium were purchased from Sangon Biotech Co., Ltd. All chemicals were analytical reagents and used without further purification. Deionized ultrapure water (18.2 MΩ.cm) was used throughout the experiments.
Synthesis of Ag NPs
The stocked yeast cells were inoculated into Luria-Bertani (LB) medium and shaken at approximately 150 rpm overnight at 25 °C for activation. Then, the activated yeast cells were washed with 0.9% saline solution and dispersed in 2% sucrose solution with shaking at approximately 150 rpm for 6 h at 25 °C. Finally, the cell-free yeast extract was collected for the biosynthesis of Ag NPs by centrifugation at 2000 rpm for 5 min. During the biosynthetic process, the pH value of yeast extract was adjusted to 10 with a NaOH solution, and then, the AgNO3 solution was gradually added to the above solution under vigorously magnetic stirring. At last, the obtained Ag NPs were dialyzed with 1 kDa dialysis membranes for 5 days and freeze-dried for further characterization.
Characterizations
Transmission electron microscopy (TEM) images of Ag NPs were observed on JEM-2100 microscope with an accelerating voltage of 200 kV (JEOL, Japan). Scanning electron microscopy (SEM) images were obtained on a Carl Zeiss ULTRA plus scanning electron microscope (Carl Zeiss, Germany) equipped with an energy dispersive spectrometer (EDS) operated at 20 kV. Ultraviolet-visible (UV-Vis) absorption spectra were recorded on a Lambda 950 UV/Vis/NIR spectrophotometer (Perkin-Elmer, USA). X-ray powder diffraction (XRD) patterns were obtained using a D8 Advance instrument (Bruker, Germany). Fourier transform infrared spectroscopy (FTIR) was recorded from 4000-500 cm−1 with samples prepared as KBr pellets on a Vertex 70 FTIR spectrometer (Bruker, Germany). The zeta-potential of Ag NPs was measured with a Malvern Zeta Nano ZS-90 (Malvern, United Kingdom) at 25 °C. The surface elements on Ag NPs were identified by X-ray photoelectron spectroscopy (XPS) using a Kratos AXIS Ultra DLD instrument with a monochromatic Al Kα source (1486.6 eV) (Shimadzu, Japan). The amino acid components were analyzed with an L-8900 high-speed amino acid analyzer (Hitachi, Japan).
Cell Cytotoxicity Assay
To explore the biocompatibility of the prepared Ag NPs, an MTS assay was employed to evaluate the cell cytotoxicity of the Ag NPs [13]. Cos-7 cells were cultured in DMEM supplemented with 10% FBS complete medium in a humidified atmosphere incubator containing 5% CO2 at 37 °C. The cells were plated into 96-well flat-bottomed plates at a density of 10000 cells per well and cultured for 24 h. Then, the growth medium was replaced with fresh DMEM medium containing different concentrations of Ag NPs. After incubation for another 24 h, the relatively viable cells were determined by MTS. The absorbance was measured at 490 nm using a SpectraMax® M5 microplate reader (Molecular Devices, USA). Nontreated cells in DMEM medium were used as a control.
SDS-PAGE Assay
Standard SDS-PAGE was performed with a 10% (w/v) separating gel and a 4% stacking gel. The samples were boiled for 5 min with 1 × NuPAGE® LDS Sample Buffer and centrifuged at 12000 rpm for 5 min before application to the gels. The standard protein marker was used as a reference control. The gels were stained with 0.5% Coomassie Blue. Images of gels were recorded with GelDoc XR+ gel imaging systems (Bio-Rad, USA).
Antimicrobial Activity Studies
To determine the antimicrobial activity, the synthesized Ag NPs were tested for bactericidal activity against E. coli [14]. A single colony of E. coli was grown overnight at 37 °C in LB medium on an orbital shaker at 150 rpm. Colonies were adjusted to an OD of 0.01–0.02 at 600 nm with fresh LB medium. Then, 100 μL of serial dilutions of Ag NPs were filled onto 96-well microplates. The microplates were then inoculated with 100 μL of diluted E. coli solution and incubated for 16 h at 37 °C. The viability of E. coli was determined by the measurement of the absorbance at 600 nm with a SpectraMax® M5 microplate reader (Molecular Devices, USA). A time-course analysis was performed to evaluate the antibacterial sensitivity against E. coli over time. Finally, 100 μL of E. coli solution was added to sterile tubes containing 10 and 20 μg/mL Ag NPs, respectively. The absorbance at 600 nm was measured with a SpectraMax® M5 microplate reader (Molecular Devices, USA) after 1, 2, 4, and 6 h.
Colony-forming unit assay was introduced to investigate the Ag NPs against the antibiotic-resistant bacterial cells. E. coli stably expresses pcDNA3.4 plasmid containing the β-lactamase gene which confers resistance to ampicillin as a model. When the ampicillin-resistant E. coli (E. coli-Amp+) cells reached log phase growth, the E. coli-Amp+ cells were grown in the LB agar plate in the treatment with ampicillin alone or in the combinational treatment with Ag NPs and incubated at 37 °C for 18 h. The number of E. coli-Amp+ colonies formed on LB plates was calculated. All assays were performed at least three times.
