Abstract
Clinically relevant human induced pluripotent stem cell (hiPSC) derivatives require efficient protocols to differentiate hiPSCs into specific lineages. Here we developed a fully defined xeno-free strategy to direct hiPSCs toward osteoblasts within 21 days. The strategy successfully achieved the osteogenic induction of four independently derived hiPSC lines by a sequential use of combinations of small-molecule inducers. The induction first generated mesodermal cells, which subsequently recapitulated the developmental expression pattern of major osteoblast genes and proteins. Importantly, Col2.3-Cherry hiPSCs subjected to this strategy strongly expressed the cherry fluorescence that has been observed in bone-forming osteoblasts in vivo. Moreover, the protocol combined with a three-dimensional (3D) scaffold was suitable for the generation of a xeno-free 3D osteogenic system. Thus, our strategy offers a platform with significant advantages for bone biology studies and it will also contribute to clinical applications of hiPSCs to skeletal regenerative medicine.
Keywords: Mesoderm, Wnt, Hedgehog, Helioxanthin, Three-dimensional culture
1. Introduction
Induced pluripotent stem cells (iPSCs) have emerged as a promising option for disease modeling, drug screening, and cell-based therapies. The prompt translation of iPSC technology to various tissues depends largely on the development of efficient protocols for iPSC differentiation, which must be safe and facile while producing functional cells. In particular, defined protocols directing pluripotent stem cells (PSCs) toward specific lineages are required to achieve not only clinical grade cells but also to stably reproduce tissue formation processes and disease status in vitro; batch-to-batch variation of undefined culture components, e.g., fetal bovine serum (FBS), possibly affects the cell fate decision and the outcome of the culture.
At present, several strategies to generate clinically relevant human iPSC (hiPSC) derivatives under xeno-free conditions have been described. Induced lineages include retinal cells [1,2], hepatocytes [3], cardiomyocytes [4], endothelial cells [5], neuronal cells [6], and more. In the skeletal field, several strategies have been reported to direct human PSCs (hPSCs) including hiPSCs toward osteoblasts, which are responsible for bone formation and maintenance [[7], [8], [9], [10], [11]]. Although they successfully induce osteoblastic cells from hPSCs, there may still be a room for improvement in terms of definiteness of culture components and simplification of procedures.
Osteoblast differentiation is a sequential process regulated by local and systemic signaling molecules. A number of signaling pathways have been shown to play roles during osteoblast specification and differentiation, such as hedgehog (Hh) [[12], [13], [14], [15]], Wnt [[16], [17], [18], [19]], bone morphogenetic proteins (BMP) [20], and fibroblast growth factor (FGF) [21]. However, the complex interactions and the stage-specific actions of those pathways present a challenge to the design of effective protocols for directing hPSCs into osteoblasts. In addition, the contribution of those pathways to osteoblast development was revealed mainly by mouse genetic studies, which may not accurately reflect human skeletal development, raising a concern about species-related discrepancies between protocols intended for osteoblast differentiation.
Based on the accumulated knowledge about key signaling pathways for osteoblast development, we developed a small molecule-mediated induction protocol for directing PSCs toward osteoblasts [22]. The protocol enabled the successful osteoblast differentiation of mouse PSCs (mPSCs) via mesoderm formation under defined conditions by sequential treatment with combinations of four small-molecule inducers [22]. However, the induction ability of the protocol in hiPSCs was limited. In the present study, we optimized the osteogenic molecular pathways to differentiate hiPSCs into osteoblasts under fully defined xeno-free conditions, and we propose an optimized strategy for generating osteoblasts from hiPSCs.
2. Materials and methods
2.1. Adaptation and maintenance of hiPSCs under xeno-free conditions
Four independently derived hiPSC lines (hiPSC-1, hiPSC-2, hiPSC-3, and hiPSC-4) were used in this study. Human neonatal fibroblast-derived iPSCs (hiPSC-1) were established from commercially available human neonatal dermal fibroblasts (Lonza, Basel, Switzerland) by transducing the human POU5F1, SOX2, KLF4, and MYC genes [22]. 201B7 (hiPSC-2; HPS0063) [23] and Nips-B2 (hiPSC-3; HPS0223) [24] were obtained from RIKEN BioResource Research Center (Tsukuba, Japan). To establish the Col2.3-Cherry hiPSC line (hiPSC-4), human skin fibroblasts were initially reprogrammed with POU5F1, SOX2, KLF4, and MYC using Sendai virus vectors (provided by DNAVEC/ID Pharma, Tsukuba, Japan). Site-specific integration of the Col2.3-RFPcherry reporter into the “safe harbor” AAVS1 locus was then achieved as described previously [25], except that the CRIPSR/Cas approach was used to target the AAVS1 locus instead of zinc finger nucleases. The hiPSCs were maintained on mouse embryonic fibroblasts (MEFs) in DMEM/F12 supplemented with 20% (vol/vol) KnockOut Serum Replacement (KSR) (#10828-028; Invitrogen, Carlsbad, CA, USA), 0.1 mM MEM non-essential amino acids (#11140-050; Gibco, Grand Island, NY, USA), 0.1 mM 2-mercaptoethanol (#21985-023; Gibco), and 5 ng/mL recombinant human FGF2 (#064-04541; Wako, Osaka, Japan). For the osteoblast induction, the hiPSCs were first adapted and maintained in a commercially available xeno-free culture system (E8/VTN) using Essential 8™ medium (E8; Thermo Fisher Scientific, Waltham, MA, USA) and recombinant human vitronectin (VTN) (#A14700; Gibco)-coated dishes (5 μg/mL). Typically, hiPSCs were well adapted after 6–10 passages. For the dissociation of the cells, we used 0.5 mM EDTA (#15575-020; Gibco).
2.2. Differentiation of hiPSCs into osteoblasts under serum-free conditions
hiPSCs adapted to E8/VTN were maintained on six-well plates up to 70% confluency (day 0). Mesoderm differentiation was achieved by 3-day treatment (from day 0 to day 3) with two small-molecules: CHIR99021 (20 μM, #039-20831; Wako) and cyclopamine (5 μM, #BML-GR334; Enzo Life Sciences, New York, NY, USA) in the basal differentiation medium (BM) consisting of DMEM/F12 with HEPES and l-glutamine (#11330-032; Gibco), 0.1 mM MEM non-essential amino acids (#11140-050; Gibco), 0.1 mM 2-mercaptoethanol (#21985-023; Gibco), B-27 Serum-Free Supplement (#17504-044; Gibco), ITS+1 Liquid Media Supplement (#I2521; Sigma–Aldrich, St. Louis, MO, USA), and 1% penicillin-streptomycin solution (#P4458; Sigma–Aldrich). The medium was changed every day. As a comparison, mesoderm differentiation in hiPSC-1 and hiPSC-2 was also induced by 3-day culture in STEMdiff™ Mesoderm Induction Medium (#05220; Stemcell Technologies, Grenoble, France). Following the mesoderm induction, the osteogenic program was initiated on day 3 with 1 μM SAG (#566660; Calbiochem, Darmstadt, Germany) and 1 μM TH (a helioxanthin derivative, 4-(4-Methoxyphenyl)thieno[2,3-b:5,4-c']dipyridine-2-carboxamide: #M3085; Tokyo Chemical Industry Co., Ltd., Tokyo, Japan) [22,26] in the osteogenic (OG) medium, which was the BM supplemented with 50 μg/mL ascorbic acid phosphate (AsAP) (#A4034; Sigma–Aldrich), 10 mM β-glycerophosphate (β-GP) (#G9422; Sigma–Aldrich), 0.1 μM dexamethasone (Dex) (#41-18861; Wako), and 200 ng/mL human recombinant FGF2 (#064-04541; Wako). On day 4, the cells were dissociated using Accutase (#AT104; Innovative Cell Technologies, San Diego, CA, USA) and passed to VTN-coated 12-well plates (2.5–3 × 105 cells/well). The medium was changed every day. Beginning on day 10, the hiPSC-derived osteoprogenitor cells were differentiated into osteoblasts by the 4-day treatment with 8 μM CHIR and 1 μM TH in the OG medium. Osteoblast maturation was subsequently achieved by culturing the cells for an additional 7 days (from day 14 to day 21) in the OG medium without any inducers. The medium was changed every day throughout the culture.
