Abstract
Gastrointestinal colonisation by commensal microbiota is essential for the health and well-being of the host. We aimed to evaluate the influence of a reduced bacterial load environment on microbiota development and maturation, and the possibility of targeted colonisation via at-hatch administration of a selected bacterial strain. Japanese quail (Coturnix japonica) were inoculated within 1 h of hatch with a freshly grown culture of a Lactobacillus agilis isolate derived from a healthy adult quail. Hatchlings were kept in a mouse isolator for one week and then housed between one and four weeks of age, with a flock of normally grown adult quail to expose the bacteria-restricted birds to normal commensal quail bacteria. The bacterial isolate used to inoculate the birds was found to completely dominate the microbiota of the intestine of L.agilis at-hatch inoculated birds. Despite 3 weeks of co-housing of the test birds with an adult flock harbouring normal rich gut microbiota, neither the Lactobacillus inoculated nor PBS inoculated birds reached the level of bacterial diversity seen in birds raised under normal conditions. Neither PBS nor Lactobacillus inoculated birds were able to adopt normal quail microbiota after one week of restricted exposure to bacteria, indicating that contact with diverse microbiota during the early days of gut development in birds is critical for the establishment of healthy intestinal community. Very early intervention in the form of a suitable bacterial probiotic inoculant immediately post-hatch protected birds grown in extreme hygiene conditions from developing anomalous gut microbiota and intestinal damage. Our data shows that it is possible to induce dominance of desired strain using simple timed manipulation.
Keywords: Biotechnology, Microbiology, Animal science, Gastrointestinal system, Infectious disease, Probiotic administration, Microbiota manipulation, Excessive hygiene
Biotechnology; Microbiology; Animal science; Gastrointestinal system; Infectious disease; Probiotic administration; Microbiota manipulation; Excessive hygiene.
1. Introduction
Development of healthy gastrointestinal tract (GIT) commensal microbiota is essential for the homeostasis, health, and wellbeing of the host, and has been shown to be of pronounced biological importance for an organism. The commensal GIT microbiota plays a protective role by excluding harmful pathogen colonisation and will actively resist change by producing antibacterial molecules [1]. The first exposure of microbiota has been shown to be essential for the establishment and regulation of both adaptive and innate immunity [2] as well as for the optimum development and the morphology of the intestinal tract [3] as the host matures. These observations have been elucidated through experiments using germ-free animals [4].
The internal component of avian eggs were previously thought to be near-sterile at hatch [5]. In mammals, inoculation with the first gut microbiota occurs during passage through the birth canal. This journey exposes the newborn to both vaginal and faecal microbes during the birthing process [6]. In birds, however, it is thought that with naturally brooded eggs the hatching chicks get their first inoculum through microbes inhabiting the eggshell and nesting material and exposure to adult birds [5, 7, 8]. In some birds the hatchlings are also inoculated by vertical transmission of bacteria through regurgitated food [9, 10]. Modern poultry hatcheries commonly disinfect and fumigate eggs prior to hatching and the hatchlings do not have any maternal contact. This often results in highly variable microbiota recruitment from environmental sources within the hatchery, bird transport system, and initial placement environment, rather than microbial community adopted directly from adult birds [11]. Similar disturbance of the human infant's microbiota is recorded during birth via Caesarean section where, despite the mothers' continual presence, the lack of usual vaginal inoculation can have permanent consequences on the gut development and overall health [12]. Caesarean section birth has been linked to susceptibility to pathogens and skin conditions [13], allergies and asthma [14, 15]. Thus the first microbial inoculum and the first days of the formation of the gut microbiota community is a time of great vulnerability and high health risk.
In humans, after an initial period of dynamic microbiota formation, the microbial gut community assumes a more fixed and stable state with an ability to resist change [16, 17], even as severe as the administration of high doses of antibiotics [18]. The formation of stable mature GIT microbiota takes approximately 3–5 years [19]. In contrast, the bird gut matures faster and the chicken microbiota becomes stable and diverse around the first or second week post-hatch [20]. This indicates that in birds the window to manipulate gut microbiota is very short and that administration of probiotics to the mature microbiota birds will likely have little success and limited consequence for bird health. The first bacterial colonisers in the GIT shape microbiota for life. Thus, the time before microbiota maturation is crucial for health but also represents the window within which controlled microbiota manipulation may be most readily achieved.