Results and Discussion
Synthesis of Ag NPs
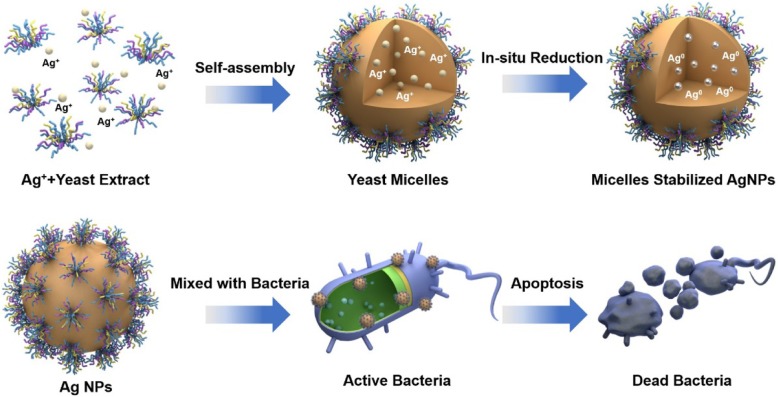
As schematically illustrated in Scheme 1, the preparation of Ag NPs started with the self-assembly of biomolecules in the yeast extract to form yeast micelles. Then, Ag+ was reduced in situ by the reducing agents in the yeast extract, including amino acids, vitamins, and carbohydrates. The formed Ag nanoparticles were stabilized by the biomolecules. The surface coating on Ag NPs enhanced the affinity towards the bacterial membrane, increasing the permeability of the cell wall. The interaction between Ag NPs and peptidoglycan changed the configuration of peptidoglycan, finally leading to the apoptosis process to damage the bacteria.
Scheme 1.
Proposed schematic illustration of the biosynthesis of Ag NPs
Structural Characterization of Ag NPs
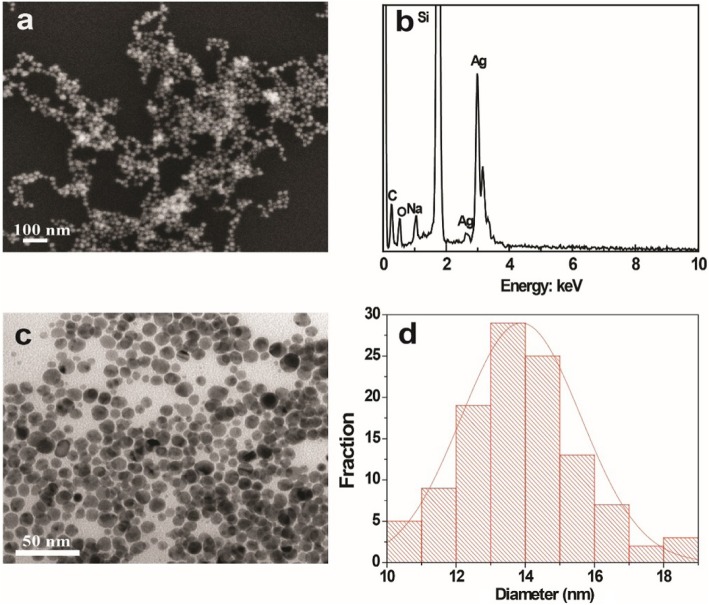
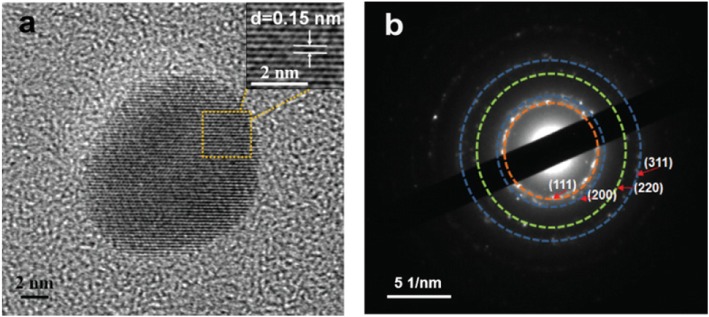
As shown in Fig. 1a, the typical SEM image showed that the synthesized Ag NPs have a spherical shape and fine size. The EDX confirmed the formation of Ag NPs (Fig. 1b). A strong optical absorption peak was observed at approximately 3 keV, which is a typical optical absorption peak of silver nanocrystallites for surface plasmon resonance. The minor amounts of oxygen and carbon could be attributed to the thin layer of organic capping on the synthesized Ag NPs. The reaction of AgNO3 solution with NaOH leads to the formation of a small amount of Ag2O. Therefore, a small amount of O can also be attributed to the presence of Ag2O. The morphology and size of the Ag NPs were further characterized by high-resolution TEM (HRTEM). The Ag NPs ranged in diameter from 10.3 to 18.9 nm (Fig. 1c), with an average size of 13.8 nm (Fig. 1d). The size, shape, and surface chemistry of Ag NPs showed an important effect on the antimicrobial activity. The smaller size and higher surface area allowed the Ag NPs to better interact with the bacterial membrane for further enhanced antimicrobial activity [15–17]. The clear lattice fringes in the HRTEM image showed a fringe spacing of 0.15 nm (Fig. 2a), which corresponds to the (220) planes of silver. As shown in Fig. 2b, the crystalline nature of the Ag NPs was demonstrated by the typical selected-area diffraction (SAED) patterns, where the bright circular rings correspond to the (311), (220), (200), and (111) planes [18, 19].