2.3. Differentiation of hiPSCs into osteoblasts under xeno-free conditions
hiPSCs adapted to E8/VTN were maintained on six-well plates up to 70% confluency (day 0). Mesoderm differentiation was achieved by the 3-day treatment with CHIR (15 μM) and Cyc (5 μM) in the xeno-free BM. The xeno-free conditions of the BM were achieved by substituting B-27 Serum-Free Supplement with B-27 Xeno-Free Supplement (#A1486701; Gibco), and ITS+1 Liquid Media Supplement with ITS Liquid Media Supplement (#I3146-5 ML; Sigma–Aldrich) in the BM. The other components were the same as those used in the BM. On day 3, the osteogenic program was initiated by 7-day treatment with 1 μM SAG and 1 μM TH in the xeno-free osteogenic medium (xeno-free OG medium), which was the xeno-free BM supplemented with 50 μg/mL AsAP, 10 mM β-GP, 0.1 μM Dex, and 100 ng/mL human recombinant FGF2. On day 4, the cells were dissociated using Accutase and passed to VTN-coated 12-well plates (2.5–3 × 105 cells/well). Beginning on day 10, the cells were differentiated into osteoblasts by the 2-day treatment with 4.5 μM CHIR and 1 μM TH in the xeno-free OG medium. Osteoblast maturation was subsequently achieved by culturing the cells for an additional 9 days (from day 12 to day 21) in the xeno-free OG medium without any inducers. The culture medium was changed every day.
2.4. Differentiation of hiPSCs into osteoblasts in 2D and 3D cultures under xeno-free conditions
hiPSC-1 was maintained in the E8/VTN system and differentiated into osteoblasts according to the xeno-free strategy described above with the following modifications. After mesoderm induction (day 4), 2.5 × 105 cells dissociated from a well were sub-cultured under 2D and 3D conditions. Temperature-responsive 12-well plates (#174899; Thermo Fisher Scientific) were used for the 2D culture and porous scaffolds were used for the 3D culture. The scaffolds were fabricated from a commercially available human type I collagen-based recombinant peptide (Cellnest™; Fujifilm, Tokyo, Japan) and were shaped upon our request as follows: discs (dia. 4 mm, height 1 mm) with pores ranging from 200 μm to 400 μm.
For detailed experimental procedures, see Supplementary information.
3. Results
3.1. Optimization of the mesoderm induction under serum-free conditions
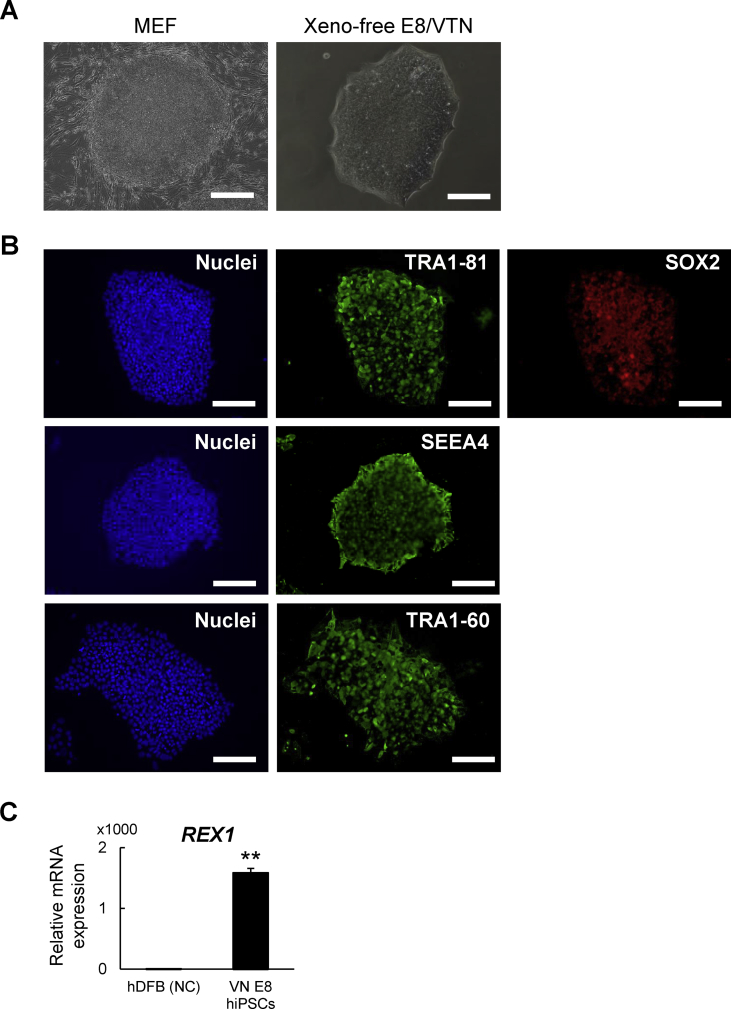
We initially adapted hiPSCs to Essential 8 medium on vitronectin (VTN)-coated plates, referred to as “the E8/VTN system” hereafter. The E8/VTN system is a fully defined xeno-free culture system [27]. The adaptation was successfully achieved within 10 passages, as shown by the typical morphology of the hiPSC colonies (Fig. 1A) with strong expression of pluripotency markers (Fig. 1 B, C).
Fig. 1.
Characterization of hiPSCs maintained under the xeno-free condition. (A) Colony morphology in hiPSC-1 maintained on MEFs (left) or under feeder-free and xeno-free conditions (right). Scale bars: 200 μm. (B) Immunostaining for pluripotency markers (TRA1-81, SOX2, SEEA4, and TRA1-60) in hiPSC-1 maintained under the xeno-free conditions with DAPI staining of nuclei. Scale bars: 200 μm. (C) The mRNA expression of REX1 by RT-qPCR analysis in hiPSC-1 maintained under the xeno-free conditions and human dermal fibroblasts (hDFBs, negative control). Data are the means ± SD from three independent experiments. **P < 0.01 vs. negative control.
To develop a protocol for differentiating hiPSCs into the osteoblast lineage under defined xeno-free conditions, we set out to follow the small molecule-based stepwise differentiation strategy that we developed [22], and to optimize each step for hiPSCs. The strategy consists of (1) the mesoderm induction of PSCs, (2) osteoblast induction from the iPSC-derived mesodermal cells, and (3) osteoblast maturation [22]. In that strategy, the activation of canonical Wnt signaling with 30 μM CHIR99021 (CHIR) and the suppression of Hh signaling with 5 μM cyclopamine (Cyc) allowed us to obtain mesodermal cells from hiPSCs within 5 days [22]. However, the strategy requires the use of plates coated with Matrigel, which is not a fully defined reagent [28], to maintain the cell viability. In the present study, we therefore examined whether treatment with a lower concentration of CHIR in combination with 5 μM Cyc and a shorter period of treatment would improve the cell survival and induce the mesoderm differentiation of hiPSCs on VTN-coated plates.
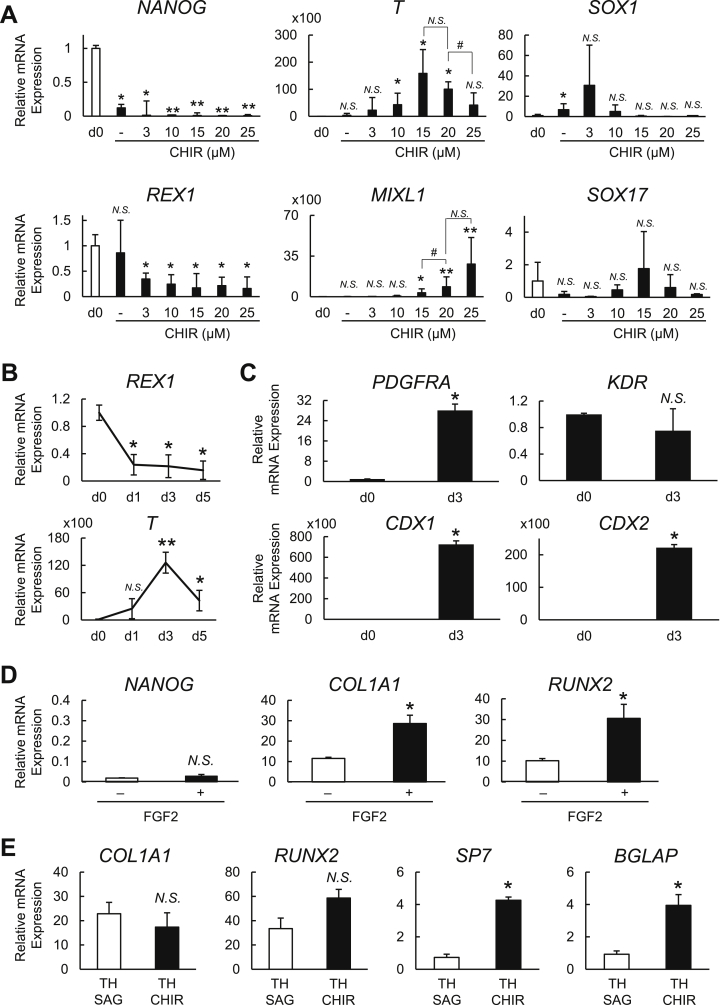
We observed that relative to day 0, the CHIR treatment downregulated the pluripotency-related genes Nanog homeobox (NANOG) and ZFP42 zinc finger protein (ZFP42; also known as REX1) at all concentration tested (Fig. 2A). The expressions of the mesoderm-related markers T-box transcription factor T (TBXT; also known as T or Brachyury) and Mix paired-like homeobox (MIXL1) were higher at both 15 μM and 20 μM CHIR than at lower concentrations. However, the MIXL1 expression was significantly higher in the cells treated with 20 μM compared to those with 15 μM CHIR (Fig. 2A). The cells treated with 25 μM CHIR in turn showed a decreased expression of T and a comparable expression of MIXL1 to 20 μM. We also examined SRY-box transcription factor 1 (SOX1) and SRY-box transcription factor 17 (SOX17), which are ectoderm and endoderm marker genes, respectively. SOX1 was not upregulated at CHIR concentrations >10 μM, and SOX17 was not significantly altered under any of the tested conditions (Fig. 2A). Therefore, we chose 20 μM CHIR and 5 μM Cyc for the mesoderm induction. Regarding the period of treatment, we found that a 3-day treatment was optimal, as the expression of T was higher on day 3 (d3) than on day 1 (d1) or day 5 (d5), whereas no significant difference was found in REX1 between those periods (Fig. 2B).
