Abstract
Background and Purpose
Whether and how circadian clock proteins regulate drug detoxification are not known. Here, we have assessed the effects of CLOCK (a core circadian clock protein) on drug metabolism and detoxification.
Experimental Approach
Regulation by CLOCK protein of drug‐metabolizing enzymes was assessed using Clock knockout (Clock −/−) mice and Hepa‐1c1c7/AML‐12 cells. The relative mRNA and protein levels were determined by qPCR and Western blotting respectively. Toxicity and pharmacokinetic experiments were performed with Clock −/− and wild‐type mice after intraperitoneal injection of coumarin or cyclophosphamide. Transcriptional gene regulation was investigated using luciferase reporter, mobility shift, and chromatin immunoprecipitation (ChIP) assays.
Key Results
Clock deletion disrupted hepatic diurnal expressions of a number of drug‐metabolizing enzymes in mice. In particular, CYP2A4/5 expressions were markedly down‐regulated, whereas CYP2B10 was up‐regulated. Positive regulation of Cyp2a4/5 and negative regulation of Cyp2b10 by CLOCK were confirmed in Hepa‐1c1c7 and AML‐12 cells. Based on a combination of luciferase reporter, mobility shift, and ChIP assays, we found that CLOCK activated Cyp2a4/5 transcription via specific binding to E‐box elements in promoter region and repressed Cyp2b10 transcription through REV‐ERBα/β (two target genes of CLOCK and transcriptional repressors of Cyp2b10). Furthermore, Clock ablation sensitized mice to coumarin toxicity by down‐regulating CYP2A4/5‐mediated metabolism (a detoxification pathway) and to cyclophosphamide toxicity by up‐regulating CYP2B10‐mediated metabolism (generating the toxic metabolite 4‐hydroxycyclophosphamide).
Conclusion and Implications
CLOCK protein regulates metabolism by the cytochrome P450 family and drug detoxification. The findings improve our understanding of the crosstalk between circadian clock and drug detoxification.
What is already known
Circadian clock machinery is suggested to regulate drug metabolism and detoxification.
What this study adds
Our experiments showed some of the molecular links between CLOCK protein and drug metabolism.
CLOCK protein regulates time‐dependent toxicities of coumarin and cyclophosphamide.
What is the clinical significance
Our findings would facilitate the practice of chronotherapeutics with clinical drugs such as cyclophosphamide.
Abbreviations
- 4‐OH‐CPA
4‐hydroxycyclophosphamide
- ChIP
chromatin immunoprecipitation
- CYP
cytochrome P450 family
- DECP
3‐dechloroethylcyclophosphamide
- RevRE
Rev‐erb response element
- siRNA
short interfering RNA
- ZT
zeitgeber time
1. INTRODUCTION
Many physiological processes and behaviours of biological organisms are subject to circadian rhythms (a 24‐hr oscillation pattern; Bozek et al., 2009). Disruption of circadian rhythms is associated with various types of disorders such as depression, diabetes, cancers, and cardiovascular diseases (Gale et al., 2011; Martino et al., 2008; Salgado‐Delgado, Tapia Osorio, Saderi, & Escobar, 2011; Savvidis & Koutsilieris, 2012). Circadian rhythms are generated by circadian genes and proteins that are regulated by biological clocks, also known as circadian clocks. The circadian clock system comprises the central and peripheral clocks (Dibner, Schibler, & Albrecht, 2010). The former resides in the suprachiasmatic nucleus (SCN) of the brain, whereas the latter is present in the peripheral organs. At the molecular level, the circadian clock is a transcriptional–translational feedback loop system consisting of positive (including circadian locomotor output cycles kaput [https://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=872#2649], neuronal PAS domain protein 2 [NPAS2], and brain and muscle ARNT‐like 1 [BMAL1]) and negative (including period [PER] and cryptochrome [CRY]) limbs (Alexander, Cidlowski, et al., 2017; Reppert & Weaver, 2002). CLOCK or NPAS2 heterodimerizes with BMAL1 to activate the transcription of clock‐controlled genes including PER and CRY (Zmrzljak & Rozman, 2012). When a critical concentration is reached , PER and CRY proteins inhibit CLOCK/NPAS2‐BMAL1 activity and down‐regulate their own expression and the expression of other clock‐controlled genes (Zmrzljak & Rozman, 2012).
CLOCK is a member of the basic helix–loop–helix PER–ARNT–SIM (PAS) transcription factor family and an integral component of the circadian clock, as noted above (Antoch et al., 1997; King et al., 1997). Mice with a mutation in the Clock gene become arrhythmic and have a disrupted pattern of circadian gene expression (Oishi et al., 2003; Vitaterna et al., 1994). CLOCK binds to its specific response element (known as an E‐box element) in the promoter regions and activates transcription of target genes (Yoshitane et al., 2014). In addition to regulating circadian gene expression, CLOCK plays important roles in regulation of many physiological processes such as cell cycle, lipid metabolism, glucose metabolism, and immune responses (Cretenet, Le Clech, & Gachon, 2010; Feillet et al., 2014; Spengler et al., 2012). NPAS2 is another basic helix–loop–helix PAS transcription factor and is reported to have the same role as CLOCK in regulation of circadian rhythms in tissues such as the SCN and vasculature (DeBruyne, Weaver, & Reppert, 2007; Hogenesch, Gu, Jain, & Bradfield, 1998; McNamara et al., 2001). However, Npas2 knockout mice have mild phenotypes in circadian gene expression and behaviour (Dudley et al., 2003; Reick, Garcia, Dudley, & McKnight, 2001).
Xenobiotic (drug) detoxification generally involves three processes, namely, Phase I modification, Phase II conjugation, and Phase III excretion (Xu, Li, & Kong, 2005). Cytochromes P450 (https://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=242s) are a superfamily of enzymes that catalyse Phase I oxidation and reduction of xenobiotics and endogenous substances (Nebert & Dalton, 2006). In fact, CYPs are responsible for metabolism of up to 75% of clinically used drugs (Evans & Relling, 1999). CYP metabolism usually converts drug molecules to biologically inactive metabolites (a detoxification mechanism) although, in some important instances, CYP metabolism causes toxicities because the generated metabolites are toxic, such as https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5239 (acetaminophen) and aflatoxin B1 (He et al., 2006; J. Zhang, Huang, Chua, Wei, & Moore, 2002). Many orthologous CYP genes have been identified for humans and mice (e.g., human https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1324 vs. mouse Cyp2b10, human https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1321 vs. mouse Cyp2a5 and human CYP7A1 vs. mouse Cyp7a1; Alexander, Fabbro, et al., 2017; Hrycay & Bandiera, 2009; Nelson et al., 2004). CYP enzymes are highly expressed in the liver. This supports the liver as a major organ for drug detoxification, particularly when drugs are intravenously delivered.
There is accumulating evidence that drug detoxification is under the control of the circadian clock (Claudel, Cretenet, Saumet, & Gachon, 2007; Lévi & Okyar, 2011). The Circadian clock appears to regulate drug detoxification through modulating the expression of drug‐metabolizing enzymes (DMEs) and efflux transporters. Such regulation leads to a dependency of drug tolerance and toxicity on the dosing time. For instance, CYP2B10‐mediated detoxification of https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5480 and https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7154 is modulated by three clock output genes D‐site binding protein (DBP), hepatic leukaemia factor (HLF), and thyrotroph embryonic factor (TEF; Gachon, Olela, Schaad, Descombes, & Schibler, 2006). The circadian gene small heterodimer partner (SHP) participates in regulating toxicities of paracetamol, aflatoxin B1, and https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7242 (T. Zhang et al., 2018). Also, the two clock output genes HLF and E4BP4 control the diurnal rhythms of expression of the intestinal drug transporter https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=768 and of https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4726 accumulation (Murakami, Higashi, Matsunaga, Koyanagi, & Ohdo, 2008). However, whether and how the central clock genes such as CLOCK and BMAL1 regulate DMEs and transporters remains largely unknown.
https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=596/https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=597 (NR1D1/2) are nuclear haem receptors best known as transcriptional repressors (Alexander, Cidlowski, et al., 2017; Solt, Kojetin, & Burris, 2011). They bind to the specific response element (Rev‐erb response element [RevRE]) in target genes and repress gene transcription by recruiting the co‐repressors NR corepressor 1 and https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2617 (Duez & Staels, 2008; Ercolani et al., 2015; Raghuram et al., 2007). REV‐ERBα/β are involved in regulation and maintenance of circadian rhythms by repressing the transcription of BMAL1 (Yin et al., 2007). In addition, REV‐ERBα/β play important roles in regulation of many other physiological processes including cell differentiation, inflammation, lipid, and glucose metabolism (Cho et al., 2012; Gibbs et al., 2012; J. Wang & Lazar, 2008). Our recent studies also suggest a critical role for REV‐ERBα in regulation of DMEs including CYP2B10 and carboxylesterases (Zhang et al., 2018; Zhao, Zhang, Yu, Guo, & Wu, 2018).
To clarify the effects of CLOCK protein on liver detoxification, we established a Clock knockout (Clock −/−) mouse strain in this study. Deletion of Clock disrupted diurnal expressions of an array of CYP enzymes including CYP2A4/5 and CYP2B10. Cell‐based assays confirmed positive regulation of Cyp2a4/5 and negative regulation of Cyp2b10 by CLOCK. Clock deletion sensitized mice to coumarin and cyclophosphamide toxicities due to altered metabolism. Mechanistic studies revealed that CLOCK transactivated Cyp2a4/5 transcription whereas it indirectly repressed Cyp2b10 transcription via REV‐ERBα/β. Collectively, our study for the first time establishes the molecular links between Clock gene and CYP metabolism and has implications in the practice of chronotherapeutics.