Fig. 1.
a Field emission SEM image of Ag NPs, b EDX spectrum of Ag NPs, c TEM image of Ag NPs, and d size distribution of Ag NPs
Fig. 2.
a Lattice fringes of Ag NPs in the HR-TEM image, b circular rings of Ag NPs from the typical selected-area diffraction (SAED) patterns
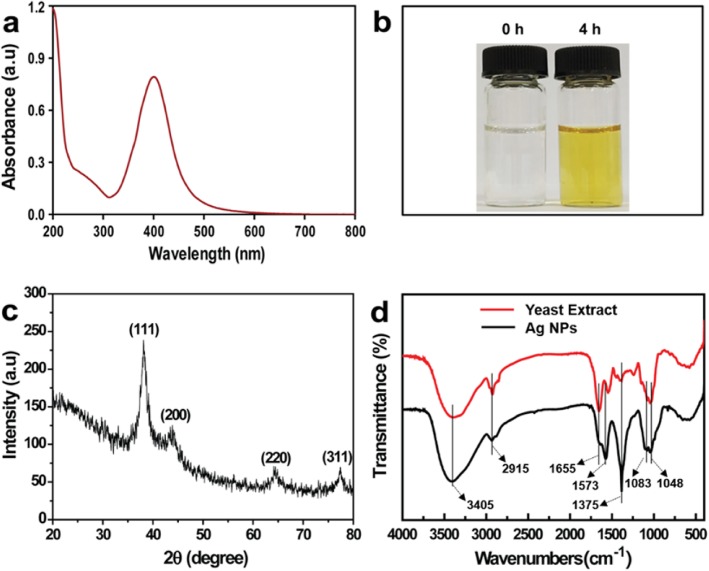
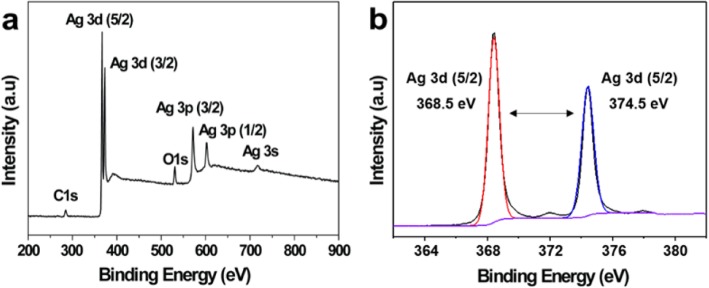
The UV-Vis spectrum of Ag NPs exhibited a strong peak at 418 nm, which was due to surface plasmon resonance (Fig. 3a). A yellow solution of synthesized Ag NPs is shown in Fig. 3b, which indicates the formation of Ag NPs. The XRD pattern analysis of the synthesized Ag NPs showed four intense peaks at 77.36°, 64.30°, 43.52°, and 38.16°, corresponding to the (311), (220), (200), and (111) planes for silver, respectively (Fig. 3c). The data was confirmed by standard silver data from JCPDS card No. 04-0783 [20]. The XRD pattern demonstrated the crystalline nature of the synthesized Ag NPs, in agreement with a previous report [21]. FTIR analysis was employed to characterize and identify the potential biomolecules on the synthesized Ag NPs (Fig. 3d). The broad band at 3405 cm−1 corresponds to −OH stretching [22]. The weaker peak at 2915 cm−1 is assigned to the −CH2 stretching vibration. The band at 1655 cm−1 in the yeast extract is due to the C=O stretching vibration of carboxyl moieties, and this band shifts to 1573 cm−1 in Ag NPs, due to the interaction between carboxyl moieties and Ag NPs [23]. The sharp peak at 1375 cm−1 is attributed to the C–N stretching vibration. The bands at 1048 cm−1 and 1083 cm−1 are assigned to the stretching vibrations of C–O–C and C–OH, respectively [24, 25]. These results demonstrated that biomolecules of the yeast extract were responsible for the biosynthesis of Ag NPs. The surface coating on Ag NPs affected the affinity towards the bacterial membrane [26, 27]. The states of Ag NPs were further characterized by XPS. As shown in Fig. 4a, the full scan of the XPS spectrum with clear peaks was attributed to C 1s, Ag 3d, Ag 3p, Ag 3s, and O 1s. The Ag 3d (5/2) and Ag 3d (3/2) peaks were observed at binding energies of approximately 368.5 and 374.5 eV, respectively (Fig. 4b). This energy splitting value of 6.0 eV demonstrated the formation of Ag NPs [28, 29].
Fig. 3.
a UV-Vis spectrum of Ag NPs, b photo of synthesized Ag NPs, c XRD pattern of Ag NPs, and d FTIR spectrum of Ag NPs and yeast extract
Fig. 4.
a The full scan of XPS spectrum of Ag NPs and b the Ag 3d XPS spectrum
The surface charge of Ag NPs was determined by Malvern Zeta Nano ZS-90 instrument, which is an important parameter of stability and dispersion of the colloidal solutions. The zeta potential is the surface electrostatic potential at the boundary between the diffuse layer and compact layer of nanoparticles, and which is an indicator for applications of biomedical polymers [30]. As shown in Additional file 1: Fig. S1, at a lower pH value of 3, the zeta potential of Ag NPs revealed a slightly negative charge (− 3.2 mV). The zeta potential of Ag NPs decreased monotonically from − 12.1 mV at pH 7.0 to − 24.4 mV at pH 11.0, which confirmed the negatively charged groups on the surface of Ag NPs. Gao et al. reported that the dispersion and stability of Ag NPs mainly attribute to the surface charge [31]. The presence of negatively charged groups improves the stability and dispersion of Ag NPs in aqueous solutions [32].