Fig. 2.
Optimization of the protocol for mesoderm induction and osteoblast differentiation in hiPSCs. (A) The mRNA expression of pluripotency (NANOG and REX1), mesoderm (T and MIXL1), ectoderm (SOX1), and endoderm (SOX17) markers determined by RT-qPCR analysis in hiPSC-1 treated for 3 days with 5 μM cyclopamine (Cyc) and CHIR at different concentrations. Data are the means ± SD from four independent experiments. *P < 0.05 and **P < 0.01 vs. all others. #P < 0.05 as indicated. (B) The mRNA expression of REX1 and T determined by RT-qPCR analysis in hiPSC-1 before (d0) and after the treatment with 5 μM Cyc and 20 μM CHIR for 1 day (d1), 3 days (d3), and 5 days (d5). Data are the means ± SD from three independent experiments. *P < 0.05 and **P < 0.01 vs. d0. (C) The mRNA expression of mesoderm-related genes (PDGFRA, KDR, CDX1, and CDX2) determined by RT-qPCR analysis in hiPSC-1 before (d0) and after treatment with 5 μM Cyc and 20 μM CHIR for 3 days (d3). Data are the means ± SD from four independent experiments. *P < 0.01 vs. d0. (D) The mRNA expression of pluripotency (NANOG) and osteoblast markers (COL1A1 and RUNX2) on day 10 determined by RT-qPCR analysis in hiPSC-1-derived mesodermal cells treated with TH and SAG in the presence (+) or absence (−) of FGF2 for 7 days (from day 4 to day 10). Data analyzed relative to day 0 are shown as the means ± SD from three independent experiments. *P < 0.05 vs. FGF2 (−) group. (E) The mRNA expression of osteoblast markers on day 14 determined by RT-qPCR analysis in hiPSC1-derived osteogenic cells treated with TH and SAG or TH and CHIR for 4 days (from day 10 to day 14) in the presence of FGF2. Data analyzed relative to day 0 are shown as the means ± SD from three independent experiments. *P < 0.05 vs. TH + SAG group.
We further evaluated the expression of other mesoderm-related genes in hiPSC cultured by the optimized mesoderm induction strategy (Fig. 2C). Platelet derived growth factor receptor alpha (PDGFRA), caudal type homeobox 1 (CDX1), and caudal type homeobox 2 (CDX2) were upregulated, whereas kinase insert domain receptor (KDR), a proposed marker for the mesodermal subpopulation committed to hematoendothelial and cardiovascular rather than mesenchymal lineages [29,30], was not significantly altered compared to day 0. KDRlow/PDGFR-αhigh populations were reported to display paraxial mesodermal phenotypes, which specifically exhibit osteogenic, chondrogenic, and myogenic differentiation potential in vitro [31]. These data suggest that our optimized strategy is able to generate hiPSC-derived mesodermal cells, and that it likely proceeds at least partly via a process similar to the paraxial mesoderm specification.
3.2. Optimization of the osteoblast differentiation under serum-free conditions
The specification of the hiPSC-derived mesodermal cells into osteoblast precursors following the mesoderm induction was achieved by the treatment with 1 μM concentrations of two small molecules, the Smoothened agonist SAG and the helioxanthin derivative TH, for the next 7 days (from day 4 to day 10) in the osteogenic medium (see the Experimental Procedures) as done in our previous strategy [22]. Since FGF2 has been positively implicated in human osteoblastic population [32,33], we examined the combinatorial effect of FGF2 and SAG + TH. On day 10, the FGF2-treated cells showed significantly higher expression of early osteoblast marker genes (RUNX2 and COL1A1) compared to the untreated cells, whereas NANOG expression was unaltered between the two conditions (Fig. 2D).
Canonical Wnt signaling is necessary for the transition of Runx2-positive osteoblast precursors to Bglap-positive osteoblasts through Sp7-positive precursors [[16], [17], [18], [19]]. In addition, Hh signaling was shown to be dispensable for the terminal differentiation of osteoblasts [19], and sustained Hh signaling was actually found to suppress the osteoblast number and activity [34]. Based on this knowledge, we then evaluated whether the CHIR-mediated stimulation of canonical Wnt signaling and the cancellation of Hh activation following the 7-day early osteoblast differentiation phase further induced the expression of markers for early and late stage osteoblasts. The treatment with 8 μM CHIR in the presence of TH from day 10 to day 14 induced upregulations of SP7 and BGLAP expression compared to the treatment with SAG + TH on day 14, whereas no significant difference was found in RUNX2 or COL1A1 expression (Fig. 2E). This result suggests that the hiPSC-derived osteoblast precursors are further differentiated into osteoblasts by the subsequent CHIR treatment.
3.3. Characterization of the optimized protocol for the osteoblast differentiation of hiPSCs
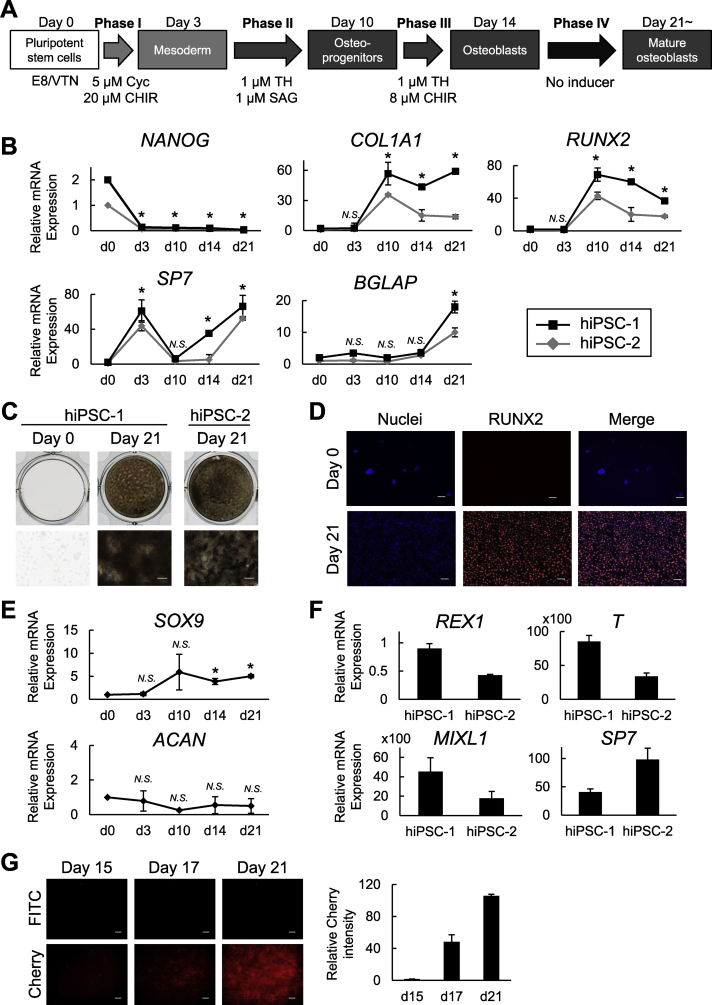
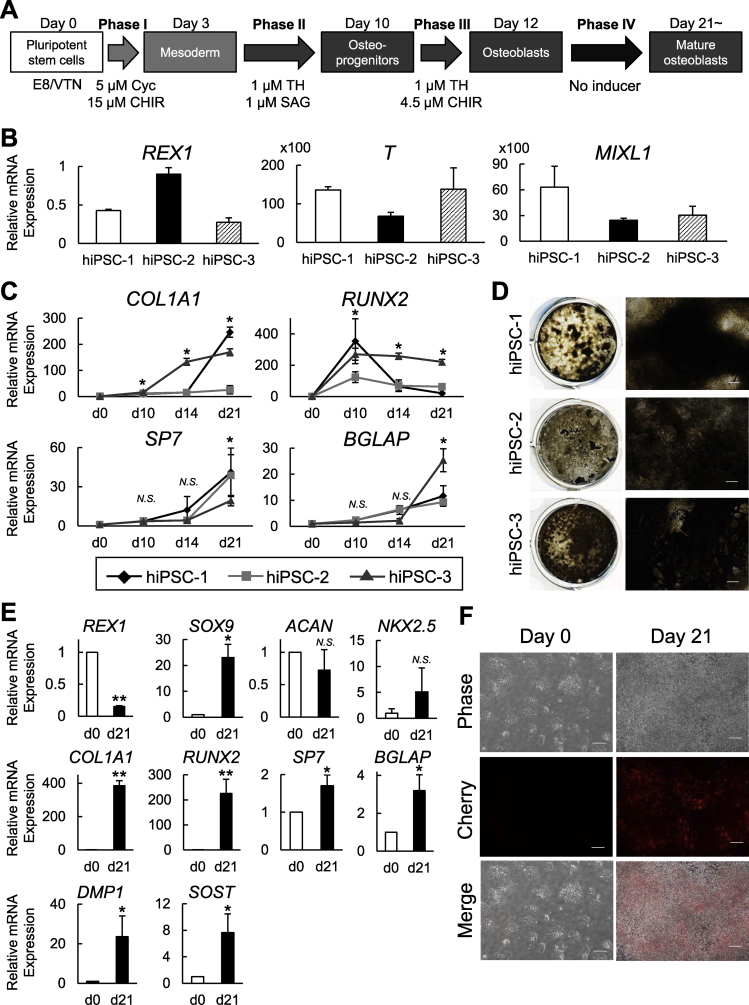
As described above, we optimized the protocol for differentiating hiPSCs into osteoblast (Fig. 3A). The protocol includes three phases that we optimized here: a 3-day mesoderm induction (phase I), a 7-day osteoblast specification (phase II), and a 4-day osteoblast differentiation (phase III), followed by an osteoblast maturation for 7 days or more (phase IV) that was included in our previous strategy. The additional culture without inducers was proposed to allow further maturation of the osteogenic cell population derived from hiPSCs [22].
