Abstract
Ovarian cancer is the malignant tumor of the female reproductive system with the highest fatality rate. Tolerance to chemotherapeutic drugs such as paclitaxel (PTX) occurring in the very early stage is one of the important factors of the poor prognosis of ovarian cancer. Herein, we aim to study the dysregulation of a particular circular RNA (circRNA), circCELSR1 (hsa_circ_0063809), and its role in the progression and PTX resistance of ovarian cancer. The high expression of circCELSR1 in PTX-resistant tissues of ovarian cancer and PTX-resistant ovarian cancer cells (SKOV3/PTX and HeyA-8/PTX) was determined by microarray analyses and quantitative real-time PCR. Cell Counting Kit-8 (CCK-8) assays were performed to investigate the effect of circCELSR1 on PTX sensitivity of ovarian cancer cells. Flow cytometer assays were used to detect cell cycle and apoptosis of ovarian cancer cells. The effect of circCELSR1 on ovarian cancer cells was assessed in vitro and in vivo. The microRNA (miRNA) sponge mechanism of circRNAs was demonstrated using dual-luciferase reporter and RNA immunoprecipitation assays. By microarray (5 PTX-resistant ovarian cancer tissues νs 5 PTX-sensitive ovarian cancer tissues) and qRT-PCR (36 normal ovarian tissues and ovarian cancer tissues) we identified circCELSR1 to be dramatically highly expressed in ovarian cancer samples and correlated with PTX resistance. Compared with sensitive cell lines, circCELSR1 was also highly expressed in PTX-resistant ovarian cancer cell lines, and circCELSR1 silencing enhanced PTX-induced cytotoxicity in ovarian cancer cells. Meanwhile, the inhibition of circCELSR1 also caused ovarian cancer cell G0/G1 arrest and an increase in apoptosis. In vivo studies revealed that circCELSR1 was stably inhibited in a xenograft mouse model and inhibited the growth of ovarian cancer. Furthermore, we demonstrated that circCELSR1 acts as a sponge for miR-1252 and verified that forkhead box 2 (FOXR2) is a novel target of miR-1252. In this study, we explored the specific mechanisms of PTX resistance and tumor progress of ovarian cancer due to circCELSR1; presented the circCELSR1-miR-1252-FOXR2 axis and its role in ovarian cancer drug sensitivity and progression; and suggest that the results may provide an experimental basis for clinical application.
Keywords: circular RNA, CELSR1, microRNA, resistance, growth ovarian cancer
Introduction
Ovarian cancer has the second highest incidence of gynecological malignant tumors, and it is the leading cause of cancer-related mortality in the female reproductive system.1 Due to untypical early symptoms and unreliable screening approaches, most ovarian cancer patients are diagnosed at an advanced stage and present a poor response to currently available therapeutic interventions, with a 5-year survival rate of 15%–30%.2,3 The first-line therapy for ovarian cancer includes cytoreductive surgery and combined paclitaxel (PTX)-based chemotherapy.4 Although improvement in median survival has been observed in recent years, tumor recurrence frequently occurs following the development of multidrug resistance (MDR).5 Resistance to PTX, a frontline chemotherapeutic agent with long-term clinical applications in the therapy of ovarian cancer, continues to be one of the primary causes of treatment failure.6 Evidence has shown that PTX resistance is a process with multifactorial participation that may originate through a series of modifications;7 however, the mechanism responsible for chemoresistance in ovarian cancer remains poorly understood. Therefore, it is imperative to identify an effective therapeutic target that can sensitize ovarian cancer to PTX and elucidate the molecular mechanism of drug resistance in ovarian cancer.