Being of small size, the Japanese quail (Coturnix japonica) are versatile as a laboratory animal; they can be housed in existing specialised equipment, such as mouse or rat isolators, and adapt to a wide range of husbandry conditions. For example, unlike in the painstaking process of generating germ-free mice, where the pups have to be delivered by Caesarean section under sterile conditions and antibiotic usage, quail eggs are easily disinfected to achieve near germ-free conditions and hatchlings can immediately feed themselves, hence do not require nursing or parenting. As the GIT of birds hatched under near sterile conditions carry very little or no bacterial load, the Japanese quail presents an ideal model to examine the dynamics of bacterial colonisation under very clean conditions.
We hypothesised that the first inoculum with a suitable bacterial isolate would establish within the quail GIT if administered at hatch and that the restriction of exposure to normal quail microbiota during the early post-hatch period would result in a low diversity, anomalous, microbial community that would stabilise and resist further colonisation. Our main aim was to determine the importance of the first week of life in microbiota development and assess what consequences this may have for classic probiotic administration, later in life, or any other intervention targeting gut microbiota after maturation and stabilisation of microbiota composition. An appropriate inoculum would be expected to stimulate optimal intestinal and immune system development, enhance bird health, and decrease mortality. Lactobacillus species are considered an important component of the healthy poultry GIT microbiota and some strains have been reported to improve weight gain in chickens [21]. Here we demonstrate that 1) restricting access to other bacteria during the first week post-hatch can produce disproportionate and permanent colonisation with desired inoculated strain; 2) birds kept in low bacterial load conditions during the first week of life do not recover from low diversity when reintroduced to normal microbiota; and 3) a prolonged low bacterial load in the immediate environment can result in visible anomalies in gut morphology and microbiota.
2. Methods
2.1. Isolation of native Lactobacillus from quail
Lactobacillus sp. were cultured from ileum contents of Japanese quail. The birds that were sampled for the strain isolation were from a commercial facility in which the birds had never been exposed to antibiotics or egg disinfection procedures. The objective was to isolate and use a strain that is well adapted to survival within the quail GIT. Diluted ileal content was plated and grown on De Man, Ragosa and Sharpe (MRS) agar plates under aerobic conditions. Individual colonies were picked and streaked on MRS to ensure a clonal population. Bacterial DNA was isolated and the 16S rRNA gene was PCR amplified and sequenced. The most abundant ileal isolate based on 100% sequence identity of 16S rRNA gene sequence was selected for use in the quail inoculation experiment. The isolate was most similar to Lactobacillus agilis strain BF-26 16S ribosomal RNA gene, partial sequence (100% ID) using blastn. Since 16S analysis on its own is not sufficient to reliably infer species level, and a range of molecular, microbiological and metabolomics techniques are required in addition to 16S sequence, we are referring to the strain we used as provisional quail L. agilis isolate. The amplified 16S rRNA gene sequence for this isolate was TAGGGAATCTTCCACAATGGGCGCAAGCCTGATGGAGCAACGCCGCGTGAGTGAAGAAGGTCTTCGGATCGTAAAACTCTGTTGTTAGAGAAGAACATGCAGGAGAGTAACTGTTCTTGTATTGACGGTATCTAACCAGAAAGCCACGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGTGGCAAGCGTTGTCCGGATTTATTGGGCGTAAAGGGAACGCAGGCGGTCCTTTAAGTCTGATGTGAAAGCCTTCGGCTTAACCGAAGAATTGCATTGGAAACTGGAGGACTTGAGTGCAGAAGAGGAGAGTGGAACTCCATGTGTAGCGGTGAAATGCGTAGATATATGGAAGAACACCAGTGGCGAAAGCGGCTCTCTGGTCTGTAACTGACGCTGAGGTTCGAAAGTGTGGGTAGCAAACAGG.
2.2. Animal trial
Fertilised Japanese quail eggs were sourced from Banyard Game Birds, Toowoomba, Queensland, an organic supplier that has practised relatively natural husbandry and hatchery operations for decades, without the use of antibiotics, growth promoters, or egg disinfection and fumigation procedures.
A clean hatching room within the laboratory facility was prepared by washing the walls, door, floor, ceiling, window and electrical sockets with bleach diluted in water 4 fold, and commercial disinfectants. The incubator (IM70, WA Poultry Equipment) used had a fully controlled, external accessed, humidity chamber. The incubator was placed inside a class 2 biosafety cabinet and both were thoroughly washed inside and out with 70% ethanol. The quail eggs were wiped with 70% ethanol and placed under UV in the laminar flow for 5 min inside a rolling tray that rotated eggs to ensure equal surface exposure to the UV light. The eggs were then loaded into the incubator and the whole room was fumigated using a ClO2 gas steriliser (#HAD-209A, Greewin) with the door of the incubator left open to allow fumigation of the eggs. After the fumigation process the incubator was closed and the incubation process started.