Fig. 3.
Characterization of hiPSC-derived cells induced by the present strategy under serum-free conditions. (A) A schematic of the four-phase strategy for osteoblast differentiation of hiPSCs using small molecule inducers under serum-free conditions. (B) Time course of the mRNA expression determined by an RT-qPCR analysis in two independently derived hiPSC lines (hiPSC-1 and hiPSC-2) cultured using the present strategy for osteoblast differentiation. Data are the means ± SD from four independent experiments. *P < 0.05 at the indicated time points (d3, d10, d14, and d21) vs. day 0 in both hiPSC-1 and hiPSC-2. (C) Representative pictures of von Kossa staining in the osteogenic culture of the two hiPSC lines. Scale bars: 200 μm. (D) Immunostaining for RUNX2 in the osteogenic culture of hiPSC-1 on days 0 (negative control) and 21 with DAPI staining of nuclei. Scale bars: 50 μm. (E) Time course of the mRNA expression of SOX9 and ACAN determined by RT-qPCR analysis in hiPSC-1 cultured under the serum-free strategy for osteoblast differentiation. Data are the means ± SD from four independent experiments. *P < 0.05 at the indicated time points (d3, d10, d14, and d21) vs. day 0. (F) The mRNA expression of pluripotency (REX1), mesoderm (T and MIXL1), and osteoblast markers (SP7) determined by RT-qPCR analysis in hiPSC-1 treated with a commercially available mesoderm induction medium for 3 days (relative to day 0). Data are the means ± SD from three independent experiments. (G) Representative pictures of Cherry expression in Col2.3-Cherry hiPSCs (scale bars: 100 μm) and the time-line quantification of the Cherry fluorescence intensity. Data are the means ± SD from three independent experiments.
Fig. 3B shows the time course of the osteoblast-related marker expression in two independently derived hiPSC lines, hiPSC-1 and hiPSC-2 (see Materials and methods), cultured under the protocol. Both lines showed similar gene expression patterns. RUNX2 was upregulated at the end of the osteoblast specification phase (day 10), and decreased as the differentiation advanced. COL1A1 was also upregulated on day 10, and the expression at later points trended down to varying degrees between the two lines. SP7 was upregulated during the mesoderm induction phase (from day 0 to day 3; Phase I) and downregulated during the osteoblast specification phase (Phase II); it was then upregulated again during the osteoblast differentiation phase (Phase III) and finally reached a peak at Phase IV. BGLAP was progressively upregulated during the osteoblast maturation phase (from day 14 to day 21; Phase IV). In addition, matrix calcification was largely observed at the Phase IV (on day 21) as was indicated by von Kossa staining (Fig. 3C). The immunohistochemical examination of hiPSC-1 revealed the nuclear localization of RUNX2 in most of the induced cells at the Phase IV (86 ± 6% on day 21) (Fig. 3D). We also evaluated the time course of the expressions of SOX9 and ACAN in hiPSC-1, which are makers for osteo-chondroprogenitors and committed chondrocytes, respectively [35]. SOX9 was slightly upregulated after the mesodermal induction, with an increase of fivefold compared to day 0 or day 3, whereas ACAN did not show upregulation at any time point (Fig. 3E). These data suggest that our strategy can reproduce the expression pattern of major osteogenic markers in osteoblast development but not those in chondrocyte development. One exception was SP7; our strategy induced a transient upregulation of SP7 at the mesoderm induction stage (Phase I), although SP7 has not been implicated in the early mesodermal population. This phenomenon is unlikely to be specific to our strategy, since hiPSC-1 and hiPSC-2 cultured in a commercially available mesoderm induction medium for 3 days (see Materials and methods) also showed the upregulations of SP7 and mesoderm markers with a downregulation of the pluripotency marker REX1 (Fig. 3F).
To further examine the in vivo relevance of the hiPSC-derived osteoblastic population generated by the present strategy, we cultured Col2.3-Cherry hiPSCs (see Materials and methods) carrying a transgene in which the red fluorescent protein (RFP) Cherry was driven by a 2.3-kb rat Col1a1 promoter fragment, and we monitored the Cherry-derived florescence signal throughout the culture. Cherry expression driven by the activation of the fragment can be used as a bona-fide indicator of “in vivo-functional osteoblasts” for the following three reasons. First, the green fluorescent protein (GFP) signal driven by the Col1a1 promoter fragment was observed exclusively in osteoblasts and osteocytes in Col2.3-GFP transgenic mouse lines [36]. Second, when a Col2.3-GFP human embryonic stem cell (hESC)-derived mesenchymal population or Col2.3-GFP human bone marrow mesenchymal stem cells (MSCs) were transplanted to a critical-sized mouse calvarial bone defect, GFP-positive cells contributed to orthotopic bone formation in vivo [25,37]. Third, the Col2.3-GFP transgene has successfully enabled the monitoring of osteoblast differentiation in mouse ESCs [22,38,39].