2. METHODS
2.1. Mice
All animal care and experimental procedures were performed using protocols approved by the Institutional Animal Care and Use Committees of Jinan University (Guangzhou, China). At the end of the experiments, the mice were killed using CO2. The authors declare that this work followed the BJP guidelines and that every effort was made to minimize the number of animals used and their level of suffering. Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny, Browne, Cuthill, Emerson, & Altman, 2010) and with the recommendations made by the British Journal of Pharmacology. Clock −/−, Npas2 −/−, Bmal1 −/−, Rev‐erbα −/−, and Rev‐erbβ −/− (on a C57BL/6 background) mice were obtained from Cyagen Biosciences Inc (Guangzhou, China). All mice were housed and bred at the Institute of Laboratory Animal Science, Jinan University (Guangzhou, China). Mice were synchronized to a standard lighting condition of LD12:12, with lights on from 7 a.m. (zeitgeber time [ZT] 0) to 7 p.m. (ZT12) prior to experiments.
Mice were grouped randomly (n = 5 per group) for the experiment. Sample size calculations for the toxicity experiments were based on the ability to detect a 50% difference in ALT and AST levels between drug and vehicle groups at a power of 0.8, α = .05, and an assumed SD of 30% of group means according to our pilot study. A sample size of at least four mice per group was derived using the power and sample size calculation software. We therefore used a sample size of five mice for the proposed experiments. Sample size calculations for the pharmacokinetic experiments were based on the ability to detect a 30% difference in drug exposure between Clock +/+ and Clock −/− mice at a power of 0.8, α = .05, and an assumed SD of 20% of group means. According to power calculation, a sample size of five mice per group was determined.
2.2. qPCR assay
Total RNA was isolated from liver and cell samples using RNAiso Plus reagent (Takara Bio Inc., Shiga, Japan). cDNA was synthesized using PrimeScript™ RT reagent kit (TaKaRa). qPCR reactions were performed as described in our previous study (Zhao et al., 2018). Hydroxymethylbilane synthase was used as an internal control. Primers are provided in Table S1.
2.3. Western blotting
The immuno‐related procedures used comply with the recommendations made by the British Journal of Pharmacology (Alexander et al., 2018) and was described previously (Wang et al., 2018). In brief, mouse liver proteins were extracted using RIPA buffer (Beyotime Biotechnology). The samples were subject to SDS‐PAGE (10% gel) and transferred to PVDF membranes. The membranes were sequentially incubated with primary antibody and HRP‐conjugated secondary antibody. After adding enhanced chemiluminescence, protein bands were visualized by Omega Lum G imaging system (Aplegen). Band density was analysed by Quantity One software (Bio‐Rad). GAPDH was used as a loading control.
2.4. Liver CYP microsomal metabolism assay
Mouse liver microsomes were prepared by sequential centrifugation as previously described (Zhao et al., 2018). https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2858, coumarin, and pentoxyresorufin were used as respective specific substrates to determine the activities of CYP2A4, CYP2A5, and CYP2B10. Each of these substrates was incubated with liver microsomes and NADPH as described in our previous study (Zhang et al., 2018). The reaction was terminated by adding ice‐cold acetonitrile. The resulting mixture was centrifuged at 15,000 g (4°C) for 15 min, and the supernatant was blindly subjected to quantitative analyses by UHPLC–MS/MS (for testosterone and coumarin) and by microplate spectrophotometer (Synergy HTX, BioTek; for pentoxyresorufin).
2.5. Toxicity experiments
Toxicity experiments were performed for coumarin and cyclophosphamide. Clock +/+ (wild‐type) and Clock −/− mice (male, 8–11 weeks of age, n = 5) received a single dose of coumarin (50 mg·kg−1) or cyclophosphamide (300 mg·kg−1) by intraperitoneal injection. Hepatotoxicity was blindly assessed by determining plasma alanine aminotransferase (ALT), aspartate aminotransferase (AST), and https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6737 levels at 4 hr post drug administration. ALT, AST, and GSH were quantified using their respective assay kits according to the manufacturer's instructions.
2.6. Pharmacokinetic experiments
Coumarin (i.p., 30 mg·kg−1) or cyclophosphamide (i.p., 100 mg·kg−1) was administered to wild‐type and Clock −/− mice (male, 8–11 weeks of age). Blood samples were collected by retro‐orbital bleeding at predetermined time points (5 min, 10 min, 0.5 hr, and 1 hr for coumarin and 0.25, 0.5, 1, and 2 hr for cyclophosphamide [n = 5 per time point]). The plasma samples were prepared as previously described (Liu et al., 2014; Sadagopan, Cohen, Roberts, Collard, & Omer, 2001). Of note, the cyclophosphamide metabolite, 4‐hydroxycyclophosphamide (4‐OH‐CPA) is unstable in the plasma (Sadagopan et al., 2001). To quantify 4‐OH‐CPA, the samples were treated with O‐methylhydroxylamine to transform 4‐OH‐CPA into a stable product hydroxycyclophosphamide O‐methyloxime as previously performed (Sadagopan et al., 2001). Chemicals were quantified in a blinded manner by UHPLC/MS/MS analysis.
2.7. UHPLC–MS/MS analysis
Quantification of drugs and metabolite was performed using a UHPLC–MS/MS system consisting of a Dionex ultimate 3000 UHPLC system and AB 3500 triple‐quadrupole mass spectrometer (AB Sciex, Ontario, Canada). Chromatographic separations were performed on an ACE Ultra‐Core 2.5 Super C18 column (Phenomenex, Torrance, CA) using a gradient elution with mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile). The gradient programme consisted of 10% B (0–2 min), 10–90% B (2–6 min), and 10% B (6–8 min). The flow rate was set at 0.3 ml·min−1. The mass transition ion‐pair was selected as m/z 147.0 → 103.1 for coumarin, m/z 163.1 → 107.0 for 7‐hydroxycoumain, m/z 288.9 → 96.8 for testosterone, m/z 305.0 → 287.3 for 6β‐hydroxytestosterone, m/z 260.9 → 140.1 for cyclophosphamide, m/z 306.1 → 221.1 for hydroxycyclophosphamide‐O‐methyloxime, m/z 199.0 → 171.0 for 3‐dechloroethylcyclophosphamide (DECP), m/z 275.2 → 109.0 for 19‐nortestosterone (an internal standard for testosterone and 6β‐hydroxytestosterone), m/z 261.9 → 154.1 for ifosfamide (an internal standard for cyclophosphamide and hydroxy‐ cyclophosphamide‐O‐methyloxime), and m/z 165.4 → 149.0 for 3′,4′‐(methylenedioxy)acetophenone (an internal standard for coumarin and 7‐hydroxycoumain).
2.8. Luciferase reporter assays
NIH3T3 cells were seeded into 48‐well plates. After 24 hr, the cells were transfected with Cyp reporter plasmid [Cyp2a5 (2000)‐Luc, Cyp2a5 (1600)‐Luc, Cyp2a5 (800)‐Luc, Cyp2b10 (2500)‐Luc, Cyp2b10 (2000)‐Luc, or Cyp2b10 (1000)‐Luc], pRL‐TK vector, expression plasmid (pcDNA‐Clock, pcDNA‐Rev‐erbα, or pcDNA‐Rev‐erbβ), and/or siClock using the jetPRIME® transfection reagent; 24 hr later, the luciferase activity was assayed using the Dual‐Luciferase reporter assay system (Promega) and expressed as relative luciferase unit.
2.9. EMSA
EMSA assays were performed as previously described (Lu, Wang, Xie, Guo, & Wu, 2017). Briefly, NIH3T3 cells were transfected with pcDNA‐Clock or pcDNA‐Rev‐erbβ plasmids. The nuclear extracts were prepared using the transfected cells and cytoplasmic protein extraction reagents (Beyotime Biotech, Shanghai, China). Biotin‐labelled probes were incubated with the extracts using the chemiluminescent EMSA kit (Beyotime Biotech). Unlabelled probes or mutated probes were added to the incubation mixture to validate specific DNA–protein interaction. Samples were then loaded onto a 4% non‐denaturing polyacrylamide gel. The gel was electrophoresed at 200 V for 30 min. The products were transferred to a positively charged nylon membrane. The membrane was treated with enhanced chemiluminescence and visualized by Omega Lum G imaging system (Aplegen). Probe sequences are provided in Table S2.
2.10. Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assays were performed as described in our previous publication (Wang et al., 2018). In brief, mouse liver samples were cross‐linked in 1% formaldehyde. The cross‐linked samples were digested with micrococcal nuclease and sonicated to reduce the DNA length (a final size of 150–900 bp). The sheared chromatin was incubated with anti‐CLOCK (Abcam, Cambridge, MA), anti‐REV‐ERBα (CST, Beverly, MA), or control rabbit IgG at 4°C overnight. Samples were de‐cross‐linked, and DNAs were purified, followed by qPCR analysis (the primers are listed in Table S3).