At day 15, the rolling of eggs was stopped and the birds hatched between days 17–19. Chicks were given time to dry in the incubator after hatch (~1 h) and were then orally gavaged with the phosphate-buffered saline (PBS) suspended Lactobacillus agilis inoculum (0.5 ml, OD600 = 2) or with PBS only. The two groups of birds were then transferred to sterilised mouse isolator cages under HEPA filtered sterile airflow (GM500, Tecniplast) with one bird per cage and total of 5 birds for the PBS control and 6 birds for the inoculated group. The temperature in the mouse isolator was maintained at 30 °C. Brooding lamps were used for the first 3 days. Sterilised food and water were supplied ad libitum. Each day, to minimise the exposure of birds to faecal material, the birds were moved into fresh autoclaved cages with a supply of sterile food and water sufficient for one day.
On day 7 chicks were moved from the clean room isolator into a poultry room and housed with the mature quail (>1 year old) from the same quail parental line, that had been raised under conventional conditions. To avoid issues with pecking order the experimental birds were in conjoined cages, but physically separated from the mature conventional birds. From day 7, food and water were supplied ad libitum but did not undergo any sterilisation procedures. Feed supplied was a commercial turkey starter (Barastoc) (22% protein, 2.5% fat, 5% fibre, 0.3% salt, 1% calcium, 8 mg/kg copper and 0.3 mg/kg selenium) [22].
We started the experiment with 15 eggs per treatment, aiming to get n = 10 birds. However, following surface cleaning and fumigation of the eggs, there was low hatchability rate and during the trial 2 birds were lost from each group. These bird mortalities occurred after the birds left the clean environment and were placed in the room with conventionally raised quail. The final microbiota analysis was carried out on three PBS gavaged birds and four birds that had been inoculated with provisional L. agilis. Due to technical issues with maintaining germ-free conditions and a limited number of cages available in isolators, germ-free and gnotobiotic trials in medical research are often performed on the similar or lower number of mice [23].
Faecal samples were collected from the cage at day 0, 3, 5, 7, 14, 21 and 28 prior to experiment termination. All birds were euthanised by intraperitoneal injection of pentobarbital sodium. Contents of duodenum, ileum and caecum were collected for analysis at day 28 (4 weeks of age) and stored at -80 °C.
2.3. Sample collection and microscopy
Samples for histological investigation were collected from duodenum, ileum, and caecum. All samples were obtained within 1 h of dissection and washed in phosphate-buffered saline and stored in 10% buffered neutral formalin at 20 °C until processed. For light microscopy, stored tissues were cut to appropriate dimensions, placed in tissue cassettes and processed in an automated processing device overnight (Tissue-Tek V.I.P. Tissue processor). Tissues were further processed by embedding in paraffin blocks for storage and sectioning. Selected samples were prepared using a Leica (RM 2125 RTS) microtome at 5um and Haematoxylin-Eosin stained. Slides were imaged at the TRI Microscopy core facility (Brisbane) using a Nikon Brightfield, Olympus VS120 slide scanner and analysed using ImageJ and Olympus microscopy software, Olivia.
2.4. Microbiota and analysis
DNA from the faecal samples was extracted using a Bioline Isolate Fecal DNA kit, cat.no#BIO-52082. Primers were selected to amplify the V3–V4 region of 16S rRNA genes: forward ACTCCTACGGGAGGCAGCAG and reverse GGACTACHVGGGTWTCTAAT, and also contained barcodes, spacer and Illumina sequencing linker sequences as detailed previously [24]. Sequencing was performed on the Illumina MiSeq platform using 2x300 bp paired-end sequencing. A total of 87 microbiota samples were sequenced and used in the analysis.
Analysis of microbial communities was completed using QIIME v.1.9.1 [25] and QIIME default parameters unless stated otherwise. Operational Taxonomic Units (OTUs) were clustered at 97% similarity using UCLUST [26], Pintail was used to inspect for chimeric sequences [27] and taxonomic assignments were performed against the GreenGenes database [28]. OTUs with relative abundance of less than 0.01% were removed from the subsequent analysis. The complete annotated sequence dataset is publically available at the MG-RAST database under ID mgm4663621.3. The data were visualised and further analysed using Calypso (http://bioinfo.qimr.edu.au/calypso/) [29].
3. Ethics statement
Animal ethics approvals for both sampling the birds for culturing of isolates and for the animal trial were obtained from the Animal Ethics Committee of Central Queensland University under project IDs A14/03-309 and A14/11-324.