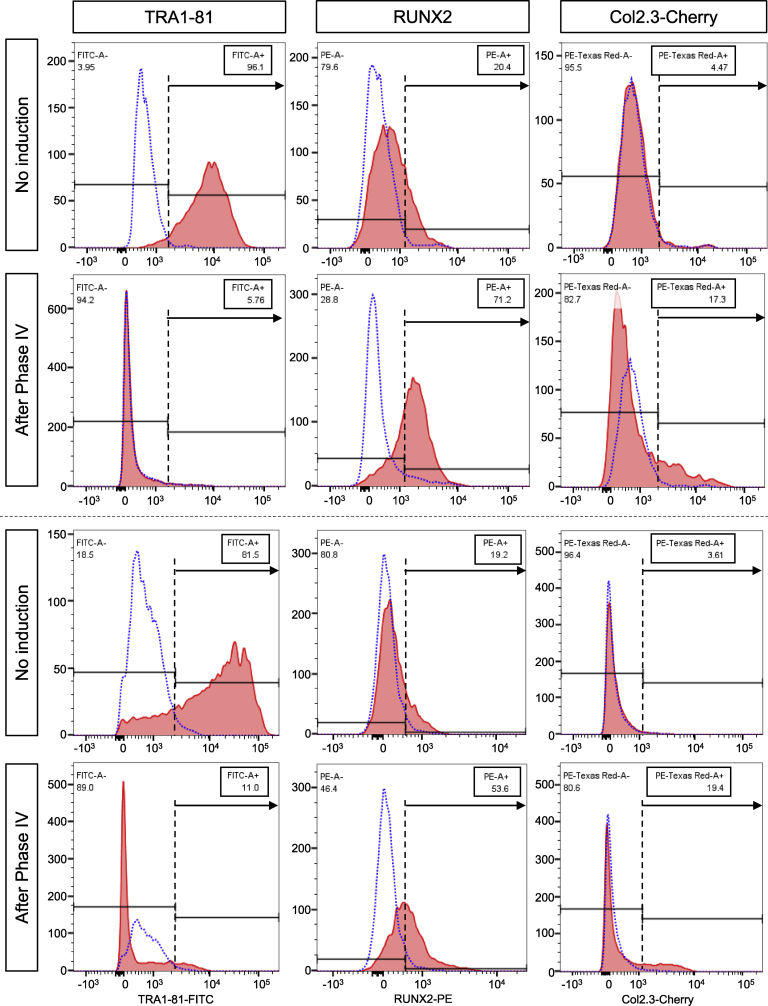
In the present experiments, Cherry fluorescence started to appear at the onset of phase IV (osteoblast maturation phase, day 15). It then gradually increased during the culture and reached high levels throughout the Phase IV (Fig. 3G). Col2.3-Cherry iPSCs further enabled us to assess differentiation efficiency of the present strategy by flow cytometry. The flow cytometry data from biological duplicate supported our findings so far in a reproducible manner (Fig. 4). The Phase IV population contained a larger percentage of RUNX2-positive early osteoblastic cells and Cherry-positive osteoblastic cells than the non-induced population (Runx2+ cells: 71.2% and 53.6% in the Phase IV vs. 20.4% and 19.2% in the non-induced; Cherry+ cells: 17.3% and 19.4% in the Phase IV vs. 4.5% and 3.6% in the non-induced). Consistent with the increase in the differentiated population, cells positive for the pluripotency marker TRA1-81 were substantially reduced in the Phase IV population compared to the non-induced one. These findings suggest that the present strategy achieves the lineage specification of hiPSCs into mesodermal cells, the subsequent differentiation of the hiPSC-derived mesodermal cells into osteoblasts, and eventually the generation of mature osteoblasts, which are potentially functional in vivo.
Fig. 4.
Flow cytometric analysis on Col2.3-Cherry hiPSC-derived cells induced by the present strategy under serum-free conditions. Cells expressing TRA1-81 (left), RUNX2 (middle), or Cherry driven by the Col2.3 promoter fragment (right) were analyzed before the induction (No Induction) and after the Phase IV (After Phase IV). Data from biological duplicate are shown in upper and lower panels separated by the dotted line. Dotted and solid lines show signal from isotype controls (negative control) and specific antibodies, respectively. Y-axes indicate cell counts. Numbers in rectangular boxes indicate percentages of cells with positive signal.
3.4. Optimization of the present strategy under xeno-free conditions
Considering the potential applications of the present strategy to cell-based therapies, we next attempted to expand the present strategy to xeno-free conditions. These conditions were achieved by substituting the supplements with their xeno-free equivalents in the basal medium (BM), and by slightly modifying the concentration and period of treatment of small-molecule inducers to improve the cell viability that was decreased by the substitution of supplements. Since the expressions of T and MIXL1 were higher at both 15 μM and 20 μM CHIR (Fig. 2A), we decreased the CHIR concentration during the Phase I from 20 μM to 15 μM. We also reduced the period of the CHIR treatment during the Phase III from 4 days to 2 days and decreased the CHIR concentration from 8 μM to 4.5 μM. Fig. 5A shows the modified osteoblast differentiation protocol for the fully defined xeno-free conditions.
Fig. 5.
Characterization of hiPSC-derived cells induced by the present strategy under the fully defined xeno-free conditions. (A) A schematic of the four-phase strategy for the osteoblast differentiation of hiPSCs under the fully defined xeno-free condition. (B) The mRNA expression of pluripotency (REX1) and mesoderm (T and MIXL1) markers determined by RT-qPCR analysis in three independently derived hiPSC lines on day 3 (relative to day 0). (C) Time course of the mRNA expression determined by RT-qPCR analysis in hiPSC-1, hiPSC-2, and hiPSC-3 cultured under the present xeno-free strategy for osteoblast differentiation. Data are the means ± SD from four independent experiments. *P < 0.05 at the indicated time points (d10, d14, and d21) vs. day 0 in all of hiPSC-1, hiPSC-2 and hiPSC-3. (D) Representative pictures of von Kossa staining in the osteogenic culture of the three hiPSC lines on day 21. Scale bars: 100 μm. (E) The mRNA expression determined by RT-qPCR analysis in Col2.3-Cherry hiPSCs before (d0) and after the osteogenic differentiation (d21). Data are the means ± SD from three experiments. *P < 0.05 at d21 vs. d0. (F) Representative pictures of Cherry expression in Col2.3-Cherry hiPSCs before (Day 0) and after the osteogenic differentiation (Day 21). Scale bars: 100 μm.
The 3-day mesoderm induction with 15 μM CHIR and 5 μM Cyc under the xeno-free conditions strongly induced the expression of mesoderm marker genes (T and MIXL1) and the downregulation of REX1 in three hiPSC lines (Fig. 5B). When hiPSC-derived mesodermal cells were then differentiated into osteoblasts (phases II–IV) under the xeno-free conditions, the gene expression profiles were similar to those under the serum-free conditions (compare hiPSC-1 and hiPSC-2 between Fig. 3, Fig. 5C), and the three hiPSC lines showed strong matrix calcification on day 21 (Fig. 5D). In addition, the Col2.3-Cherry hiPSCs that were differentiated under the xeno-free conditions showed the upregulation of not only the osteoblast markers, but also osteocyte markers on day 21 (Fig. 5E), with extensive expressions of Col2.3-Cherry (Fig. 5F). Consistent with our previous findings in hiPSC-1, a slight upregulation of SOX9, but not ACAN, was detected at day 21 compared to day 0. Moreover, NKX2.5, a marker largely expressed in lateral plate mesoderm as well as cardiomyocytes, was not significantly upregulated. These results indicate that the improved strategy can successfully direct the osteoblast differentiation of four hiPSC lines via mesoderm formation under the fully defined xeno-free conditions.
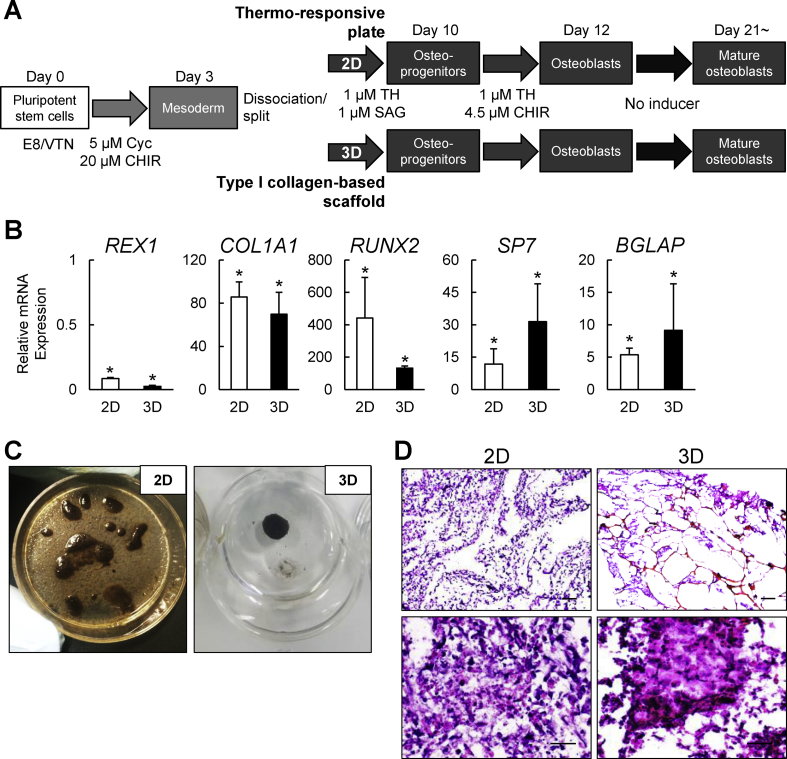
Since three-dimensional (3D) culture systems are evolving as a convenient approach for both regenerative medicine and in vitro tissue models [[39], [40], [41]], we lastly examined whether our proposed xeno-free strategy is well-suited for a 3D culture (see Materials and methods). Mesodermal cells derived from hiPSC-1 were differentiated into osteoblasts on thermo-responsive plates (2D) or with human type I collagen-based scaffolds (3D). We compared the differentiation status of the line between the two culture conditions. The experimental design is shown in Fig. 6A.
Fig. 6.