Cytotoxicity of Ag NPs and Analysis of Biomolecules
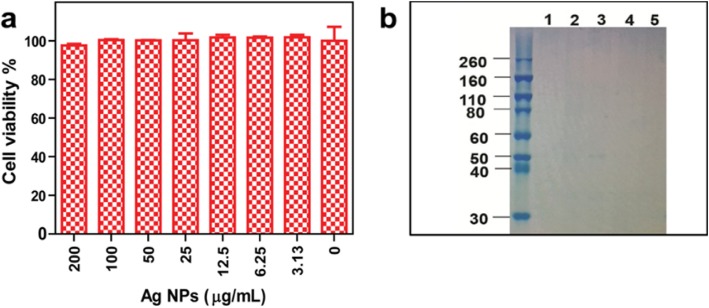
The biocompatibility of the synthesized Ag NPs is important for their further biomedical application. To investigate the cytotoxicity of the Ag NPs, the cell viability of Cos-7 cells was detected by the MTS assay. The Cos-7 cells were incubated with Ag NPs at different concentrations for 24 h. As shown in Fig. 5a, no significant cytotoxicity was revealed when cells were treated with the Ag NPs at concentrations as high as 200 μg/mL. It can be concluded that the Ag NPs showed negligible cytotoxicity and good biocompatibility towards Cos-7 cells.
Fig. 5.
a Cytotoxicity of Ag NPs in Cos-7 cells and, b SDS-PAGE analysis. Lane 1: loading buffer control. Lanes 2–4: synthesized Ag NPs. Lane 5: yeast extract centrifuged with 8000 rpm
To explore the synthetic mechanisms of the synthesized Ag NPs, we analyzed biomolecules on the surface of Ag NPs and yeast extract. As shown in Fig. 5b, the SDS-PAGE analysis showed no detectable or marginal protein on the surface of the synthesized Ag NPs or in the yeast extract. We further determined the biomolecules in the yeast extract with a high-speed amino acid analyzer. As summarized in Additional file 1: Table S1 of supporting information, there are approximately 22 kinds of amino acids in the yeast extract that are rich in glutamic acid, γ-aminobutyric acid, ornament, and alpha-linolenic acid. The isoelectric point of these amino acids is approximately 6, except those of lysine and arginine are approximately 10~11. In addition, a variety of components containing −NH2, such as urea, ammonia, asparagine, and glutamine, could be found. The biomolecules of reductive amino acids, alpha-linolenic acid, and carbohydrates in the yeast extract have a significant role in the formation of Ag NPs. It was reported that NADH-dependent reductase [33, 34] or the nitrate reductase enzyme is involved in the reduction process [35–37] in the biosynthesis of Ag NPs via the microorganism extract.
Biomolecules of the yeast extract play a decisive part in the formation of Ag NPs by protecting them from aggregation. Stabilizers of biomolecules help to prevent redundant reactions between Ag NPs [38]. The amphoteric molecules of amino acids contain both basic and acidic groups. The net charge of these amino acid compounds can be negative or positive depending on the pH changes of the yeast extract solution, which further affects the binding ability during the synthesis of Ag NPs [39]. In the alkaline solution, amino acids on the surface of Ag NPs carry net negative charges which maximize the electrostatic repulsion interactions [40–42]. The biomolecules from the yeast extract act as a capping agent and play a key role in controlling the size distribution, shape, and morphology in the formation of Ag NPs. The value of pH is an important factor with an effect on the controlled synthesis of Ag NPs in the study. When the pH value is below 7, nucleation occurs at a low rate. Ag NPs can be formed in a few minutes at higher pH values, and the particle size decreases with the increasing pH values of the solution. The optimal balance was demonstrated between the growth processes and nucleation [43]. The unstable and agglomerated Ag NPs always presented in the reduction process of solutions with extreme pH values (> 11) [44].
Antibacterial Activity
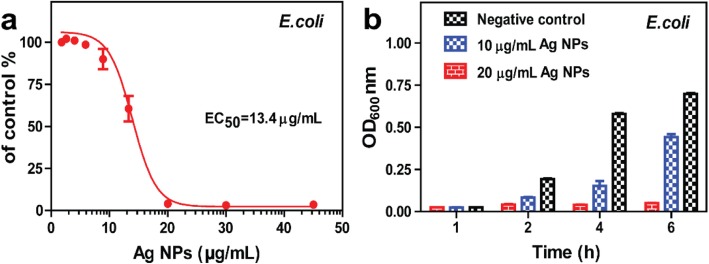
E. coli has been extensively evaluated for the antimicrobial activity of Ag NPs. The growth of E. coli in the presence or absence of Ag NPs proves the antimicrobial ability. As shown in Fig. 6a, the synthesized Ag NPs exhibited significant antibacterial activity in a concentration-dependent manner against E. coli. The growth inhibition assay demonstrated a complete reduction in E. coli at Ag NP concentrations above 20.0 μg/mL compared to the negative control. The half inhibitory concentration (EC50) of Ag NPs was 13.4 μg/mL. The dose of 20.0 μg/mL Ag NPs exhibited a significant antibacterial effect against E. coli throughout the tested time, while the 10.0 μg/mL Ag NPs showed a partial inhibitory effect (Fig. 6b).
Fig. 6.
a The growth inhibition of E. coli and b time course analysis of the antibacterial effect
In order to investigate if the Ag NPs really affects the antibiotic-resistant bacterial cells, we evaluated the antibacterial activity of Ag NPs against ampicillin-resistant E. coli by colony-forming unit assay. E. coli-Amp+ stably expresses a high copy number of pcDNA3.4 plasmid containing the β-lactamase gene which confers ampicillin-resistance to E. coli [45]. The E. coli-Amp+ cells were grown in the LB agar plate in the treatment with ampicillin alone or in the combinational treatment with Ag NPs. The inhibitory activity of the prepared Ag NPs is presented in Fig. 7. It was noted that the Ag NPs in combination treatment with ampicillin displayed superior antibacterial activity compared to ampicillin alone. In contrast, the treatment of ampicillin alone has no inhibitory activity on E. coli-Amp+. Combination therapy of antibiotics and Ag NPs provides a complementary strategy to overcome antibiotic-resistant bacterial cells, which further improves the current therapeutic approaches. The overall results presented in this study contribute to the development of alternative antibacterial inhibitors to treat bacterial infections caused by multidrug-resistant bacterial strains.