4. Results
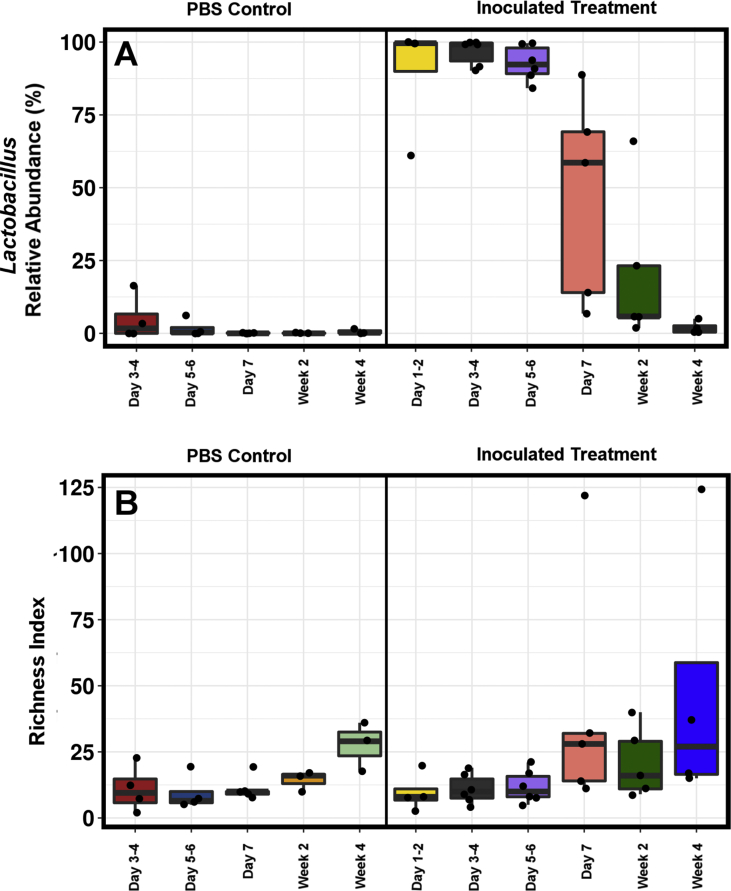
There were no significant differences in growth and weights between control and inoculated groups at any stage of the experiment with birds reaching mean weight of 192 g at 4 weeks of age. The Lactobacillus isolate chosen to inoculate the birds was identified by 16S rRNA gene sequencing as most similar to L. agilis (100%). The orally inoculated isolate had almost no competition from other Lactobacillus species originating from the cohoused conventionally raised quail stock, at necropsy 99.38% of all Lactobacillus sequences in the dataset were assigned by QIIME and confirmed with blastn as 100% identical to the inoculated L. agilis isolate. Thus, effectively no other Lactobacillus species were able to colonise to any significant level after the birds were moved from the clean environment to the poultry room. The Lactobacillus isolate was able to colonise the inoculated birds to very high relative abundance from day 1 (Figure 1A). Lactobacillus comprised 100% abundance in several of the inoculated birds’ faecal samples from day 1 to day 4, remaining high during the clean room isolation week. The faecal samples from sterile-PBS inoculated control birds, days 0–2, were too low in bacterial DNA to be successfully sequenced. Moreover, enumerating the bacterial load using classic microscopy and haemocytometer revealed low bacterial density in control samples. The Lactobacillus inoculated birds were clearly dominated with Lactobacillus during the time they spent in the clean isolator, but less so after 3 weeks of co-housing with the conventionally raised mature quail flock.
Figure 1.
Relative abundance (%) of the genus Lactobacillus (A) was significantly different between the groups (P = 1.4e−15). 99.38% of all Lactobacillus sequences were assigned to L. agilis. There were no significant differences in Richness Index (B) between the groups. Both control and inoculated birds showed low faecal richness and diversity.