Characterization of hiPSC-derived cells induced by the present strategy in the two-dimensional (2D) thermo-responsive plate culture or the three-dimensional (3D) collagen scaffold culture under the fully defined xeno-free condition. (A) A schematic of the osteoblast differentiation strategy in osteogenic 2D and 3D cultures under the fully defined xeno-free condition. (B) The mRNA expression determined by RT-qPCR analysis in hiPSC-1 differentiated in the osteogenic 2D and 3D cultures at day 21 (d21). Data are the means ± SD from three independent experiments. *P < 0.05 at d21 in 2D and 3D cultures vs. d0. (C) Representative pictures of von Kossa staining of hiPSC-1 on day 21 in the osteogenic 2D and 3D cultures. (D) Representative H&E staining pictures in the 2D and 3D osteogenic cultures of hiPSC-1. Scale bars: 200 μm (upper panels) and 50 μm (lower panels).
The cells differentiated in the 3D culture showed mRNA expressions of the osteoblast-related markers (RUNX2, COL1A1, SP7 and BGLAP) on day 21 that were comparable to those of the cells cultured in the 2D culture, whereas no significant difference was found in the mRNA expression of REX1 between the two conditions (Fig. 6B). Calcification in the 2D and 3D cultures was confirmed by von Kossa staining (Fig. 6C) and the histological features were examined with H&E staining on cell sheet (2D) and scaffold (3D) sections (Fig. 6D). Although the cell distribution and morphology of the extracellular matrix (ECM) were similar between the two culture conditions, the ECM in the 3D culture exhibited a denser structure. Thus, the developed xeno-free protocol is likely to be suitable for the development of relevant osteogenic 3D cultures, and it can also be applied to the cell-sheet strategy with the thermo-responsive plate, supporting its potential advantages in hiPSC-derived osteoblast sheet-based therapies.
4. Discussion
We have established a fully defined, xeno-free, and stepwise strategy in which hiPSCs are maintained and differentiated into mature osteoblasts in vitro. As mentioned in the Results section, an educated guess based on developmental biology led us to test crucial regulators for osteoblast development in order to optimize each differentiation step.
The results presented here suggest that the signaling pathways that we need to manipulate for in vitro osteoblast differentiation are different between human and mouse iPSCs. Whereas the mesoderm induction and osteoblast specification (Phases I and II) were achieved in hiPSCs with the same set of small molecule inducers as used in mPSC differentiation, the osteoblast differentiation (Phase III) in hiPSCs required additional exogenous signaling stimulation as well as cancellation of the one signaling activated in the Phase II.
The initial treatment with CHIR and Cyc induced the differentiation of hiPSCs and mPSCs [22] into mesodermal cells. The activation of canonical Wnt signaling induces the specification of mESCs and hESCs into primitive streak (PS), a progenitor population for both the mesoderm and endoderm [42,43]. On the other hand, the inhibition of Hh signaling has been shown to prevent neuroectoderm derivatives from PSCs [44]. In our strategy, the combinatorial effect of CHIR and Cyc in hiPSCs resulted in a population that specifically expressed a wide range of paraxial mesoderm markers, whereas the key endoderm and ectoderm markers were not upregulated.
In contrast to mPSC cultures [22], the hiPSC-derived mesodermal cells showed a transient upregulation of SP7, which was considered to be an osteoblast marker. The mechanisms that govern SP7 expression remain to be fully identified. Although mouse genetic studies demonstrated that Sp7 acts genetically downstream of Runx2 in skeletal tissues [45,46], human SP7 was detected in several mesoderm-derived human tissues other than skeletal tissues, such as testis, heart, lung, spleen, and placenta [47]. In line with this, we observed in the present study that the SP7 expression in the hiPSC-derived mesodermal cells was not accompanied by RUNX2 expression, which suggests that there may be RUNX2-independent gene regulatory machinery for SP7 in human mesoderm tissues.
Indeed, Sp7 expression can be directly regulated by signaling pathways such as BMP [48] and transcription factors including Msx [48], Dlx [49], and NFκB [50]. Since our strategy used CHIR to induce the mesoderm differentiation, it is also possible that Wnt-mediated mechanisms underlie the SP7 expression in that phase. Although further detailed analyses are required to understand the gene regulatory mechanisms and to gain functional insights into SP7 regulation in human mesodermal cells, our data together with those of other studies suggest that SP7 functions may vary among species in this context.
The subsequent treatment of hiPSC-derived mesodermal cells with SAG and TH in the presence of FGF2 successfully induced the expression of early osteoblasts markers. SAG and TH exerted similar effects in mPSCs and hiPSCs; the rationale for using SAG and TH in this phase was supported by our previous findings [22,51] and mouse genetic studies [12,13]. The effects of FGF2 on osteogenic cultures of human versus mouse cells are a matter of controversy; FGF2 has been shown to have roles in both the differentiation and proliferation of osteoblasts in both mouse and human cells [32,33,[52], [53], [54]]. Since FGF2 is also known to play a key role in the pluripotency maintenance of hESCs and hiPSCs [55], but not in that of their mouse counterparts, we infer that hiPSC-derived mesodermal cells undergoing osteogenic specification may rely more on the FGF signal than mesodermal cells derived from mPSCs (at least under our culture conditions), possibly by regulating the proliferation of the progenitor pools or by directing the population toward the osteoblast lineage.
Flow cytometry and immunostaining revealed that 20–30% of cells in our culture expressed little or no RUNX2 at day 21. We consider two possibilities regarding the fates of the RUNX2-negative (RUNX2low) cells. The first possibility is that the culture contains osteoblastic cells with different differentiation stages, each of which is characterized by different expression levels of RUNX2. During osteoblast development, RUNX2 protein is first detected in preosteoblasts; the expression increases in immature osteoblasts, but decreases in mature osteoblasts [45,56,57]. Based on this fact, RUNX2low cells in our culture may reflect mature osteoblasts that weakly express RUNX2. The idea may be consistent with the time-line of RUNX2 mRNA expression in our culture, in which RUNX2 decreased as the differentiation advanced (day 21), and also be supported by the difference in the percentages between RUNX2-positive cells and mCherry-positive cells (Fig. 4). RUNX2 expression occurs earlier than activation of the 2.3-kb Col1a1 promoter fragment, i.e. the mCherry-positive stage in osteoblast differentiation. The promoter fragment became active in harmony with IBSP expression, and the activation is restricted to osteoblasts within differentiated nodules in vitro [58] and to periosteal mature osteoblasts and osteocytes located deep in the cortical bone in vivo [36]. The second possibility regarding the fates of the RUNX2-negative (RUNX2low) cells is that they are not osteoblastic population; they may contain pluripotent cells remaining in the induction culture, mesodermal cells that have not yet committed to the osteoblastic lineage, or cells that committed to other lineages. Given that heterogeneity in our system underlies both of the above possibilities, sorting RUNX2-positive cells after day 10 could potentiate our strategy.
We and others have shown that 3D culture can significantly improve the differentiation of mPSCs [[39], [40], [41]]. Particularly, mPSC-derived mesodermal cells showed accelerated osteogenic differentiation in the 3D culture, giving rise to functional osteoblast-osteocyte populations that were not observed in 2D cultures at the same time period of the cultures [39]. In contrast, we show in this study that molecular signature of hiPSC-derived osteoblastic population in 3D cultures are comparable to that in 2D cultures; hiPSCs were unlikely to express osteocyte markers even in the 3D culture. The timeline of events in mouse and human embryogenesis may also be reflected in the characteristics of PSCs derived from the two species, leading to the difference in their differentiation potential under certain conditions. Typically, hPSC-derived cells are difficult or need prolonged culture periods to acquire fully mature phenotypes, compared to mPSC-derived ones.
We cannot exclude the possibility that responsiveness to different external stimuli depends on an inherent capacity of each hiPSC line. Indeed, we found differences in the mRNA expression level of key osteoblast markers as well as the calcification level between different hiPSC lines. Both the differentiation potential and the response to external stimuli such as the dimensionality may vary across different lines. Substantial progress has been made in understanding of whether and how the genetic origin of donors, the type of original cells, and the reprogramming method affect the differentiation potential of iPSCs to specific lineages. For instance, epigenetic dissimilarities were found to affect the ability of hPSCs to commit to the osteogenic lineage; methylation status in PAX7 and TWIST1 regions was suggested to play a determining role in osteoblast development from hPSCs [59]. Extensive studies unraveling regulatory landscape on the genome during human osteoblast development are needed to further clarify this point, and our present hiPSC differentiation system potentially contributes to the studies.