Fig. 7.
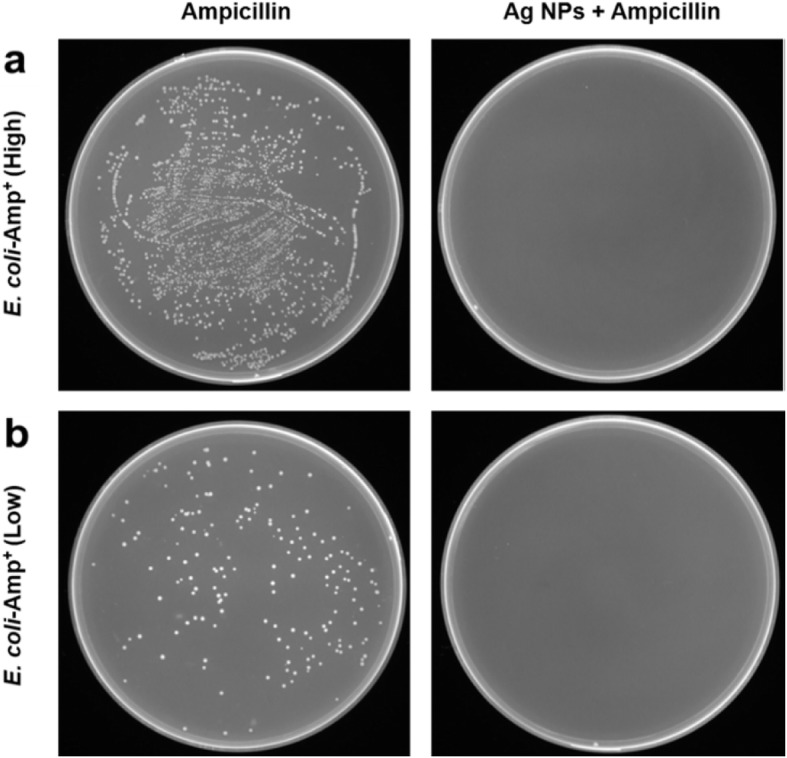
The growth of E. coli-Amp+ in treatment with ampicillin alone (50 μg/mL) or in combination with Ag NPs (25 μg/mL). a High density and b low density of E. coli-Amp+ cells
There is a great need for novel drugs with different mechanisms to combat bacterial resistance. Due to their potent antimicrobial activity, Ag NPs have been used in medical products, personal care products and textiles. There are multiple mechanisms by which Ag NP combat microbial resistance [46]. Ag NPs accumulated on the bacterial membrane surface, increasing the permeability of the cell wall. The interaction between Ag NPs and peptidoglycan changed the configuration of peptidoglycan and thus damaged the bacterial membrane [47]. The characteristics of shape, surface structure, morphology, dispersity, and biocompatibility of Ag NPs have a significant role in their antimicrobial activity.
Conclusions
Herein, we report a novel biosynthetic method for the preparation of Ag NPs using the yeast extract. The yeast micelles formed when the Ag+ solution was mixed with the yeast extract. Bioreducing biomolecules play a major role in the reduction of Ag+. In addition, the biomolecules provide favorable stability, monodispersity, and controllable size distribution for the synthesized Ag NPs, exhibiting good stability for more than a year without precipitation. The high-speed amino acid analysis revealed that the yeast extract is rich in biomolecules, including amino acids, alpha-linolenic acid, and aminobutyric acid. The Ag NPs exhibited significant antibacterial activity in a concentration-dependent manner against E. coli. The growth inhibition assay demonstrated a complete reduction in E. coli at concentrations of Ag NPs above 20.0 μg/mL. The Ag NPs in combination treatment with ampicillin exhibit superior antibacterial activity compared to ampicillin alone against ampicillin-resistant E. coli (E. coli-Amp+) cells. The surface coatings on Ag NPs enhanced the affinity towards the bacterial membrane and increased the permeability of the cell wall. The interaction between Ag NPs and peptidoglycan changed the configuration of peptidoglycan and finally led to the apoptosis of bacteria. Furthermore, these Ag NPs stabilized by the biomolecules exhibited low cytotoxicity and good biocompatibility toward Cos-7 cells.
Supplementary information
Additional file 1: Figure S1 Zeta potential of Ag NPs versus pH value. Table S1 Summary of of biomolecules in yeast extract.
Acknowledgements
We acknowledge the analysis support from the Instrumental Analysis Center of Shanghai Jiao Tong University and the Center for Advanced Electronic Materials and Devices of Shanghai Jiao Tong University.