2.11. Data and statistical analysis
The data and statistical analysis comply with the recommendations of the British Journal of Pharmacology on experimental design and analysis in pharmacology (Curtis et al., 2018). Experiments were carried out at n = 5, where n = number of independent experiments. Data are presented as raw data (mean ± SD) and tested for normality with a Shapiro–Wilk test, outliers were included in data analysis. Student's t‐test was used to test for statistical differences between two groups. One‐way or two‐way ANOVA followed by Bonferroni post hoc test (only when the F value attained P < .05) was used for multiple group comparisons. There was no significant inhomogeneity of variances. All statistical analyses were performed using Graphpad Prism software version 7.00 (GraphPad Software, Inc., California, USA, RRID:SCR_002798), and the level of significance was set at P < .05.
2.12. Materials
Testosterone, coumarin, 7‐hydroxycoumain, resorufin, and methoxyamine were purchased from Aladdin Chemicals (Shanghai, China). 6β‐hydroxyltestosterone was obtained from Steraloids (Wilton, NH). 19‐nortestosterone, 3′,4′‐(methylenedioxy)acetophenone, and https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7201 were purchased from Sigma‐Aldrich (St. Louis, MO). Pentoxyresorufin was purchased from ApexBio Technology (Houston, TX). Cyclophosphamide was purchased from J&K Scientific (Beijing, China). Anti‐CYP2A and anti‐CLOCK antibodies were purchased from Abcam, anti‐CYP2B10 from OriGene (Rockville, MD), anti‐REV‐ERBα from Sigma‐Aldrich, and anti‐REV‐ERBβ from Abnova (Tebu‐bio, France). SimpleChIP® Plus Enzymatic Chromatin IP kit was purchased from Cell Signaling Technology (Beverly, MA). ALT, AST, and GSH assay kits were purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). Cyp2a5 (2000)‐Luc, Cyp2a5 (1600)‐Luc, Cyp2a5 (800)‐Luc, Cyp2b10 (2500)‐Luc, Cyp2b10 (2000)‐Luc, Cyp2b10 (1000)‐Luc, pcDNA‐Clock, siClock (short interfering RNA [siRNA] targeting Clock), siRev‐erbc (siRNA targeting Rev‐erbα), and siRev‐erbβ (siRNA targeting Rev‐erbβ) were obtained from TranSheep Bio‐Tech (Shanghai, China). pGL4.11 and pRL‐TK vectors were purchased from Promega (Madison, WI). pcDNA‐Rev‐erbα and pcDNA‐Rev‐erbβ were obtained from Biowit Technologies (Shenzhen, China).
2.13. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 (Alexander, Cidlowski, et al., 2017; Alexander, Fabbro, et al., 2017; Alexander, Kelly et al., 2017)
3. RESULTS
3.1. Clock deletion disrupts the rhythmicity of CYP expression in mouse liver
We created a Clock‐deficient mouse model by deleting the exons 9 to 13 using the CRISPR/Cas9 technique (Figure 1a). It was confirmed that hepatic Clock transcripts were absent in these genetic mice (Figure 1a). As expected, Clock deficiency led to down‐regulation of its target gene Dbp (Figure S1; Ripperger, Shearman, Reppert, & Schibler, 2000; Triqueneaux et al., 2004). qPCR analyses showed that many xenobiotic‐detoxifying genes in mouse liver (e.g., Cyp2a4, Cyp2a5, Cyp2b10, Cyp3a11, Cyp3a25, Ugt1a1, Ugt1a9, Sult1a1, and Sult2a7) were regulated by CLOCK (Figure 1b). Of note, Cyp2a4 and Cyp2a5 were two genes down‐regulated the most in Clock‐deficient mice, whereas Cyp2b10 was up‐regulated the most (Figure 1b). Moreover, Clock ablation blunted diurnal rhythms of hepatic Cyp2a4, Cyp2a5, and Cyp2b10 mRNAs (Figure 1c). Likewise, diurnal rhythms of hepatic CYP2A4/5 and CYP2B10 proteins were disrupted in Clock −/− mice (Figure 1d). In addition, Clock −/− mice showed altered liver microsomal CYP activities that agreed well with the changes in CYP protein expression (Figure 1e). Collectively, these data indicated that CLOCK played a critical role in regulation of CYP enzymes, particularly CYP2A4/5 and CYP2B10.
Figure 1.
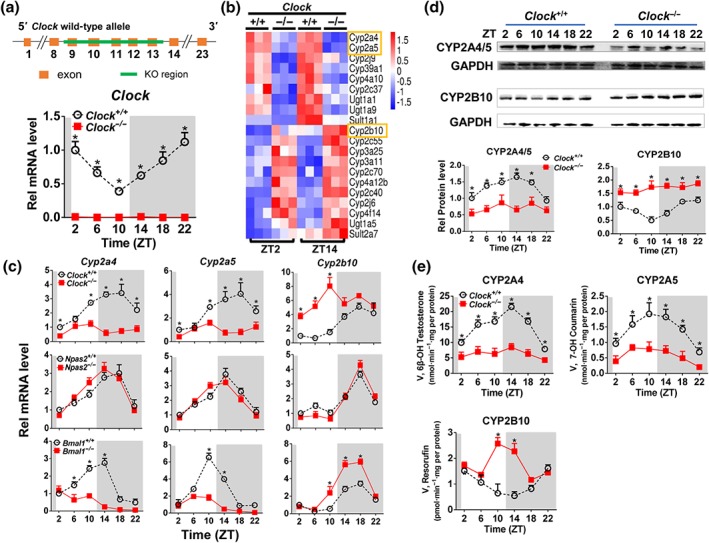
Clock deletion disrupts the rhythmicity of CYP expression in mouse liver. (a) Validation of Clock knockout (Clock −/−) mice by determining Clock transcript. (b) Heatmap showing relative mRNA expression of drug‐metabolizing enzymes at ZT2 and ZT14 in Clock +/+ versus Clock −/− mice. (c) Diurnal mRNA expression of Cyp2a4/5 and Cyp2b10 in the livers from Clock +/+ and Clock −/− mice; Npas2 +/+ and Npas2 −/− mice; and Bmal1 +/+ and Bmla1 −/− mice. (d) Diurnal protein expression of CYP2A4/5 and CYP2B10 in the livers from Clock +/+ and Clock −/− mice. (e) CYP activities derived from liver microsomes of Clock +/+ and Clock −/− mice. The specific substrates used to determine the activities of CYPs were testosterone for CYP2A4, coumarin for CYP2A5, and pentoxyresorufin for CYP2B10. In Panels a, c, d, and e, data are mean ± SD (n = 5). * P < .05, significant differences between two genotypes; two‐way ANOVA with Bonferroni post hoc test
NPAS2 has been reported to perform similar functions as CLOCK does in some organs (e.g., SCN and vasculature). We thus determined the potential effects of NPAS2 on Cyp2a4/5 and Cyp2b10 expressions using Npas2 knockout (Npas2 −/−) mice. Npas2 ablation did not alter hepatic diurnal expressions of Cyp2a4/5 or Cyp2b10 (Figure 1c), suggesting no or negligible contribution of NPAS2 to regulation of Cyp2a4/5 and Cyp2b10. As BMAL1 is a partner of CLOCK in regulation of diurnal gene expression by acting on E‐box elements (Reppert & Weaver, 2002), we also determined the effects of BMAL1 on Cyp2a4/5 and Cyp2b10 expressions using Bmal1 −/− mice. Bmal1 ablation resulted in reduced expression of Cyp2a4/5 and up‐regulated expression of Cyp2b10 in the liver (Figure 1c). Our data show regulation of CYP2A4/5 and CYP2B10 by the E‐box‐binding proteins BMAL1 and CLOCK.
3.2. CLOCK transcriptionally regulates Cyp2a4/5 expression
The regulatory effects of CLOCK on Cyp2a4/5 expression were assessed using mouse hepatoma Hepa‐1c1c7 cells and AML‐12 mouse hepatocytes. Overexpression of Clock led to significant increases in Cyp2a4/5 expressions in Hepa‐1c1c7 and AML‐12 cells, whereas knockdown of Clock (by siRNA) resulted in reduced Cyp2a4/5 expressions, supporting positive regulation of Cyp2a4/5 by CLOCK (Figure 2a). In luciferase reporter assays, Clock dose dependently induced Cyp2a5 promoter activity, whereas Clock siRNA decreased the promoter activity, indicating a transcriptional control of Cyp2a5 by CLOCK (Figure 2b). Sequence analysis revealed three potential E‐box (a motif for CLOCK binding) elements (i.e., E‐box 1, E‐box 2, and E‐box 3) in Cyp2a5 promoter (Figure 2c). Truncation analysis showed that the E‐box 1 element located at −1,706/−1,700 bp region was actually responsible for CLOCK binding (Figure 2c). A direct interaction of CLOCK with E‐box 1 element was confirmed by EMSA experiment (Figure 2d). ChIP assay further indicated significant recruitment of hepatic CLOCK protein to E‐box 1 element of Cyp2a5 in wild‐type mice (Figure 2e). However, such recruitment was lost in Clock −/− mice (Figure 2e). It is noted that Cyp2a4 and Cyp2a5 share identical E‐box elements in their promoter regions, indicating the same mechanism for CLOCK regulation of Cyp2a4 (Figure 2f). Taken together, CLOCK transactivates Cyp2a4/5 via direct binding to E‐box elements in promoter regions.
Figure 2.