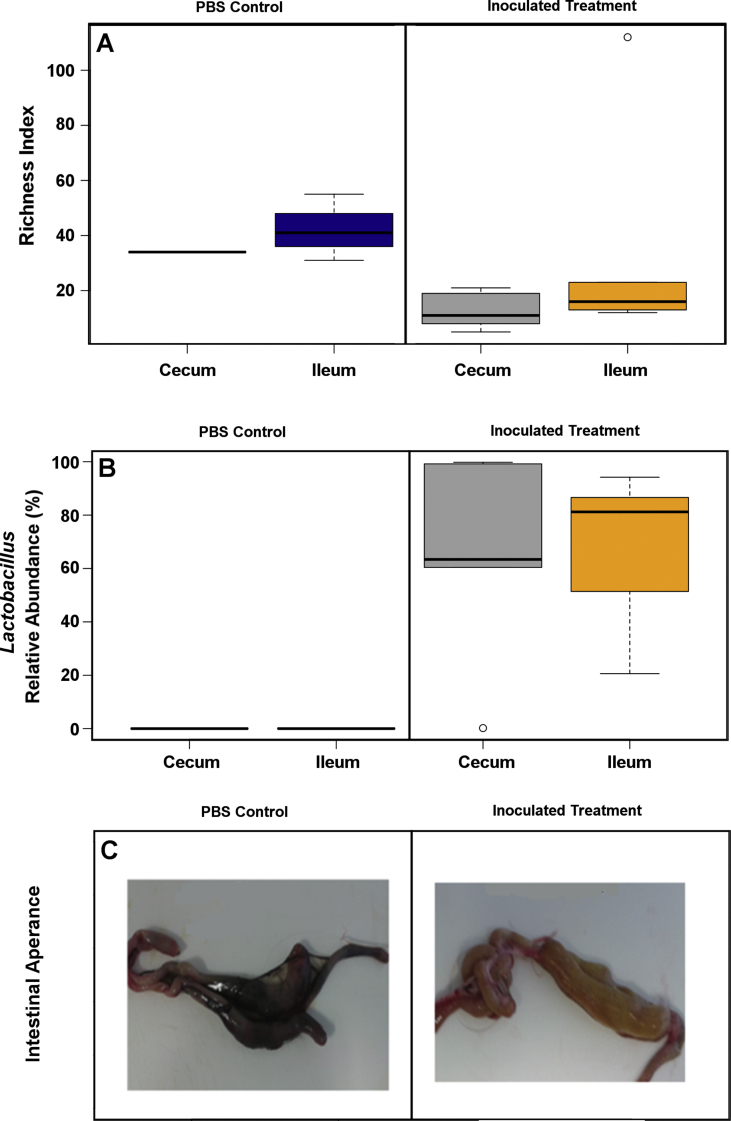
Both the Lactobacillus inoculated and PBS gavaged bird groups had low diversity (Figure 1B) of fecal microbiota than conventionally raised birds. The total number of OTUs in the excreta of these birds was 20–30 for PBS inoculated birds (Figure 1B) with surprisingly even less OTUs in the cecum (Figure 2). There were no significant differences in faecal microbiota richness between PBS gavaged and Lactobacillus inoculated groups by either Chao1 or Richness indices. The PBS treated birds had approximately triple the median Richness at week 4 in both caecum and ileum, compared to the Lactobacillus treated birds (Figure 2A). Lactobacillus strongly dominated both ileum and caecum in the Lactobacillus inoculated birds at 4 weeks of age and this apparently prevented new colonisation from environmental sources in the cohousing arrangement with the conventionally reared birds. No trace of Lactobacillus was detected in the ileum or caecum of the PBS treated birds (Figure 2B), indicating that early exposure is essential for healthy bacterial colonisation of ileum and cecum. This extreme difference in ileum and cecum was also evident in the gut appearance (Figure 2C), with strong and consistent difference in gut colour and morphology, as well as in the appearance of intestinal content, that was almost black for the PBS treated birds despite eating the same feed.
Figure 2.
The caecum and ileum of PBS treated birds showed higher Richness Index than Lactobacillus inoculated birds (A) likely due to the strong dominance of Lactobacillus shown in panel B in both caeca and ileum of inoculated birds (P=0.026). The different treatments affected the appearance of the intestinal content (C) and influenced gut morphology (C).