Abbreviations
- Ag NPs
Silver nanoparticles
- DMEM
Dulbecco’s modified Eagle medium
- E. coli
Escherichia coli
- EDS
Energy dispersive spectrometer
- FBS
Fetal bovine serum
- FTIR
Fourier transform infrared spectroscopy
- HRTEM
High-resolution TEM
- LB
Luria-Bertani
- SAED
Selected-area diffraction
- SEM
Scanning electron microscopy
- TEM
Transmission electron microscopy
- UV-Vis
Ultraviolet-visible
- XRD
X-ray powder diffraction
Authors’ Contributions
MS and FH designed and performed the study. MS, FH, and FG supervised the conceptual framework and wrote the manuscript. ZL, XZ, YM, and ZZ gave technical support and discussed the results. MZ discussed the results and revised the manuscript. ZY supervised the projects. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (61971284, 61671299, and 21171117), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (GZ2016005), the Center of Hydrogen Science at SJTU, the Startup Fund for Youngman Research at SJTU (SFYR at SJTU), and Medical-Engineering Crossover Fund of SJTU (YG2016MS26 and YG2019QNA13).
Availability of Data and Materials
The datasets supporting the conclusions of this current study are available from the corresponding authors upon reasonable request.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mengjun Shu and Fengjiao He contributed equally to this work.
Contributor Information
Zhi Yang, Email: zhiyang@sjtu.edu.cn.
Feng Gao, Email: fgao200906@126.com.
Min Zeng, Email: minzeng@sjtu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s11671-019-3244-z.
References
- 1.Mestrovic T, Ljubin-Sternak S. Molecular mechanisms of chlamydia trachomatis resistance to antimicrobial drugs. Front Biosci. 2018;23:656–670. doi: 10.2741/4611. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Gong Y, Yang T, Song X, Li J, Gan Y, Yin X, Lu Z. Antimicrobial resistance in Neisseria gonorrhoeae in China: a meta-analysis. BMC Infect Dis. 2016;16:108. doi: 10.1186/s12879-016-1435-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ukah UV, Glass M, Avery B, Daignault D, Mulvey MR, Reid-Smith RJ, Parmley EJ, Portt A, Boerlin P, Manges AR. Risk factors for acquisition of multidrug-resistant Escherichia coli and development of community-acquired urinary tract infections. Epidemiol Infect. 2018;146:46–57. doi: 10.1017/S0950268817002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masi M, Réfregiers M, Pos KM, Pagès JM. Mechanisms of envelope permeability and antibiotic influx and efflux in Gram-negative bacteria. Nat Microbiol. 2017;2:17001. doi: 10.1038/nmicrobiol.2017.1. [DOI] [PubMed] [Google Scholar]
- 5.Baym M, Stone LK, Kishony R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science. 2016;351:6268. doi: 10.1126/science.aad3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietro PD, Zaccaro L, Comegna D, Gatto AD, Saviano M, Snyders R, Cossement D, Satriano C, Rizzarelliaf E. Silver nanoparticles functionalized with fluorescent cyclic RGD peptide: a versatile integrin targeting platform for cells and bacteria. RSC Adv. 2016;6:112381–112392. doi: 10.1039/C6RA21568H. [DOI] [Google Scholar]
- 7.Karunamuni R, Tsourkas A, Maidment AD. Exploring silver as a contrast agent for contrast-enhanced dual-energy X-ray breast imaging. Br J Radiol. 2014;87:20140081. doi: 10.1259/bjr.20140081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atabaev TS. Plasmon-enhanced solar water splitting with metal oxide nanostructures: a brief overview of recent trends. Front Matera Sci. 2018;12:207–213. doi: 10.1007/s11706-018-0413-4. [DOI] [Google Scholar]
- 9.Brown AN, Smith K, Samuels TA, Lu J, Obare SO, Scott ME. Nanoparticles functionalized with ampicillin destroy multiple-antibiotic-resistant isolates of Pseudomonas aeruginosa and Enterobacter aerogenes and methicillin-resistant Staphylococcus aureus. Appl Environ Microbiol. 2012;78:2768–2774. doi: 10.1128/AEM.06513-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasithevar M, Periakaruppan P, Muthupandian S, Mohan M. Antibacterial efficacy of silver nanoparticles against multi-drug resistant clinical isolates from post-surgical wound infections. Microb Pathog. 2017;107:327–334. doi: 10.1016/j.micpath.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 11.Kumar CG, Sujitha P. Green synthesis of Kocuran-functionalized silver glyconanoparticles for use as antibiofilm coatings on silicone urethral catheters. Nanotechnology. 2014;25:325101. doi: 10.1088/0957-4484/25/32/325101. [DOI] [PubMed] [Google Scholar]
- 12.Cheng Z, Li Z, Xu J, Yao R, Li Z, Liang S, Cheng G, Zhou Y, Luo X, Zhong J. Morphology-controlled fabrication of large-scale dendritic silver nanostructures for catalysis and SERS applications. Nanoscale Res Lett. 2019;14:89. doi: 10.1186/s11671-019-2923-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng L, Shu M, Chen Y, Yang D, He Q, Zhao H, Feng Z, Liang C, Yu K. A novel lead compound CM-118: antitumor activity and new insight into the molecular mechanism and combination therapy strategy in c-Met- and ALK-dependent cancers. Cancer Biol Ther. 2014;15:721–734. doi: 10.4161/cbt.28409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng C, Vishwakarma A, Li Z, Miyoshi T, Bartonb HA, Joy A. Modification of a conventional polyurethane composition provides significant anti-biofilm activity against Escherichia coli. Polym Chem. 2018;9:3195. doi: 10.1039/C8PY00492G. [DOI] [Google Scholar]