One may point out two potential limitations of this study: (1) unidentified mechanisms underlying the effects of TH on osteoblast differentiation, and (2) the lack of in vivo evidence of the function of generated osteoblasts. We have been studying the first point and will report our findings in the near future. Regarding the second point, we need to optimize an appropriate scaffold system to maximize the in vivo function of in vitro-generated cells. Although the xeno-free human type I collagen-based scaffold allowed us to expand the osteogenic culture to a clinically relevant 3D system, it may require further optimization for in vivo use. We have been working on hydrogels for medical applications [60] and will report the combinatorial effect of the present hiPSC differentiation strategy and the newly developed 3D scaffold system on in vivo bone regeneration in the future. In addition, we obtained around 17–19% cells positive for Col2.3-Cherry in our induction culture. As mentioned in the Result section, we have shown that Col2.3-GFP hESC-derived GFP+ mesenchymal population or Col2.3-GFP human MSC-derived GFP+ cells were able to contribute to orthotopic bone formation in vivo [25,37]. Thus, the Col2.3-Cherry fluorescence in our culture at least partly supports in vivo osteogenic potential of the population that we generated from hiPSCs. This logic may not be in harmony with the report by Phillips et al., which showed that in vitro osteoblastic features of hiPSC-derived cells did not always predict their in vivo osteogenic potential [7]. However, given that they assessed in vivo potential by an ectopic bone formation model (subcutaneous transplantations of induced cells), their finding may not be the case with orthotopic bone formation models that are more clinically-relevant than ectopic bone formation models.
5. Conclusions
We have generated a robust platform to differentiate hPSCs into osteoblasts by sequentially manipulating signaling pathways involved in osteoblast specification and maturation. In contrast to the current platforms for osteoblast differentiation of hPSCs [[7], [8], [9], [10], [11]], we extensively describe the differentiation process, characterize the cells in every stage, and compare the differentiation status in multiple cell lines. This fully defined xeno-free strategy, at least to some extent, reproduces osteoblast development with confounding factors minimized, providing significant advantages for disease modeling, drug screening, and developmental studies in the skeletal field. It will also contribute to clinical applications of hPSCs to skeletal regenerative medicine.
Author contributions
D. Zujur, K. Kanke, and S. Ohba designed the project. D. Zujur, K. Kanke, S. Onodera, S. Tani, J. Lai, X. Xin, and H. Masaki performed the experiments. D. Zujur, T. Azuma, A.C. Lichtler, D.W. Rowe, T. Saito, S. Tanaka, H. Nakauchi, U. Chung, H. Hojo, and S. Ohba analyzed and interpreted the data. D. Zujur, U. Chung, H. Hojo, and S. Ohba wrote the manuscript. S. Ohba supervised the project and approved the final version of the manuscript.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgements
We thank Drs. Fumiko Yano, Yasuyuki Sakai, Horacio Cabral, and Takashi Ushida for the helpful discussions, and Katsue Morii, Rie Yonemoto, Akiko Nakamichi, and Nozomi Nagumo for providing technical assistance. This work utilized the core research facility of Center for Disease Biology and Integrative Medicine, The University of Tokyo, which was organized by The University of Tokyo Center for NanoBio Integration entrusted by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) Japan. This work was supported by JSPS KAKENHI Grant Numbers JP26713054, JP15K15732, JP16H06312, JP17H04403, and JP18K19635; the Rising Star Award Grant from the American Society for Bone and Mineral Research (to S. Ohba) and the Japan Science and Technology Agency (JST) Center of Innovation Program (to U.C.). D.Z. and J.L. was supported by Japanese Government (MEXT) Scholarship and UTokyo Amgen Scholars Program, respectively.
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.reth.2019.12.010.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Sridhar A., Steward M.M., Meyer J.S. Nonxenogeneic growth and retinal differentiation of human induced pluripotent stem cells. Stem Cells Transl Med. 2013;2(4):255–264. doi: 10.5966/sctm.2012-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tucker B.A., Anfinson K.R., Mullins R.F., Stone E.M., Young M.J. Use of a synthetic xeno-free culture substrate for induced pluripotent stem cell induction and retinal differentiation. Stem Cells Transl Med. 2013;2(1):16–24. doi: 10.5966/sctm.2012-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farzaneh Z., Pakzad M., Vosough M., Pournasr B., Baharvand H. Differentiation of human embryonic stem cells to hepatocyte-like cells on a new developed xeno-free extracellular matrix. Histochem Cell Biol. 2014;142(2):217–226. doi: 10.1007/s00418-014-1183-4. [DOI] [PubMed] [Google Scholar]
- 4.Masumoto H., Ikuno T., Takeda M., Fukushima H., Marui A., Katayama S. Human iPS cell-engineered cardiac tissue sheets with cardiomyocytes and vascular cells for cardiac regeneration. Sci Rep. 2014;4:6716. doi: 10.1038/srep06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao X., Lian X., Dunn K.K., Shi M., Han T., Qian T. Chemically-defined albumin-free differentiation of human pluripotent stem cells to endothelial progenitor cells. Stem Cell Res. 2015;15(1):122–129. doi: 10.1016/j.scr.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai Y., Cutts J., Kimura A., Varun D., Brafman D.A. A chemically defined substrate for the expansion and neuronal differentiation of human pluripotent stem cell-derived neural progenitor cells. Stem Cell Res. 2015;15(1):75–87. doi: 10.1016/j.scr.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Phillips M.D., Kuznetsov S.A., Cherman N., Park K., Chen K.G., McClendon B.N. Directed differentiation of human induced pluripotent stem cells toward bone and cartilage: in vitro versus in vivo assays. Stem Cells Transl Med. 2014;3(7):867–878. doi: 10.5966/sctm.2013-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeon O.H., Panicker L.M., Lu Q., Chae J.J., Feldman R.A., Elisseeff J.H. Human iPSC-derived osteoblasts and osteoclasts together promote bone regeneration in 3D biomaterials. Sci Rep. 2016;6:26761. doi: 10.1038/srep26761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang P., Ma T., Guo D., Hu K., Shu Y., Xu H.H.K. Metformin induces osteoblastic differentiation of human induced pluripotent stem cell-derived mesenchymal stem cells. J Tissue Eng Regen Med. 2018;12(2):437–446. doi: 10.1002/term.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang H., Shih Y.R., Nakasaki M., Kabra H., Varghese S. Small molecule-driven direct conversion of human pluripotent stem cells into functional osteoblasts. Sci Adv. 2016;2(8) doi: 10.1126/sciadv.1600691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bilousova G., Jun du H., King K.B., De Langhe S., Chick W.S., Torchia E.C. Osteoblasts derived from induced pluripotent stem cells form calcified structures in scaffolds both in vitro and in vivo. Stem Cells. 2011;29(2):206–216. doi: 10.1002/stem.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.St-Jacques B., Hammerschmidt M., McMahon A.P. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13(16):2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long F., Chung U.I., Ohba S., McMahon J., Kronenberg H.M., McMahon A.P. Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development. 2004;131(6):1309–1318. doi: 10.1242/dev.01006. [DOI] [PubMed] [Google Scholar]
- 14.Ohba S. Hedgehog signaling in endochondral ossification. J Dev Biol. 2016;4(2):20. doi: 10.3390/jdb4020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohba S., Kawaguchi H., Kugimiya F., Ogasawara T., Kawamura N., Saito T. Patched1 haploinsufficiency increases adult bone mass and modulates Gli3 repressor activity. Dev Cell. 2008;14(5):689–699. doi: 10.1016/j.devcel.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Day T.F., Guo X., Garrett-Beal L., Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8(5):739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 17.Hill T.P., Spater D., Taketo M.M., Birchmeier W., Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8(5):727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Hu H., Hilton M.J., Tu X., Yu K., Ornitz D.M., Long F. Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development. 2005;132(1):49–60. doi: 10.1242/dev.01564. [DOI] [PubMed] [Google Scholar]
- 19.Rodda S.J., McMahon A.P. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133(16):3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 20.Bandyopadhyay A., Tsuji K., Cox K., Harfe B.D., Rosen V., Tabin C.J. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2(12):e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marie P.J. Fibroblast growth factor signaling controlling osteoblast differentiation. Gene. 2003;316:23–32. doi: 10.1016/s0378-1119(03)00748-0. [DOI] [PubMed] [Google Scholar]