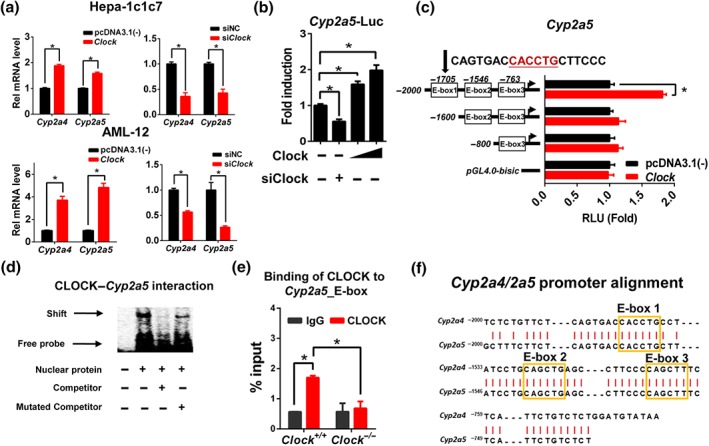
CLOCK transactivates Cyp2a4/5 through direct binding to E‐boxes. (a) qPCR analyses of Cyp2a4/5 in Hepa‐1c1c7 and AML‐12 cells treated with siNC or siClock (50 nM) or pcDNA3.1(‐) or Clock expression plasmid (800 ng). (b) CLOCK induces Cyp2a5 luciferase reporter activity. NIH3T3 cells were transfected with Cyp2a5‐Luc reporter (100 ng) and Clock expression plasmids (100 or 200 ng) or siClock (50 nM); 24 hr later, the cells were collected for luciferase activity measurement. (c) Luciferase reporter assays with different versions of Cyp2a5‐Luc reporter constructs, indicating that −1,706/−1,700 bp region was responsible for CLOCK binding. (d) EMSA assay showing a direct interaction between CLOCK protein and Cyp2a5 E‐box element. The nuclear extracts were obtained from NIH3T3 cells transfected with Clock expression plasmid. (e) ChIP assay showing a recruitment of CLOCK protein to Cyp2a5 E‐box in the liver of Clock +/+ mice, and no recruitment in Clock −/− mice. (f) Sequence alignment of Cyp2a4 and 2a5 promoters (derived using Global Align from NCBI). Data are presented as mean ± SD (n = 5). In a, b, and c, * P < .05, significantly different as indicated; Student's t‐test. In e, * P < .05, significantly different as indicated; two‐way ANOVA and Bonferroni post hoc test
3.3. CLOCK regulates Cyp2b10 via REV‐ERBα/β
Consistent with a negative regulation of Cyp2b10 by CLOCK in vivo, Clock overexpression decreased Cyp2b10 expression in Hepa‐1c1c7 and AML‐12 cells and Clock knockdown increased Cyp2b10 expression (Figure 3a). As CLOCK is a transcriptional activator, an indirect mechanism involving a negative regulator was necessary for its regulation of Cyp2b10. REV‐ERBα was previously proposed as a transcriptional repressor of Cyp2b10, and Rev‐erbα/β are direct target genes of CLOCK (Cho et al., 2012; Zhang et al., 2018). We then investigated the roles of REV‐ERBα/β in CLOCK regulation of Cyp2b10. As expected, Clock knockout led to decreased Rev‐erbα/β expressions in mouse liver and blunted their diurnal rhythmicity (Figure 3b,c). Positive regulation of Rev‐erbα/β by CLOCK was confirmed in Hepa‐1c1c7 and AML‐12 cells by Clock overexpression and knockdown experiments (Figure 3d).
Figure 3.
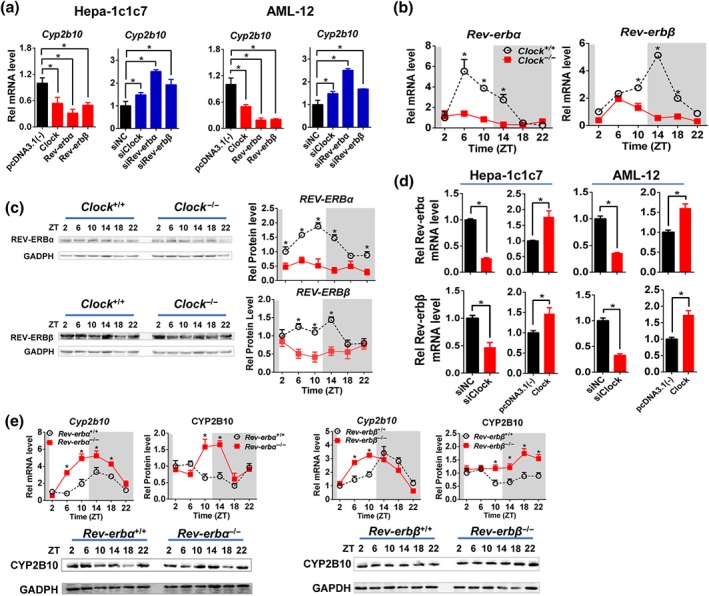
CLOCK regulates Cyp2b10 via REV‐ERBα/β. (a) qPCR analysis of Cyp2b10 in Hepa‐1c1c7 and AML‐12 cells treated with siRNA or pcDNA3.1(‐) or expression plasmids as labelled below x‐axis. * P < .05, significantly different as indicated; Student's t‐test. (b) CLOCK ablation down‐regulated Rev‐erbα/β mRNA expression and blunted their diurnal rhythmicity in mice. * P < .05, significant differences between two genotypes; two‐way ANOVA and Bonferroni post hoc test. (c) Hepatic protein expressions of REV‐ERBα/β in Clock −/− mice were down‐regulated and diurnal rhythmicities were blunted. * P < .05, significant differences between two genotypes; two‐way ANOVA and Bonferroni post hoc test (d) Positive regulation of Rev‐erbα/β by CLOCK confirmed in Hepa‐1c1c7 and AML‐12 cells. * P < .05, significantly different as indicated; Student's t‐test. (e) Rev‐erbα or Rev‐erbβ knockout caused up‐regulation of Cyp2b10 mRNA and protein in mouse liver. * P < .05, significant differences between two genotypes; two‐way ANOVA and Bonferroni post hoc test. Data are presented as mean ± SD (n = 5)
Cell‐based experiments suggested negative regulation of Cyp2b10 by REV‐ERBα/β (Figure 3a). This regulation was validated by genetic experiments in which Rev‐erbα or Rev‐erbβ knockout caused up‐regulation of Cyp2b10 mRNA and protein in mouse liver (Figure 3e). In luciferase reporter assays, REV‐ERBα/β inactivated Cyp2b10 transcription (Figure 4a). Truncation analysis identified a DNA region (−2,342/−2,326 bp) within the Cyp2b10 promoter as the binding site of REV‐ERBβ (named RevRE1) and a DNA region (−1,962/−1,951 bp) as the binding site of REV‐ERBα (named RevRE2; Figure 4b). ChIP assays revealed significant recruitment of REV‐ERBα onto the RevRE2 in Cyp2b10 promoter in wild‐type mice, whereas such recruitment was absent in Clock −/− mice (Figure 4c). Additionally, EMSA experiments confirmed a direct interaction REV‐ERBβ with the RevRE1 element (Figure 4d). Taken together, CLOCK regulates Cyp2b10 expression via REV‐ERBα/β.
Figure 4.
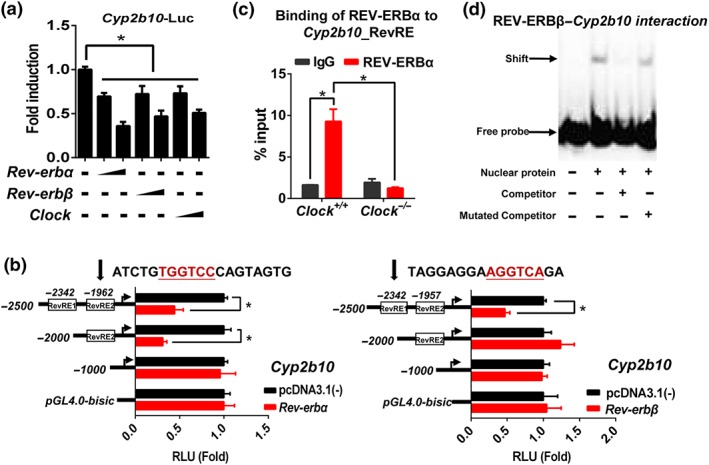
CLOCK regulates Cyp2b10 via REV‐ERBα/β. (a) REV‐ERBα/β and CLOCK repressed Cyp2b10 luciferase reporter activity. NIH3T3 cells were transfected with Cyp2b10‐Luc reporter (100 ng) and Rev‐erbα or Rev‐erbβ or Clock expression plasmid (200 or 300 ng); 24 hr later, the cells were collected for luciferase activity mesurement. * P < .05, significantly different as indicated; Student's t‐test. (b) Luciferase reporter assays with different versions of Cyp2b10‐Luc reporter constructs, indicating that −1,962/−1,951 bp region was responsible for REV‐ERBα binding and −2,342/−2,326 bp region was responsible for REV‐ERBβ binding. * P < .05, significantly different as indicated; Student's t‐test. (c) ChIP assay showing a recruitment of REV‐ERBα protein to Cyp2b10 Rev‐RE in the liver of Clock +/+ mice, and no recruitment in Clock −/− mice. * P < .05, significantly different as indicated; two‐way ANOVA and Bonferroni post hoc test (d) EMSA assay showing a direct interaction between REV‐ERBβ protein and Cyp2b10 Rev‐RE element. Data are presented as mean ± SD (n = 5)
3.4. Clock ablation blunts the diurnal rhythm of coumarin toxicity through down‐regulation of metabolism
Coumarin is detoxified to 7‐hydroxylated metabolite by CYP2A4/5 in mice (Figure 5a; Lake, 1999). Treatment of coumarin (i.p., 50 mg·kg−1, n = 5) at each of six circadian points (ZT2, 6, 10, 14, 18, and 22) induced hepatotoxicity in wild‐type mice (Figure 5b). We also observed a diurnal rhythmicity in coumarin toxicity (Figure 5b). The toxicity was more severe at ZT2/22 than that at ZT14 (Figure 5b). Clock ablation exacerbated the coumarin toxicity at both ZT2 and ZT14 as shown by higher plasma levels of ALT and AST (Figure 5c). This was supported by lower levels of plasma GSH (Figure 5c). Moreover, the time difference in toxicity ceased to exist in Clock −/− mice (Figure 5c). The higher level of coumarin toxicity in Clock −/− mice was accompanied by elevated systemic exposure of coumarin (Figure 5d). This was because 7‐hydroxylation of coumarin was reduced due to down‐regulated expression of CYP2A4/5 (Figures 5d and 1d). Additionally, dosing time‐dependent metabolism and pharmacokinetics were lost in Clock −/− mice (Figure 5d). Taken together, Clock ablation exacerbates coumarin toxicity and blunts its rhythm through down‐regulation of CYP2A4/5‐mediated metabolism.