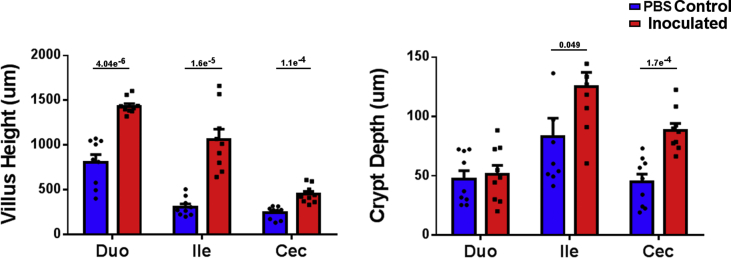
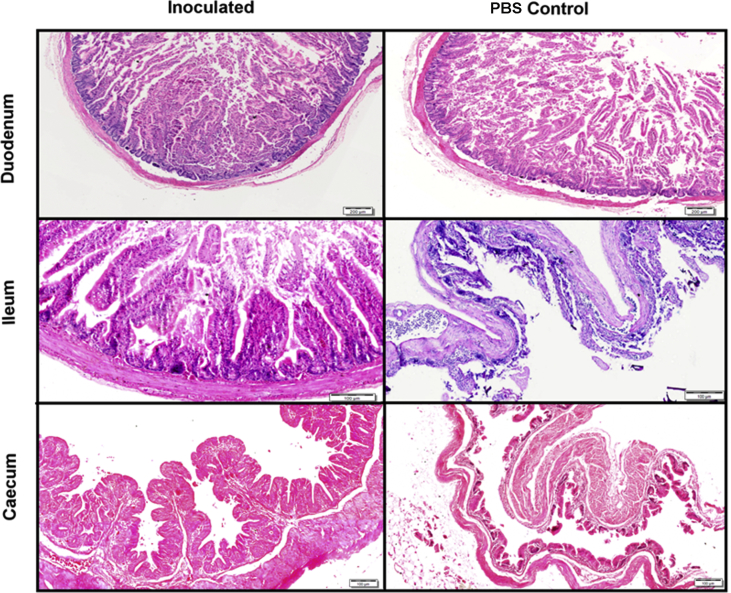
Histological analysis revealed significant differences in villus height and crypt depth (Figure 3) as well as in the gut morphology and overall histological appearance, demonstrating the poor development or damage sustained by the birds that had very low bacterial colonisation and no exposure to the Lactobacillus or any other quail commensal bacteria during the first week post-hatch (Figure 4).
Figure 3.
Villus height was significantly higher in probiotic inoculated birds across the inspected gut sections. Crypt depth was not affected in duodenum, but was significantly higher in ileum and caecum of Lactobacillus inoculated birds. P-values are shown on the figure, n = 9, SEM error bars.
Figure 4.
Photomicrographs of quail duodenum, ileum and caecum showed morphological differences between PBS and Lactobacillus treatment groups. Villi length was severely retarded, especially in the PBS treated bird ileum and caecum. Caecal mucosae had reduced thickness in the PBS treated birds. There was an absence of mucosal folds detected in the PBS treated birds while in Lactobacillus inoculated birds mucosal folds were well defined. Haematoxylin and Eosin (H&E) stain.
5. Discussion
Commensal microbiota plays a number of major roles in the health of animals. Although recognised as significant, the processes involved in the formation of the mature intestinal microbiota community is still poorly understood, not due to lack of research interest but mainly due to the complexity of mutual microbial interactions and their interactions with the host, as well as the difficulty of controlling the immediate response to even small environmental changes. Microbial load in the environment can vary significantly, and in very clean environments, it may result in colonisation of the gut with anomalous communities [30].
Our results demonstrate that restricting access to appropriate and natural host gut colonising bacteria during the early days of life can cause permanent and severe effects on gut health and development. Very early post-hatch intervention in the form of a suitable bacterial inoculant may protect birds grown in extreme hygiene conditions from developing anomalous gut microbiota. This emphasises the potentially important role that targeted probiotics introduced in early life, rather than after microbiota has formed, could have, especially when access to maternal bacteria is limited. The birds kept in low bacterial load/high hygiene conditions in the first week of life did not recover from low diversity despite co-housing and exposure to diverse bacteria from mature conventionally reared birds. This suggests possible lifelong consequences of high hygiene levels and it is relevant to both humans and animals. The outcome of this experiment, although more severe, is in agreement with data from children exposed to a high level of hygiene [31] in which susceptibility to autoimmune diseases appears to be an outcome [32]. The severity of problems that may result from excessive hygiene levels in the early period of gut maturation and the benefits in early probiotic administration under such conditions were evident via visible damage in gut morphology in PBS treated but not in Lactobacillus inoculated birds. The near-total dominance of Lactobacillus was previously reported in the ileum of chicken [33] and quail [22], however, it was unexpected in caeca, equally, the gut morphology in these birds was of healthy appearance compared to the PBS treated birds.
High diversity in intestinal microbiota generally correlates to good health while diminished diversity correlates with disease or dysbiosis [34]. Small numbers of pathogenic bacteria are generally a normal part of the gut ecosystem but dysbiosis can occur when there is an imbalance between commensal bacteria and an overgrowth of pathogenic species. Over 25 disorders or diseases have been correlated with decreased diversity of the gut microbiota [34]. To address the dysbiosis attributed to these diseases, much research has been devoted to prebiotic and probiotic bacteria to correct for these conditions [35]. In humans, faecal transplants have gained popularity since the turn of the century, providing promising results [36, 37].