- 15.Zielińska-Jurek A, Wei Z, Wysocka I, Szweda P, Kowalska E. The effect of nanoparticles size on photocatalytic and antimicrobial properties of Ag-Pt/TiO2 photocatalysts. Appl Surf Sci. 2015;353:317–325. doi: 10.1016/j.apsusc.2015.06.065. [DOI] [Google Scholar]
- 16.Dogru E, Demirbas A, Altinsoy B, Duman F, Ocsoy I. Formation of Matricaria chamomilla extract-incorporated Ag nanoparticles and size-dependent enhanced antimicrobial property. J Photochem Photobiol B. 2017;174:78–83. doi: 10.1016/j.jphotobiol.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Huang J, Guo D, Deng Z, Chen W, Wu T, Liu H, Wu J, Chen B. Mid-infrared sub-monolayer quantum dot quantum cascade photodetector monolithically grown on silicon substrate. J Lightwave Technol. 2018;36:4033–4038. doi: 10.1109/JLT.2018.2859250. [DOI] [Google Scholar]
- 18.Khan MJ, Shameli K, Sazili AQ, Selamat J, Kumari S. Rapid green synthesis and characterization of silver nanoparticles arbitrated by curcumin in an alkaline medium. Molecules. 2019;24:719. doi: 10.3390/molecules24040719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaheen Th. I., Abd El Aty Abeer A. In-situ green myco-synthesis of silver nanoparticles onto cotton fabrics for broad spectrum antimicrobial activity. International Journal of Biological Macromolecules. 2018;118:2121–2130. doi: 10.1016/j.ijbiomac.2018.07.062. [DOI] [PubMed] [Google Scholar]
- 20.Budama L, Çakır BA, Topel Ö, Hoda N. A new strategy for producing antibacterial textile surfaces using silver nanoparticles. Chem Eng J. 2013;228:489–495. doi: 10.1016/j.cej.2013.05.018. [DOI] [Google Scholar]
- 21.Anandalakshmi K, Venugobal J, Ramasamy V. Characterization of silver nanoparticles by green synthesis method using Pedalium murex leaf extract and their antibacterial activity. Appl Nanosci. 2016;6:399–408. doi: 10.1007/s13204-015-0449-z. [DOI] [Google Scholar]
- 22.Patra JK, Baek KH. Antibacterial activity and synergistic antibacterial potential of biosynthesized silver nanoparticles against foodborne pathogenic bacteria along with its anticandidal and antioxidant effects. Front Microbiol. 2017;8:167. doi: 10.3389/fmicb.2017.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ifuku S, Tsuji M, Morimoto M, Saimoto H, Yano H. Synthesis of silver nanoparticles templated by TEMPO-mediated oxidized bacterial cellulose nanofibers. Biomacromolecules. 2009;10:2714–2717. doi: 10.1021/bm9006979. [DOI] [PubMed] [Google Scholar]
- 24.Arjunan N, Kumari HL, Singaravelu CM, Kandasamy R, Kandasamy J. Physicochemical investigations of biogenic chitosan-silver nanocomposite as antimicrobial and anticancer agent. Int J Biol Macromol. 2016;92:77–87. doi: 10.1016/j.ijbiomac.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Zhang G, Du M, Li Q, Li X, Huang J, Jiang X, Sun D. Green synthesis of Au–Ag alloy nanoparticles using Cacumen platycladi extract. RSC Adv. 2013;3:1878. doi: 10.1039/C2RA22442A. [DOI] [Google Scholar]
- 26.El Badawy AM, Silva RG, Morris B, Scheckel KG, Suidan MT, Tolaymat TM. Surface charge-dependent toxicity of silver nanoparticles. Environ Sci Technol. 2011;45:283–287. doi: 10.1021/es1034188. [DOI] [PubMed] [Google Scholar]
- 27.Zhao X, Zhang Y, Zhang J, Xue P, Wang Y, Liu R, Cao R, Zhu L, Li G, Sha Z. Natural dissociation ratio of carboxyl group controlled highly dispersed silver nanoparticles on PSA microspheres and their catalytic performance. Nanoscale Res Lett. 2018;13:406. doi: 10.1186/s11671-018-2824-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maiti N., Thomas S., Debnath A., Kapoor S. Raman and XPS study on the interaction of taurine with silver nanoparticles. RSC Advances. 2016;6(61):56406–56411. doi: 10.1039/C6RA09569K. [DOI] [Google Scholar]
- 29.Chen W, Deng Z, Guo D, Chen Y, Mazur YI, Maidaniuk Y, Salamo GJ, Liu H, Wu J, Chen B. Demonstration of InAs/InGaAs/GaAs quantum dots-in-a-well mid-wave infrared photodetectors grown on silicon substrate. J Lightwave Technol. 2017;36:2572–2581. doi: 10.1109/JLT.2018.2811388. [DOI] [Google Scholar]
- 30.Al Mahrouqi D, Vinogradov J, Jackson MD. Zeta potential of artificial and natural calcite in aqueous solution. Adv Colloid Interface Sci. 2017;240:60–76. doi: 10.1016/j.cis.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 31.Leo BF, Chen S, Kyo Y, Herpoldt K, Terrill NJ, Dunlop IE, McPhail DS, Shaffer MS, Schwander S, Gow A, Zhang J, Chung KF, Tetley TD, Porter AE, Ryan MP. The stability of silver nanoparticles in a model of pulmonary surfactant. Environ Sci Technol. 2013;47:11232–11240. doi: 10.1021/es403377p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skoglund S, Blomberg E, Wallinder IO, Grillo I, Pedersen JS, Bergström LM. A novel explanation for the enhanced colloidal stability of silver nanoparticles in the presence of an oppositely charged surfactant. Phys Chem Chem Phys. 2017;19:28037–28043. doi: 10.1039/C7CP04662F. [DOI] [PubMed] [Google Scholar]