Figure 5.
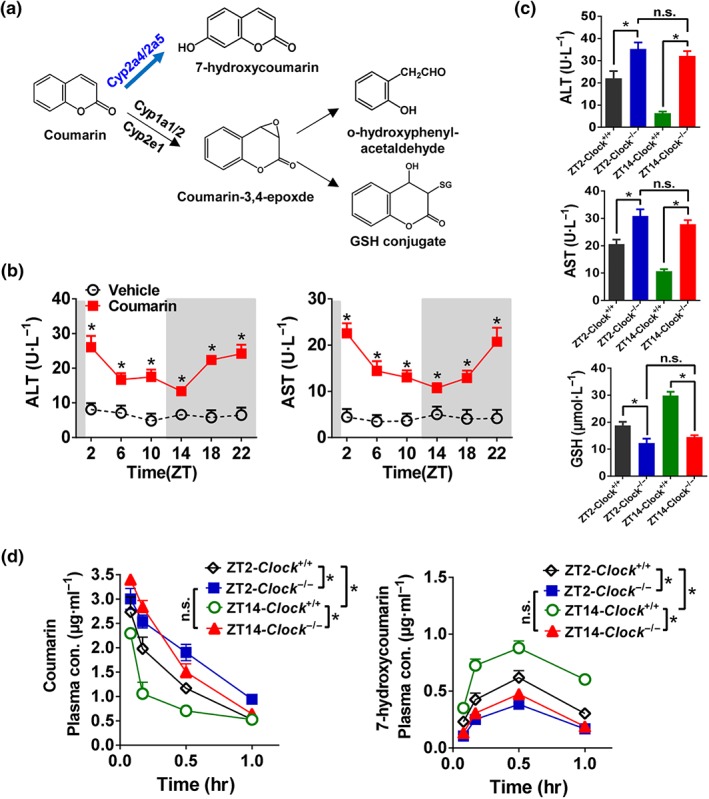
Clock ablation blunts the diurnal rhythm of coumarin toxicity through down‐regulation of metabolism. (a) Major metabolic pathways for coumarin. (b) Coumarin hepatotoxicity shows a diurnal rhythm. Plasma ALT and AST levels in Clock +/+ mice were tested 4 hr after vehicle or coumarin administration (i.p., 50 mg·kg−1) at six time points (ZT2, 6, 10, 14, 18, and 22). * P < .05, significantly different as indicated; two‐way ANOVA and Bonferroni post hoc test. (c) Clock ablation exacerbated the coumarin toxicity at both ZT2 and ZT14. Plasma ALT, AST, and GSH levels were tested in Clock +/+ and Clock −/− mice 4 hr after coumarin administration (i.p., 50 mg·kg−1) at ZT2 and ZT14. * P < .05, significantly different as indicated; two‐way ANOVA and Bonferroni post hoc test. (d) Plasma concentrations of coumarin and 7‐hydroxycoumain in Clock +/+ and Clock −/− mice at 5 min, 10 min, 0.5, and 1 hr after coumarin treatment (i.p., 30 mg·kg−1) at ZT2 and ZT14. * P < .05, significantly different as indicated; two‐way ANOVA and Bonferroni post hoc test. Data are presented as mean ± SD (n = 5). n.s., not significant
3.5. Clock ablation exacerbates cyclophosphamide toxicity and modulates its diurnal rhythm
Cyclophosphamide is widely used to treat various types of cancers including leukaemia, breast cancer, and lymphoma (Batist et al., 2001; Keating et al., 2005). Cyclophosphamide is a prodrug and bioactivated by CYP2B10 to 4‐OH‐CPA (the active form) in mice (Figure 6a; Pass et al., 2005). Cyclophosphamide also undergoes an inactivation pathway (to form the metabolite DECP) mediated by CYP3A13 in mice (Figure 6a; Pass et al., 2005). Treatment with cyclophosphamide (i.p., 300 mg·kg−1, n = 5) induced hepatotoxicity in wild‐type mice (Figure 6b) consistent with previous reports (Habibi et al., 2015; Sheweita, El‐Hosseiny, & Nashashibi, 2016). Of note, the severity of hepatotoxicity was dosing time dependent with high levels at ZT2/22 and low levels at ZT10/14 (Figure 6b). However, the time dependency of cyclophosphamide hepatotoxicity was lost in Clock −/− mice (Figure 6c). The higher level of cyclophosphamide toxicity in genetic mice at ZT14 was associated with increased formation of the toxic metabolite 4‐OH‐CPA due to up‐regulated CYP2B10 expression (Figures 6d and 1D). This was supported by reduced systemic exposure of the parent drug cyclophosphamide and unaltered formation of the inactive metabolite DECP (Figure 6d). In the meantime, cyclophosphamide metabolism and pharmacokinetics showed a dosing time dependency in wild‐type mice that was lost in Clock −/− mice (Figure 6d). Taken together, Clock deletion sensitized mice to cyclophosphamide toxicity and modulated its diurnal rhythm by up‐regulating CYP2B10 metabolism.
Figure 6.
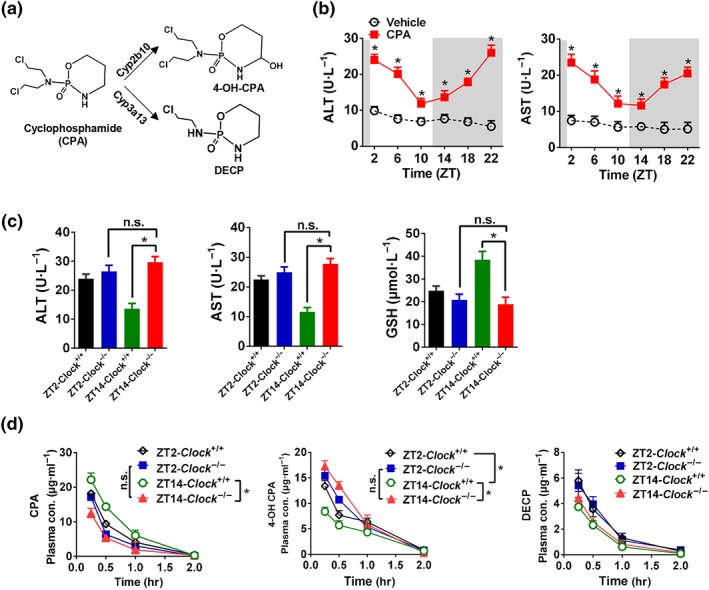
Clock ablation exacerbates cyclophosphamide toxicity and modulates its diurnal rhythm. (a) Major metabolic pathways for cyclophosphamide (CPA). (b) Cyclophosphamide hepatotoxicity shows a diurnal rhythm. Plasma ALT and AST levels in Clock +/+ mice were tested 4 hr after vehicle or cyclophosphamide administration (i.p., 300 mg·kg−1, n = 5) at six time points (ZT2, 6, 10, 14, 18, and 22). significantly different from vehicle; two‐way ANOVA and Bonferroni post hoc test (c) Plasma ALT, AST, and GSH levels in Clock +/+ and Clock −/− mice 4 hr after vehicle or cyclophosphamide administration (i.p., 300 mg·kg−1, n = 5) at ZT2 and ZT14. * P < .05, significantly different as indicated; two‐way ANOVA and Bonferroni post hoc test. (d) Plasma concentrations of cyclophosphamide, 4‐OH‐CPA, and DECP in Clock +/+ and Clock −/− mice at 0.25, 0.5, 1, and 2 hr after cyclophosphamide treatment (i.p., 100 mg·kg−1, n = 5) at ZT2 and ZT14. * P < .05, significantly different as indicated; two‐way ANOVA and Bonferroni post hoc test. Data are presented as mean ± SD. n.s., not significant
4. DISCUSSION AND CONCLUSIONS
In this study, we identified CLOCK protein as a regulator of an array of DMEs including CYP2A4/5, CYP2B10, https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2990, and SULT2A7 (Figure 1b). Contrasting with CLOCK as a transcriptional activator (e.g., it activates Cyp2a4/5 transcription), the protein was a negative regulator of certain enzymes (e.g., CYP2B10). Negative regulation of CYP2B10 by CLOCK was attained through REV‐ERBα/β (two target genes of CLOCK and transcriptional repressors of Cyp2b10; Figures 3 and 4). Therefore, CLOCK controls diurnal expression of CYPs through a two‐pronged mechanism, acting by direct transactivation, and through an intermediate CYP repressor. It was noteworthy that Clock also regulated expressions of CYP2A4/5 and CYP2B10 in female mice (Figure S2), suggesting gender‐independent diurnal control of these three CYP enzymes.