Previous strategies to control the gut health of commercial birds, on a large scale, have mostly relied on the use of in-feed antibiotic growth promoters (AGP). This practice has been banned in many countries due to concerns about antibiotic resistance in pathogens. The ban on AGP's has prompted much research into finding alternative methods for improving bird health, reducing pathogen load and mortality, and the utilisation of different strategies to improve gut health. Our results demonstrate that the control of bacterial colonisation and exposure to healthy commensal bacteria has to be of the utmost priority.
6. Conclusions
Manipulation of intestinal microbiota to enhance beneficial bacteria abundance and reduce pathogenic bacterial communities is slowly becoming a possibility as our knowledge regarding effective methods expands. Our data clearly demonstrates the importance of the first week of gut community maturation, showing the undesirable consequences of exposure to either low or inappropriate bacterial loads, as well as indicating the possibility to remodel the microbiota towards the dominance of desired bacterial strains.
Declarations
Author contribution statement
Ngare Wilkinson, Robert J. Hughes, Yadav Sharma Bajagai, William J. Aspden, Thi Thu Hao Van, Robert J. Moore: Performed the experiments; Wrote the paper.
Dragana Stanley: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by Poultry CRC, established and supported under the Australian Governments Cooperative Research Centre Program. Central Queensland University and Poultry CRC provided a post graduate scholarship for Ngare Wilkinson.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We wish to thank Erica and Clive Wiley, the owners of Banyard Game Farms who provided the birds used in this study. The data was analysed using the Isaac Newton High-Performance Computing System at Central Queensland University. We wish to acknowledge and appreciate help from Jason Bell provided in all aspects of High-Performance Computing.
References
- 1.Dethlefsen L., Huse S., Sogin M.L., Relman D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6(11) doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macpherson A.J., Slack E. The functional interactions of commensal bacteria with intestinal secretory IgA. Curr. Opin. Gastroenterol. 2007;23(6):673–678. doi: 10.1097/MOG.0b013e3282f0d012. [DOI] [PubMed] [Google Scholar]
- 3.Hooper L.V., Gordon J.I. Commensal host-bacterial relationships in the gut. Science. 2001;292(5519):1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 4.Thompson G.R., Trexler P.C. Gastrointestinal structure and function in germ-free or gnotobiotic animals. Gut. 1971;12(3):230–235. doi: 10.1136/gut.12.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Wielen P.W., Lipman L.J., van Knapen F., Biesterveld S. Spatial and temporal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb. Ecol. 2002;44:286–293. doi: 10.1007/s00248-002-2015-y. [DOI] [PubMed] [Google Scholar]
- 6.Palmer C., Bik E.M., DiGiulio D.B., Relman D.A., Brown P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook M.I., Beissinger S.R., Toranzos G.A., Arendt W.J. Incubation reduces microbial growth on eggshells and the opportunity for trans-shell infection. Ecol. Lett. 2005;8(5):532–537. doi: 10.1111/j.1461-0248.2005.00748.x. [DOI] [PubMed] [Google Scholar]
- 8.Peralta-Sanchez J.M., Moller A.P., Martin-Platero A., Soler J.J. Number and colour composition of nest lining feathers predict eggshell bacterial community in barn swallow nests: an experimental study. Funct. Ecol. 2010;24:426–433. [Google Scholar]
- 9.Godoy-Vitorino F., Goldfarb K.C., Brodie E.L., Garcia-Amado M.A., Michelangeli F., Dominguez-Bello M.G. Developmental microbial ecology of the crop of the folivorous hoatzin. ISME J. 2010;4(5):611–620. doi: 10.1038/ismej.2009.147. [DOI] [PubMed] [Google Scholar]
- 10.Kyle P.D., Kyle G.Z. An evaluation of the role of microbial flora in the salivary transfer technique for hand-rearing Chimney Swifts. Wildl. Rehabil. 1993;8:65–71. [Google Scholar]
- 11.Stanley D., Geier M.S., Hughes R.J., Denman S.E., Moore R.J. Highly variable microbiota development in the chicken gastrointestinal tract. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0084290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez-Bello M.G., Costello E.K., Contreras M., Magris M., Hidalgo G., Fierer N. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U. S. A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watson J. Community-associated methicillin-resistant Staphylococcus aureus infection among healthy newborns - Chicago and Los Angeles Country, 2004. MMWR (Morb. Mortal. Wkly. Rep.) 2006;55:329–332. [PubMed] [Google Scholar]