- 22.Kanke K., Masaki H., Saito T., Komiyama Y., Hojo H., Nakauchi H. Stepwise differentiation of pluripotent stem cells into osteoblasts using four small molecules under serum-free and feeder-free conditions. Stem Cell Rep. 2014;2(6):751–760. doi: 10.1016/j.stemcr.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 24.Ono M., Hamada Y., Horiuchi Y., Matsuo-Takasaki M., Imoto Y., Satomi K. Generation of induced pluripotent stem cells from human nasal epithelial cells using a Sendai virus vector. PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0042855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xin X., Jiang X., Wang L., Stover M.L., Zhan S., Huang J. A site-specific integrated Col2.3GFP reporter identifies osteoblasts within mineralized tissue formed in vivo by human embryonic stem cells. Stem Cells Transl Med. 2014;3(10):1125–1137. doi: 10.5966/sctm.2013-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohba S., Nakajima K., Komiyama Y., Kugimiya F., Igawa K., Itaka K. A novel osteogenic helioxanthin-derivative acts in a BMP-dependent manner. Biochem Biophys Res Commun. 2007;357(4):854–860. doi: 10.1016/j.bbrc.2007.03.173. [DOI] [PubMed] [Google Scholar]
- 27.Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8(5):424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes C.S., Postovit L.M., Lajoie G.A. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10(9):1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 29.Evseenko D., Zhu Y., Schenke-Layland K., Kuo J., Latour B., Ge S. Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc Natl Acad Sci U S A. 2010;107(31):13742–13747. doi: 10.1073/pnas.1002077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savory J.G., Bouchard N., Pierre V., Rijli F.M., De Repentigny Y., Kothary R. Cdx2 regulation of posterior development through non-Hox targets. Development. 2009;136(24):4099–4110. doi: 10.1242/dev.041582. [DOI] [PubMed] [Google Scholar]
- 31.Sakurai H., Sakaguchi Y., Shoji E., Nishino T., Maki I., Sakai H. In vitro modeling of paraxial mesodermal progenitors derived from induced pluripotent stem cells. PLoS One. 2012;7(10) doi: 10.1371/journal.pone.0047078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosetti M., Boccafoschi F., Leigheb M., Cannas M.F. Effect of different growth factors on human osteoblasts activities: a possible application in bone regeneration for tissue engineering. Biomol Eng. 2007;24(6):613–618. doi: 10.1016/j.bioeng.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 33.Zhou L., Ogata Y. Transcriptional regulation of the human bone sialoprotein gene by fibroblast growth factor 2. J Oral Sci. 2013;55(1):63–70. doi: 10.2334/josnusd.55.63. [DOI] [PubMed] [Google Scholar]
- 34.Joeng K.S., Long F. Constitutive activation of Gli2 impairs bone formation in postnatal growing mice. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0055134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozhemyakina E., Lassar A.B., Zelzer E. A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development. 2015;142(5):817–831. doi: 10.1242/dev.105536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalajzic I., Kalajzic Z., Kaliterna M., Gronowicz G., Clark S.H., Lichtler A.C. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17(1):15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 37.Yin D., Wang Z., Gao Q., Sundaresan R., Parrish C., Yang Q. Determination of the fate and contribution of ex vivo expanded human bone marrow stem and progenitor cells for bone formation by 2.3ColGFP. Mol Ther. 2009;17(11):1967–1978. doi: 10.1038/mt.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohba S., Ikeda T., Kugimiya F., Yano F., Lichtler A.C., Nakamura K. Identification of a potent combination of osteogenic genes for bone regeneration using embryonic stem (ES) cell-based sensor. FASEB J. 2007;21(8):1777–1787. doi: 10.1096/fj.06-7571com. [DOI] [PubMed] [Google Scholar]
- 39.Zujur D., Kanke K., Lichtler A.C., Hojo H., Chung U.I., Ohba S. Three-dimensional system enabling the maintenance and directed differentiation of pluripotent stem cells under defined conditions. Sci Adv. 2017;3(5) doi: 10.1126/sciadv.1602875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buttery L., Bielby R., Howard D., Shakesheff K. Osteogenic differentiation of embryonic stem cells in 2D and 3D culture. Methods Mol Biol. 2011;695:281–308. doi: 10.1007/978-1-60761-984-0_18. [DOI] [PubMed] [Google Scholar]
- 41.Caiazzo M., Okawa Y., Ranga A., Piersigilli A., Tabata Y., Lutolf M.P. Defined three-dimensional microenvironments boost induction of pluripotency. Nat Mater. 2016;15(3):344–352. doi: 10.1038/nmat4536. [DOI] [PubMed] [Google Scholar]
- 42.Gadue P., Huber T.L., Paddison P.J., Keller G.M. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103(45):16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sumi T., Tsuneyoshi N., Nakatsuji N., Suemori H. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development. 2008;135(17):2969–2979. doi: 10.1242/dev.021121. [DOI] [PubMed] [Google Scholar]
- 44.Nizzardo M., Simone C., Falcone M., Locatelli F., Riboldi G., Comi G.P. Human motor neuron generation from embryonic stem cells and induced pluripotent stem cells. Cell Mol Life Sci. 2010;67(22):3837–3847. doi: 10.1007/s00018-010-0463-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89(5):755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 46.Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J.M., Behringer R.R. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108(1):17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 47.Milona M.A., Gough J.E., Edgar A.J. Expression of alternatively spliced isoforms of human Sp7 in osteoblast-like cells. BMC Genomics. 2003;4:43. doi: 10.1186/1471-2164-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsubara T., Kida K., Yamaguchi A., Hata K., Ichida F., Meguro H. BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J Biol Chem. 2008;283(43):29119–29125. doi: 10.1074/jbc.M801774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulsamer A., Ortuno M.J., Ruiz S., Susperregui A.R., Osses N., Rosa J.L. BMP-2 induces Osterix expression through up-regulation of Dlx5 and its phosphorylation by p38. J Biol Chem. 2008;283(7):3816–3826. doi: 10.1074/jbc.M704724200. [DOI] [PubMed] [Google Scholar]
- 50.Lu X., Gilbert L., He X., Rubin J., Nanes M.S. Transcriptional regulation of the osterix (Osx, Sp7) promoter by tumor necrosis factor identifies disparate effects of mitogen-activated protein kinase and NF kappa B pathways. J Biol Chem. 2006;281(10):6297–6306. doi: 10.1074/jbc.M507804200. [DOI] [PubMed] [Google Scholar]
- 51.Maeda Y., Hojo H., Shimohata N., Choi S., Yamamoto K., Takato T. Bone healing by sterilizable calcium phosphate tetrapods eluting osteogenic molecules. Biomaterials. 2013;34(22):5530–5537. doi: 10.1016/j.biomaterials.2013.03.089. [DOI] [PubMed] [Google Scholar]
- 52.Hurley M.M., Abreu C., Harrison J.R., Lichtler A.C., Raisz L.G., Kream B.E. Basic fibroblast growth factor inhibits type I collagen gene expression in osteoblastic MC3T3-E1 cells. J Biol Chem. 1993;268(8):5588–5593. [PubMed] [Google Scholar]
- 53.Rodan S.B., Wesolowski G., Yoon K., Rodan G.A. Opposing effects of fibroblast growth factor and pertussis toxin on alkaline phosphatase, osteopontin, osteocalcin, and type I collagen mRNA levels in ROS 17/2.8 cells. J Biol Chem. 1989;264(33):19934–19941. [PubMed] [Google Scholar]
- 54.Naganawa T., Xiao L., Abogunde E., Sobue T., Kalajzic I., Sabbieti M. In vivo and in vitro comparison of the effects of FGF-2 null and haplo-insufficiency on bone formation in mice. Biochem Biophys Res Commun. 2006;339(2):490–498. doi: 10.1016/j.bbrc.2005.10.215. [DOI] [PubMed] [Google Scholar]
- 55.Chen K.G., Mallon B.S., McKay R.D., Robey P.G. Human pluripotent stem cell culture: considerations for maintenance, expansion, and therapeutics. Cell Stem Cell. 2014;14(1):13–26. doi: 10.1016/j.stem.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawane T., Komori H., Liu W., Moriishi T., Miyazaki T., Mori M. Dlx5 and Mef2 regulate a novel Runx2 enhancer for osteoblast-specific expression. J Bone Miner Res. 2014;29(9):1960–1969. doi: 10.1002/jbmr.2240. [DOI] [PubMed] [Google Scholar]
- 57.Bruderer M., Richards R.G., Alini M., Stoddart M.J. Role and regulation of RUNX2 in osteogenesis. Eur Cells Mater. 2014;28:269–286. doi: 10.22203/ecm.v028a19. [DOI] [PubMed] [Google Scholar]
- 58.Dacic S., Kalajzic I., Visnjic D., Lichtler A.C., Rowe D.W. Col1a1-driven transgenic markers of osteoblast lineage progression. J Bone Miner Res. 2001;16(7):1228–1236. doi: 10.1359/jbmr.2001.16.7.1228. [DOI] [PubMed] [Google Scholar]
- 59.Sparks N.R.L., Martinez I.K.C., Soto C.H., Zur Nieden N.I. Low osteogenic yield in human pluripotent stem cells associates with differential neural crest promoter methylation. Stem Cells. 2018;36(3):349–362. doi: 10.1002/stem.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayashi K., Okamoto F., Hoshi S., Katashima T., Zujur D.C., Li X. Fast-forming hydrogel with ultralow polymeric content as an artificial vitreous body. Nat Biomed Eng. 2017;1:44. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.