We demonstrated that CLOCK transactivates Cyp2a4/5 through direct binding to E‐box elements in the gene promoters. It is noteworthy that additional transcriptional factors DBP and PPARγ contribute to diurnal expression of Cyp2a5 in previous studies (Deng, Guo, & Wu, 2018; Lavery et al., 1999). DBP is a known target gene and PPARγ is a potential target gene of CLOCK (Oishi, Shirai, & Ishida, 2005; Ripperger et al., 2000). Therefore, in addition to direct transactivation, an indirect regulation mechanism involving DBP and PPARγ may be possible for CLOCK regulation of Cyp2a5. This is supported by the fact that the amplitude of diurnal CLOCK protein is much lower than that of diurnal Cyp2a5 (Figure S3). Additional diurnal regulators are supposed to strengthen the extent of oscillation in Cyp2a5 mRNA.
Clock −/− mice showed exacerbated cyclophosphamide toxicity consistent with a previous study by Gorbacheva et al. (2005). In the study of Gorbacheva et al. (2005), the change in cyclophosphamide toxicity was attributed to circadian control of B cell survival although the exact mechanism was unexplored. By contrast, we proposed that up‐regulated metabolic activation was a main cause of exacerbated cyclophosphamide toxicity in Clock −/− mice (Figure 6). This is also supported by apparently elevated systemic exposure of the active metabolite 4‐OH‐CPA in the study of Gorbacheva et al. (2005). Clock −/− mice showed exacerbated toxicities of both coumarin and cyclophosphamide despite a distinct nature of toxic entities (coumarin itself or the hydroxylated metabolite of cyclophosphamide). This was because the main DMEs (CYP2A4/5 for coumarin and CYP2B10 for cyclophosphamide) were altered in a different direction. CYP2A4/5 were down‐regulated, whereas CYP2B10 was up‐regulated in Clock −/− mice. Thereby, coumarin and cyclophosphamide toxicities were exacerbated due to reduced coumarin detoxification and enhanced formation of toxic metabolite respectively.
Although both REV‐ERB paralogues are involved in mediating CLOCK regulation of CYP2B10 expression, REV‐ERBα appears to play a more important role. Rev‐erbα ablation led to marked up‐regulation of CYP2B10 in mouse liver, whereas deletion of Rev‐erbβ resulted in a mild change in CYP2B10 expression (Figure 3e). The relatively weaker role of REV‐ERBβ in relation to REV‐ERBα in regulation of drug detoxification seems to be consistent with a weaker role of the former than the latter in regulation of circadian rhythms and metabolic homeostasis (Bugge et al., 2012; Cho et al., 2012).
A previous study reports that DBP regulates CYP2B10 expression through https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=607; Gachon et al., 2006). However, our data did not support the positive regulation of CAR and positive regulation of CYP2B10 by DBP in the body because (a) hepatic DBP expression was down‐regulated, but CAR was up‐regulated in Clock −/− mice (Figure S4) and (b) CYP2B10 expression was unexpectedly increased in Clock −/− mice with reduced DBP expression. Up‐regulation of hepatic CAR expression was most likely due to down‐regulated REV‐ERBα/β, two potential repressors of CAR (Kanno, Otsuka, Hiromasa, Nakahama, & Inouye, 2004). Therefore, a mediating role of CAR in CLOCK/REV‐ERBα/β regulation of CYP2B10 cannot be excluded.
Of core clock genes, Npas2 and Bmal1 expressions were up‐regulated in Clock −/− mice (Figure S5), suggesting a potential compensatory genetic mechanism for Clock. This compensation may contribute to preserved rhythms of certain genes and proteins such as Cyp2a4/5 and Cyp2b10 in CLOCK mutants (Figure 1c,d). However, Clock and Bmal1 expressions were unaffected in Npas2 knockout mice (Figure S6). The data may suggest a relatively insignificant role for Npas2 in regulating circadian rhythms in the liver compared to Clock gene.
In summary, we identified the CLOCK protein as a key regulator of an array of CYP enzymes and of diurnal coumarin and cyclophosphamide detoxification. Mechanistic studies reveal that CLOCK transactivates CYP2A4/5 via directly binding to E‐boxes in gene promoters but represses CYP2B10 expression through REV‐ERBα/β. Our findings would facilitate the practice of chronotherapeutics with clinically used drugs such as cyclophosphamide.
AUTHOR CONTRIBUTIONS
M.Z. and B.W participated in the research design. M.Z., H.Z., J.D., and L.G. conducted the experiments. M.Z. and B.W. performed the data analysis. M.Z. and B.W. wrote or contributed to the writing of the manuscript.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for https://bpspubs.onlinelibrary.wiley.com/doi/full/10.1111/bph.14207, https://bpspubs.onlinelibrary.wiley.com/doi/full/10.1111/bph.14208, and https://bpspubs.onlinelibrary.wiley.com/doi/full/10.1111/bph.14206, and as recommended by funding agencies, publishers and other organisations engaged with supporting research.
Supporting information
Table S1. Primer sequences for quantitative real‐time PCR
Table S2. Oligonucleotide sequences for EMSA assays
Table S3. Primer sequences for ChIP assays.
Figure S1. Expression of Dbp in Clock+/+ and Clock−/− mice. Data are mean ± SD (n = 5). *p < 0.05 (two‐way ANOVA with Bonferroni post hoc test).
Figure S2. Hepatic mRNA and protein expression of CYP2A4/5 and CYP2B10 in female Clock+/+ and Clock−/− mice. Data are mean ± SD (n = 5). *p < 0.05 (two‐way ANOVA with Bonferroni post hoc test).
Figure S3. Circadian expression of CLOCK protein in mouse liver. Data are mean ± SD (n = 5). *p < 0.05 (one‐way ANOVA with Bonferroni post hoc test).
Figure S4. Expression of Car in Clock+/+ and Clock−/− mice. Data are mean ± SD (n = 5). *p < 0.05 (two‐way ANOVA with Bonferroni post hoc test).
Figure S5. Core clock genes expressions in Clock+/+ and Clock−/− mice. Data are mean ± SD (n = 5). *p < 0.05 (two‐way ANOVA with Bonferroni post hoc test).
Figure S6 Core clock genes expressions in Npas2+/+ and Npas2−/− mice. Data are mean ± SD (n = 5). *p < 0.05 (two‐way ANOVA with Bonferroni post hoc test).
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (Grants 81722049 and 81573488), the Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (Grant 2017BT01Y036), the Natural Science Foundation of Guangdong Province (Grant 2017A03031387), and the Guangzhou Science and Technology Project (Grant 201904010472).