- 14.Bager P., Wohlfahrt J., Westergaard T. Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clin. Exp. Allergy. 2008;38(4):634–642. doi: 10.1111/j.1365-2222.2008.02939.x. [DOI] [PubMed] [Google Scholar]
- 15.Negele K., Heinrich J., Borte M., von Berg A., Schaaf B., Lehmann I. Mode of delivery and development of atopic disease during the first 2 years of life. Pediatr. Allergy Immunol. 2004;15(1):48–54. doi: 10.1046/j.0905-6157.2003.00101.x. [DOI] [PubMed] [Google Scholar]
- 16.Bonetti A. Assessment of the Persistence in the Human intestinal tract of two probiotic lactobacilli Lactobacillus salivarius I 1794 and Lactobacillus paracasei. Microb. Ecol. Health Dis. 2002;14(4):229–233. [Google Scholar]
- 17.Engelbrektson A.L., Korzenik J.R., Sanders M.E., Clement B.G., Leyer G., Klaenhammer T.R. Analysis of treatment effects on the microbial ecology of the human intestine. FEMS Microbiol. Ecol. 2006;57(2):239–250. doi: 10.1111/j.1574-6941.2006.00112.x. [DOI] [PubMed] [Google Scholar]
- 18.Mikkelsen K.H., Allin K.H., Knop F.K. Effect of antibiotics on gut microbiota, glucose metabolism and bodyweight regulation - a review of the literature. Diabetes Obes. Metab. 2016 doi: 10.1111/dom.12637. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez J.M., Murphy K., Stanton C., Ross R.P., Kober O.I., Juge N. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015;26:26050. doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Wielen P.W., Keuzenkamp D.A., Lipman L.J., van Knapen F., Biesterveld S. Spatial and temporal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb. Ecol. 2002;44(3):286–293. doi: 10.1007/s00248-002-2015-y. [DOI] [PubMed] [Google Scholar]
- 21.Stanley D., Hughes R.J., Geier M.S., Moore R.J. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: challenges presented for the identification of performance enhancing probiotic bacteria. Front. Microbiol. 2016;7(187) doi: 10.3389/fmicb.2016.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson N., Hughes R.J., Aspden W.J., Chapman J., Moore R.J., Stanley D. The gastrointestinal tract microbiota of the Japanese quail, Coturnix japonica. Appl. Microbiol. Biotechnol. 2016;100(9):4201–4209. doi: 10.1007/s00253-015-7280-z. [DOI] [PubMed] [Google Scholar]
- 23.Li H., Limenitakis J.P., Fuhrer T., Geuking M.B., Lawson M.A., Wyss M. The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 2015;6:8292. doi: 10.1038/ncomms9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fadrosh D.W., Ma B., Gajer P., Sengamalay N., Ott S., Brotman R.M. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2(1):6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 27.Ashelford K.E., Chuzhanova N.A., Fry J.C., Jones A.J., Weightman A.J. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 2005;71(12):7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zakrzewski M., Proietti C., Ellis J.J., Hasan S., Brion M.J., Berger B. Calypso: a user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musso G., Gambino R., Cassader M. Obesity, diabetes, and gut microbiota: the hygiene hypothesis expanded? Diabetes Care. 2010;33(10):2277–2284. doi: 10.2337/dc10-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt B., Mulder I.E., Musk C.C., Aminov R.I., Lewis M., Stokes C.R. Establishment of normal gut microbiota is compromised under excessive hygiene conditions. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0028284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada H., Kuhn C., Feillet H., Bach J.F. The 'hygiene hypothesis' for autoimmune and allergic diseases: an update. Clin. Exp. Immunol. 2010;160(1):1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanley D., Hughes R.J., Moore R.J. Microbiota of the chicken gastrointestinal tract: influence on health, productivity and disease. Appl. Microbiol. Biotechnol. 2014;98(10):4301–4310. doi: 10.1007/s00253-014-5646-2. [DOI] [PubMed] [Google Scholar]
- 34.de Vos W.M., de Vos E.A. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr. Rev. 2012;70(Suppl 1):S45–56. doi: 10.1111/j.1753-4887.2012.00505.x. [DOI] [PubMed] [Google Scholar]
- 35.Sanders M.E. Probiotics: considerations for human health. Nutr. Rev. 2003;61(3):91–99. doi: 10.1301/nr.2003.marr.91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smits L.P., Bouter K.E., de Vos W.M., Borody T.J., Nieuwdorp M. Therapeutic potential of fecal microbiota transplantation. Gastroenterology. 2013;145(5):946–953. doi: 10.1053/j.gastro.2013.08.058. [DOI] [PubMed] [Google Scholar]
- 37.Morken M.H., Valeur J., Norin E., Midtvedt T., Nysaeter G., Berstad A. Antibiotic or bacterial therapy in post-giardiasis irritable bowel syndrome. Scand. J. Gastroenterol. 2009;44(11):1296–1303. doi: 10.3109/00365520903274401. [DOI] [PubMed] [Google Scholar]