- 33.Wang G, Qian F, Saltikov CW, Jiao Y, Li Y. Microbial reduction of graphene oxide by Shewanella. Nano Res. 2011;4:563–570. doi: 10.1007/s12274-011-0112-2. [DOI] [Google Scholar]
- 34.Basu A, Ray S, Chowdhury S, Sarkar A, Mandal DP, Bhattacharjee S, Kundu S. Evaluating the antimicrobial, apoptotic, and cancer cell gene delivery properties of protein-capped gold nanoparticles synthesized from the edible mycorrhizal fungus Tricholoma crissum. Nanoscale Res Lett. 2018;13:154. doi: 10.1186/s11671-018-2561-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anil KS, Abyaneh MK, Gosavi SW, Kulkarni SK, Pasricha R, Ahmad A, Khan MI. Nitrate reductase-mediated synthesis of silver nanoparticles from AgNO3. Biotechnol Lett. 2007;29:439–445. doi: 10.1007/s10529-006-9256-7. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen NH, Padil VVT, Slaveykova VI, Černík M, Ševců A. Green synthesis of metal and metal oxide nanoparticles and their effect on the unicellular alga Chlamydomonas reinhardtii. Nanoscale Res Lett. 2018;13:159. doi: 10.1186/s11671-018-2575-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamedia S, Ghaseminezhadb M, Shokrollahzadehc S, Shojaosadatid SA. Controlled biosynthesis of silver nanoparticles using nitrate reductase enzyme induction of filamentous fungus and their antibacterial evaluation. Artif Cells Nanomed B. 2017;45:1588–1596. doi: 10.1080/21691401.2016.1267011. [DOI] [PubMed] [Google Scholar]
- 38.Zada S, Ahmad A, Khan S, Yu X, Chang K, Iqbal A, Ahmad A, Ullah S, Raza M, Khan A, Ahmad S, Fu P. Biogenic synthesis of silver nanoparticles using extracts of Leptolyngbya JSC-1 that induce apoptosis in HeLa cell line and exterminate pathogenic bacteria. Artif Cells Nanomed Biotechnol. 2018;46:471–480. doi: 10.1080/21691401.2018.1499663. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z, Li Z, Lu X, He F, Zhu X, Ma Y, He R, Gao F, Ni W, Yi Y. Controllable biosynthesis and properties of gold nanoplates using yeast extract. Nano-Micro Lett. 2017;9:5. doi: 10.1007/s40820-016-0102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diaz-Espinoza R, Nova E, Monasterio O. Overcoming electrostatic repulsions during amyloid assembly: effect of ph and interaction with divalent metals using model peptides. Arch Biochem Biophys. 2017;621:46–53. doi: 10.1016/j.abb.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Qu X, Li M, Zhang H, Lin C, Wang F, Xiao M, Zhou Y, Shi J, Aldalbahi A, Pei H, Chen H, Li L. Real-time continuous identification of greenhouse plant pathogens based on recyclable microfluidic bioassay system. ACS Appl Mater Interfaces. 2017;9:31568–31575. doi: 10.1021/acsami.7b10116. [DOI] [PubMed] [Google Scholar]
- 42.Xiao M, Chandrasekaran AR, Ji W, Li F, Man T, Zhu C, Shen X, Pei H, Li Q, Li L. Affinity-modulated molecular beacons on MoS2 nanosheets for microRNA detection. ACS Appl Mater Interfaces. 2018;10:35794–35800. doi: 10.1021/acsami.8b14035. [DOI] [PubMed] [Google Scholar]
- 43.Rafique M, Sadaf I, Rafique MS, Tahir MB. A review on green synthesis of silver nanoparticles and their applications. Artif Cells Nanomed Biotechnol. 2017;45:1272–1291. doi: 10.1080/21691401.2016.1241792. [DOI] [PubMed] [Google Scholar]
- 44.Tagad CK, Dugasani SR, Aiyer R, Park S, Sabharwal S. Green synthesis of silver nanoparticles and their application for the development of optical fiber based hydrogen peroxide sensor. Sens Actuators B Chem. 2013;183:144–149. doi: 10.1016/j.snb.2013.03.106. [DOI] [Google Scholar]
- 45.Zhang Y, Tian R, Shen X, Chen Y, Chen W, Gan L, Shen G, Ju H, Yang L, Gao F. An experimental study on use of 7 T MRI for evaluation of myocardial infarction in SD rats transfected with pcDNA 3.1(+)/VEGF121 plasmid. Am J Transl Res. 2016;8:3376–3386. [PMC free article] [PubMed] [Google Scholar]
- 46.Manna DK, Mandal AK, Sen IK, Maji PK, Chakraborti S, Chakraborty R, Islam SS. Antibacterial and DNA degradation potential of silver nanoparticles synthesized via green route. Int J Biol Macromol. 2015;80:455–459. doi: 10.1016/j.ijbiomac.2015.07.028. [DOI] [PubMed] [Google Scholar]
- 47.Quelemes PV, Araruna FB, De Faria BE, Kuckelhaus SA, Da SD, Mendonca RZ, Eiras C, Dos SSM, Leite JR. Development and antibacterial activity of cashew gum-based silver nanoparticles. Int J Mol Sci. 2013;14:4969–4981. doi: 10.3390/ijms14034969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1 Zeta potential of Ag NPs versus pH value. Table S1 Summary of of biomolecules in yeast extract.
Data Availability Statement
The datasets supporting the conclusions of this current study are available from the corresponding authors upon reasonable request.