Zhao M, Zhao H, Deng J, Guo L, Wu B. Role of the CLOCK protein in liver detoxification. Br J Pharmacol. 2019;176:4639–4652. 10.1111/bph.14828
REFERENCES
- Alexander, S. P. H. , Cidlowski, J. A. , Kelly, E. , Marrion, N. V. , Peters, J. A. , Faccenda, E. , … CGTP Collaborators (2017). The concise guide to PHARMACOLOGY 2017/18: Nuclear hormone receptors. British Journal of Pharmacology, 174, S208–S224. 10.1111/bph.13880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Marrion, N. V. , Peters, J. A. , Faccenda, E. , … CGTP Collaborators (2017). The concise guide to PHARMACOLOGY 2017/18: Enzymes. British Journal of Pharmacology, 174, S272–S359. 10.1111/bph.13877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Kelly, E. , Marrion, N. V. , Peters, J. A. , Faccenda, E. , Harding, S. D. , … CGTP Collaborators (2017). THE CONCISE GUIDE TO PHARMACOLOGY 2017/18: Transporters. British Journal of Pharmacology, 174, S360–S446. 10.1111/bph.13883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Roberts, R. E. , Broughton, B. R. S. , Sobey, C. G. , George, C. H. , Stanford, S. C. , … Ahluwalia, A. (2018). Goals and practicalities of immunoblotting and immunohistochemistry: A guide for submission to the British Journal of Pharmacology. British Journal of Pharmacology, 175(3), 407–411. 10.1111/bph.14112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoch, M. P. , Song, E. J. , Chang, A. M. , Vitaterna, M. H. , Zhao, Y. , Wilsbacher, L. D. , … Takahashi, J. S. (1997). Functional identification of the mouse diurnal Clock gene by transgenic BAC rescue. Cell, 89, 655–667. 10.1016/S0092-8674(00)80246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batist, G. , Ramakrishnan, G. , Rao, C. S. , Chandrasekharan, A. , Gutheil, J. , Guthrie, T. , … Lee, L. W. (2001). Reduced cardiotoxicity and preserved antitumor efficacy of liposome‐encapsulated doxorubicin and cyclophosphamide compared with conventional doxorubicin and cyclophosphamide in a randomized, multicenter trial of metastatic breast cancer. Journal of Clinical Oncology, 19, 1444–1454. 10.1200/JCO.2001.19.5.1444 [DOI] [PubMed] [Google Scholar]
- Bozek, K. , Relógio, A. , Kielbasa, S. M. , Heine, M. , Dame, C. , Kramer, A. , & Herzel, H. (2009). Regulation of clock‐controlled genes in mammals. PLoS ONE, 4, e4882 10.1371/journal.pone.0004882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge, A. , Feng, D. , Everett, L. J. , Briggs, E. R. , Mullican, S. E. , Wang, F. , … Lazar, M. A. (2012). Rev‐erbα and Rev‐erbβ coordinately protect the diurnal clock and normal metabolic function. Genes & Development, 26, 657–667. 10.1101/gad.186858.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H. , Zhao, X. , Hatori, M. , Yu, R. T. , Barish, G. D. , Lam, M. T. , … Evans, R. M. (2012). Regulation of circadian behaviour and metabolism by REV‐ERB‐α and REV‐ERB‐β. Nature, 485, 123–127. 10.1038/nature11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudel, T. , Cretenet, G. , Saumet, A. , & Gachon, F. (2007). Crosstalk between xenobiotics metabolism and diurnal clock. FEBS Letters, 581, 3626–3633. 10.1016/j.febslet.2007.04.009 [DOI] [PubMed] [Google Scholar]
- Cretenet, G. , Le Clech, M. , & Gachon, F. (2010). Diurnal clock‐coordinated 12 hr period rhythmic activation of the IRE1α pathway controls lipid metabolism in mouse liver. Cell Metabolism, 11, 47–57. 10.1016/j.cmet.2009.11.002 [DOI] [PubMed] [Google Scholar]
- Curtis, M. J. , Alexander, S. , Cirino, G. , Docherty, J. R. , George, C. H. , Giembycz, M. A. , … Ahluwalia, A. (2018). Experimental design and analysis and their reporting II: Updated and simplified guidance for authors and peer reviewers. British Journal of Pharmacology, 175(7), 987–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruyne, J. P. , Weaver, D. R. , & Reppert, S. M. (2007). CLOCK and NPAS2 have overlapping roles in the suprachiasmatic diurnal clock. Nature Neuroscience, 10, 543–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, J. , Guo, L. , & Wu, B. (2018). Diurnal regulation of hepatic cytochrome P450 2a5 by peroxisome proliferator‐activated receptor γ. Drug Metabolism and Disposition, 46, 1538–1545. [DOI] [PubMed] [Google Scholar]
- Dibner, C. , Schibler, U. , & Albrecht, U. (2010). The mammalian diurnal timing system: Organization and coordination of central and peripheral clocks. Annual Review of Physiology, 72, 517–549. [DOI] [PubMed] [Google Scholar]
- Dudley, C. A. , Erbel‐Sieler, C. , Estill, S. J. , Reick, M. , Franken, P. , Pitts, S. , & McKnight, S. L. (2003). Altered patterns of sleep and behavioral adaptability in NPAS2‐deficient mice. Science, 301, 379–383. [DOI] [PubMed] [Google Scholar]
- Duez, H. , & Staels, B. (2008). Rev‐erbα gives a time cue to metabolism. FEBS Letters, 582, 19–25. [DOI] [PubMed] [Google Scholar]
- Ercolani, L. , Ferrari, A. , De Mei, C. , Parodi, C. , Wade, M. , & Grimaldi, B. (2015). Diurnal clock: Time for novel anticancer strategies? Pharmacological Research, 100, 288–295. [DOI] [PubMed] [Google Scholar]
- Evans, W. E. , & Relling, M. V. (1999). Pharmacogenomics: Translating functional genomics into rational therapeutics. Science, 286, 487–491. [DOI] [PubMed] [Google Scholar]
- Feillet, C. , Krusche, P. , Tamanini, F. , Janssens, R. C. , Downey, M. J. , Martin, P. , … Rand, D. A. (2014). Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle. Proceedings of the National Academy of Sciences of the United States of America, 111, 9828–9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon, F. , Olela, F. F. , Schaad, O. , Descombes, P. , & Schibler, U. (2006). The diurnal PAR‐domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metabolism, 4, 25–36. [DOI] [PubMed] [Google Scholar]
- Gale, J. E. , Cox, H. I. , Qian, J. , Block, G. D. , Colwell, C. S. , & Matveyenko, A. V. (2011). Disruption of diurnal rhythms accelerates development of diabetes through pancreatic beta‐cell loss and dysfunction. Journal of Biological Rhythms, 26, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs, J. E. , Blaikley, J. , Beesley, S. , Matthews, L. , Simpson, K. D. , Boyce, S. H. , … Loudon, A. S. (2012). The nuclear receptor REV‐ERBα mediates diurnal regulation of innate immunity through selective regulation of inflammatory cytokines. Proceedings of the National Academy of Sciences of the United States of America, 109, 582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbacheva, V. Y. , Kondratov, R. V. , Zhang, R. , Cherukuri, S. , Gudkov, A. V. , Takahashi, J. S. , & Antoch, M. P. (2005). Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proceedings of the National Academy of Sciences of the United States of America, 102, 3407–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi, E. , Shokrzadeh, M. , Chabra, A. , Naghshvar, F. , Keshavarz‐Maleki, R. , & Ahmadi, A. (2015). Protective effects of Origanum vulgare ethanol extract against cyclophosphamide‐induced liver toxicity in mice. Pharmaceutical Biology, 53(1), 10–15. [DOI] [PubMed] [Google Scholar]
- Harding, S. D. , Sharman, J. L. , Faccenda, E. , Southan, C. , Pawson, A. J. , Ireland, S. , … NC‐IUPHAR (2018). The IUPHAR/BPS guide to PHARMACOLOGY in 2018: Updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Research, 46, D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X. Y. , Tang, L. , Wang, S. L. , Cai, Q. S. , Wang, J. S. , & Hong, J. Y. (2006). Efficient activation of aflatoxin B1 by cytochrome P450 2A13, an enzyme predominantly expressed in human respiratory tract. International Journal of Cancer, 118, 2665–2671. [DOI] [PubMed] [Google Scholar]
- Hogenesch, J. B. , Gu, Y. Z. , Jain, S. , & Bradfield, C. A. (1998). The basic‐helix–loop–helix‐PAS orphan MOP3 forms transcriptionally active complexes with diurnal and hypoxia factors. Proceedings of the National Academy of Sciences of the United States of America, 95, 5474–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycay, E. G. , & Bandiera, S. M. (2009). Expression, function and regulation of mouse cytochrome P450 enzymes: Comparison with human cytochrome P450 enzymes. Current Drug Metabolism, 10, 1151–1183. [DOI] [PubMed] [Google Scholar]
- Kanno, Y. , Otsuka, S. , Hiromasa, T. , Nakahama, T. , & Inouye, Y. (2004). Diurnal difference in CAR mRNA expression. Nuclear Receptor, 2, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating, M. J. , O'Brien, S. , Albitar, M. , Lerner, S. , Plunkett, W. , Giles, F. , … Kantarjian, H. (2005). Early results of a chemoimmunotherapy regimen of fludarabine, cyclophosphamide, and rituximab as initial therapy for chronic lymphocytic leukemia. Journal of Clinical Oncology, 23, 4079–4088. [DOI] [PubMed] [Google Scholar]
- Kilkenny, C. , Browne, W. , Cuthill, I. C. , Emerson, M. , & Altman, D. G. (2010). Animal research: Reporting in vivo experiments: The ARRIVE guidelines. British Journal of Pharmacology, 160, 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, D. P. , Zhao, Y. , Sangoram, A. M. , Wilsbacher, L. D. , Tanaka, M. , Antoch, M. P. , … Takahashi, J. S. (1997). Positional cloning of the mouse diurnal clock gene. Cell, 89, 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake, B. G. (1999). Coumarin metabolism, toxicity and carcinogenicity: Relevance for human risk assessment. Food and Chemical Toxicology, 37, 423–453. [DOI] [PubMed] [Google Scholar]
- Lavery, D. J. , Lopez‐Molina, L. , Margueron, R. , Fleury‐Olela, F. , Conquet, F. , Schibler, U. , & Bonfils, C. (1999). Circadian Expression of the Steroid 15 α‐Hydroxylase (Cyp2a4) and Coumarin 7‐Hydroxylase (Cyp2a5) Genes in Mouse Liver Is Regulated by the PAR Leucine Zipper Transcription Factor DBP. Molecular and Cellular Biology, 19(10), 6488–6499. 10.1128/mcb.19.10.6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévi, F. , & Okyar, A. (2011). Diurnal clocks and drug delivery systems: Impact and opportunities in chronotherapeutics. Expert Opinion on Drug Delivery, 8, 1535–1541. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Sun, H. , Lu, D. , Zhang, Y. , Zhang, X. , Ma, Z. , & Wu, B. (2014). Identification of glucuronidation and biliary excretion as the main mechanisms for gossypol clearance: in vivo and in vitro evidence. Xenobiotica, 44, 696–707. [DOI] [PubMed] [Google Scholar]
- Lu, D. , Wang, S. , Xie, Q. , Guo, L. , & Wu, B. (2017). Transcriptional regulation of human UDP‐glucuronosyltransferase 2B10 by farnesoid X receptor in human hepatoma HepG2 cells. Molecular Pharmaceutics, 14, 2899–2907. [DOI] [PubMed] [Google Scholar]
- Martino, T. A. , Oudit, G. Y. , Herzenberg, A. M. , Tata, N. , Koletar, M. M. , Kabir, G. M. , … Sole, M. J. (2008). Diurnal rhythm disorganization produces profound cardiovascular and renal disease in hamsters. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology, 294, R1675–R1683. 10.1152/ajpregu.00829.2007 [DOI] [PubMed] [Google Scholar]
- McNamara, P. , Seo, S. , Rudic, R. D. , Sehgal, A. , Chakravarti, D. , & FitzGerald, G. A. (2001). Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: A humoral mechanism to reset a peripheral clock. Cell, 105, 877–889. [DOI] [PubMed] [Google Scholar]
- Murakami, Y. , Higashi, Y. , Matsunaga, N. , Koyanagi, S. , & Ohdo, S. (2008). Diurnal clock‐controlled intestinal expression of the multidrug‐resistance gene mdr1a in mice. Gastroenterology, 135, 1636–1644. [DOI] [PubMed] [Google Scholar]
- Nebert, D. W. , & Dalton, T. P. (2006). The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nature Reviews. Cancer, 6, 947–960. [DOI] [PubMed] [Google Scholar]
- Nelson, D. R. , Zeldin, D. C. , Hoffman, S. M. , Maltais, L. J. , Wain, H. M. , & Nebert, D. W. (2004). Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative‐splice variants. Pharmacogenetics, 14, 1–18. [DOI] [PubMed] [Google Scholar]
- Oishi, K. , Miyazaki, K. , Kadota, K. , Kikuno, R. , Nagase, T. , Atsumi, G. , … Ishida, N. (2003). Genome‐wide expression analysis of mouse liver reveals CLOCK‐regulated diurnal output genes. The Journal of Biological Chemistry, 278, 41519–41527. [DOI] [PubMed] [Google Scholar]
- Oishi, K. , Shirai, H. , & Ishida, N. (2005). CLOCK is involved in the diurnal transactivation of peroxisome‐proliferator‐activated receptor α (PPARα) in mice. The Biochemical Journal, 386, 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pass, G. J. , Carrie, D. , Boylan, M. , Lorimore, S. , Wright, E. , Houston, B. , … Wolf, C. R. (2005). Role of hepatic cytochrome p450s in the pharmacokinetics and toxicity of cyclophosphamide: Studies with the hepatic cytochrome p450 reductase null mouse. Cancer Research, 65, 4211–4217. [DOI] [PubMed] [Google Scholar]
- Raghuram, S. , Stayrook, K. R. , Huang, P. , Rogers, P. M. , Nosie, A. K. , McClure, D. B. , … Rastinejad, F. (2007). Identification of heme as the ligand for the orphan nuclear receptors REV‐ERBα and REV‐ERBβ. Nature Structural & Molecular Biology, 14, 1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reick, M. , Garcia, J. A. , Dudley, C. , & McKnight, S. L. (2001). NPAS2: An analog of clock operative in the mammalian forebrain. Science, 293, 506–509. [DOI] [PubMed] [Google Scholar]
- Reppert, S. M. , & Weaver, D. R. (2002). Coordination of diurnal timing in mammals. Nature, 418, 935–941. [DOI] [PubMed] [Google Scholar]
- Ripperger, J. A. , Shearman, L. P. , Reppert, S. M. , & Schibler, U. (2000). CLOCK, an essential pacemaker component, controls expression of the diurnal transcription factor DBP. Genes & Development, 14, 679–689. [PMC free article] [PubMed] [Google Scholar]
- Sadagopan, N. , Cohen, L. , Roberts, B. , Collard, W. , & Omer, C. (2001). Liquid chromatography–tandem mass spectrometric quantitation of cyclophosphamide and its hydroxy metabolite in plasma and tissue for determination of tissue distribution. Journal of Chromatography. B, Biomedical Sciences and Applications, 759, 277–284. [DOI] [PubMed] [Google Scholar]
- Salgado‐Delgado, R. , Tapia Osorio, A. , Saderi, N. , & Escobar, C. (2011). Disruption of diurnal rhythms: A crucial factor in the etiology of depression. Depression Research and Treatment, 2011, 839743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvidis, C. , & Koutsilieris, M. (2012). Diurnal rhythm disruption in cancer biology. Molecular Medicine, 18, 1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheweita, S. A. , El‐Hosseiny, L. S. , & Nashashibi, M. A. (2016). Protective effects of essential oils as natural antioxidants against hepatotoxicity induced by cyclophosphamide in mice. PLoS ONE, 11(11), e0165667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt, L. A. , Kojetin, D. J. , & Burris, T. P. (2011). The REV‐ERBs and RORs: Molecular links between diurnal rhythms and lipid homeostasis. Future Medicinal Chemistry, 3, 623–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler, M. L. , Kuropatwinski, K. K. , Comas, M. , Gasparian, A. V. , Fedtsova, N. , Gleiberman, A. S. , … Antoch, M. P. (2012). Core diurnal protein CLOCK is a positive regulator of NF‐κB–mediated transcription. Proceedings of the National Academy of Sciences of the United States of America, 109, E2457–E2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triqueneaux, G. , Thenot, S. , Kakizawa, T. , Antoch, M. P. , Safi, R. , Takahashi, J. S. , … Laudet, V. (2004). The orphan receptor Rev‐erbα gene is a target of the diurnal clock pacemaker. Journal of Molecular Endocrinology, 33, 585–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna, M. H. , King, D. P. , Chang, A. M. , Kornhauser, J. M. , Lowrey, P. L. , McDonald, J. D. , … Takahashi, J. S. (1994). Mutagenesis and mapping of a mouse gene, Clock, essential for diurnal behavior. Science, 264, 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , & Lazar, M. A. (2008). Bifunctional role of Rev‐erbα in adipocyte differentiation. Molecular and Cellular Biology, 28, 2213–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Lin, Y. , Yuan, X. , Li, F. , Guo, L. , & Wu, B. (2018). REV‐ERBα integrates colon clock with experimental colitis through regulation of NF‐κB/NLRP3 axis. Nature Communications, 9, 4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C. , Li, C. Y. , & Kong, A. N. (2005). Induction of phase I, II and III drug metabolism/transport by xenobiotics. Archives of Pharmacal Research, 28, 249–268. [DOI] [PubMed] [Google Scholar]
- Yin, L. , Wu, N. , Curtin, J. C. , Qatanani, M. , Szwergold, N. R. , Reid, R. A. , … Lazar, M. A. (2007). Rev‐erbα, a heme sensor that coordinates metabolic and diurnal pathways. Science, 318, 1786–1789. [DOI] [PubMed] [Google Scholar]
- Yoshitane, H. , Ozaki, H. , Terajima, H. , Du, N. H. , Suzuki, Y. , Fujimori, T. , … Fukada, Y. (2014). CLOCK‐controlled polyphonic regulations of diurnal rhythms through canonical and non‐canonical E‐boxes. Molecular and Cellular Biology, 34, 1776–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Huang, W. , Chua, S. S. , Wei, P. , & Moore, D. D. (2002). Modulation of acetaminophen‐induced hepatotoxicity by the xenobiotic receptor CAR. Science, 298, 422–424. [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Yu, F. , Guo, L. , Chen, M. , Yuan, X. , & Wu, B. (2018). Small heterodimer partner regulates diurnal cytochromes p450 and drug‐induced hepatotoxicity. Theranostics, 8, 5246–5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M. , Zhang, T. , Yu, F. , Guo, L. , & Wu, B. (2018). E4bp4 regulates carboxylesterase 2 enzymes through repression of the nuclear receptor Rev‐erbα in mice. Biochemical Pharmacology, 152, 293–301. [DOI] [PubMed] [Google Scholar]
- Zmrzljak, U. P. , & Rozman, D. (2012). Diurnal regulation of the hepatic endobiotic and xenobiotic detoxification pathways: The time matters. Chemical Research in Toxicology, 25, 811–824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer sequences for quantitative real‐time PCR
Table S2. Oligonucleotide sequences for EMSA assays
Table S3. Primer sequences for ChIP assays.
Figure S1. Expression of Dbp in Clock+/+ and Clock−/− mice. Data are mean ± SD (n = 5). *p < 0.05 (two‐way ANOVA with Bonferroni post hoc test).
Figure S2. Hepatic mRNA and protein expression of CYP2A4/5 and CYP2B10 in female Clock+/+ and Clock−/− mice. Data are mean ± SD (n = 5). *p < 0.05 (two‐way ANOVA with Bonferroni post hoc test).
Figure S3. Circadian expression of CLOCK protein in mouse liver. Data are mean ± SD (n = 5). *p < 0.05 (one‐way ANOVA with Bonferroni post hoc test).
Figure S4. Expression of Car in Clock+/+ and Clock−/− mice. Data are mean ± SD (n = 5). *p < 0.05 (two‐way ANOVA with Bonferroni post hoc test).
Figure S5. Core clock genes expressions in Clock+/+ and Clock−/− mice. Data are mean ± SD (n = 5). *p < 0.05 (two‐way ANOVA with Bonferroni post hoc test).
Figure S6 Core clock genes expressions in Npas2+/+ and Npas2−/− mice. Data are mean ± SD (n = 5). *p < 0.05 (two‐way ANOVA with Bonferroni post hoc test).