Circular RNAs (circRNAs) are characterized by a covalent closed-loop structure with no 5′ cap or 3′ polyadenylation tail, with the property of stable structure, good conservation, tissue specificity, and expression specificity at different developmental stages in different species.8 These unique features make circRNA a research hotspot.9 It is becoming increasingly apparent that dysregulated circRNAs are implicated in carcinogenesis and progression of numerous cancers, acting as either oncogenes or tumor suppressors.10 Moreover, abnormality of circRNA levels has been shown to be associated with the development of chemoresistance in various tumors.11, 12, 13 Nonetheless, the functional role of circRNAs in PTX resistance in ovarian cancer remains unclear. In this study, we profiled the circRNA expression of PTX-resistant ovarian cancer tissues in order to improve our understanding of the precise mechanisms of PTX resistance as well as to identify potential circRNA biomarkers for ovarian cancer patients.
Using a circRNA microarray profiling, we found the top 30 upregulated circRNAs in chemoresistant ovarian cancer samples. Among these candidates, we found that the expression of circCELSR1 (hsa_circ_0063809) is markedly elevated both in chemoresistant ovarian cancer tissues and cell lines. We further demonstrated that circCELSR1 may act as a sponge of miR-1252 to upregulate the level of forkhead box 2 (FOXR2) and therefore promote ovarian cancer development. Our findings will provide new insights into the regulatory mechanisms of circCELSR1 in the progression and PTX resistance of ovarian cancer.
Results
Profile of circRNAs in PTX-Resistant Ovarian Cancer Tissues
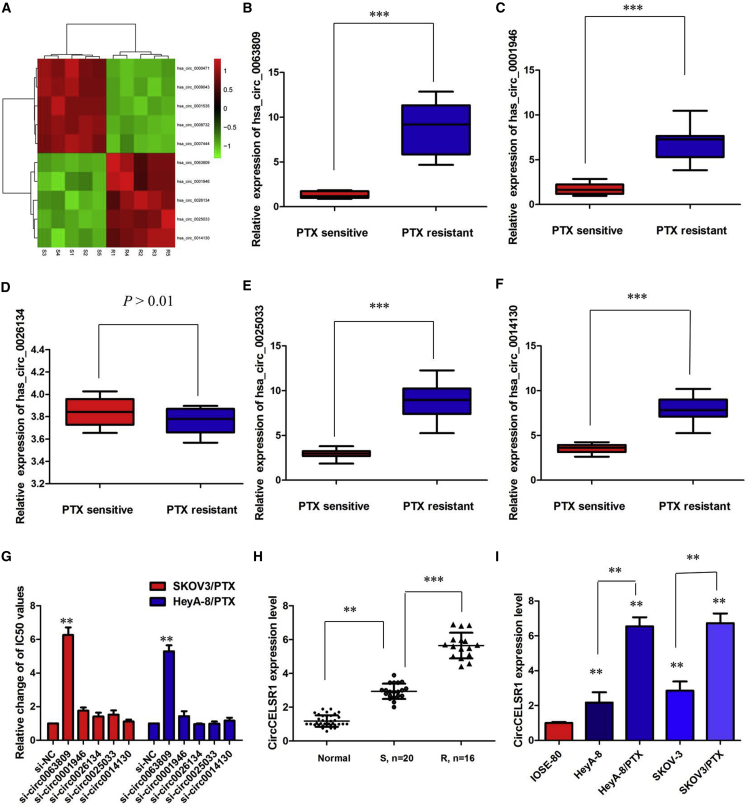
To analyze the expression pattern of circRNAs in PTX-resistant ovarian cancer tissues, we identified the expression profiles of dysregulated circRNAs in five pairs of PTX-sensitive ovarian cancer tissues and PTX-resistant ovarian cancer tissues using a high-throughput microarray assay. We found that 833 circRNAs were aberrantly expressed with a fold change of ≥2.0 and p < 0.05, of which 341 circRNAs were upregulated and 492 circRNAs were downregulated. We also explored the length distribution of circRNAs and found that most circRNAs were shorter than 1,500 bp, consistent with previous studies. Moreover, of all 833 circRNAs disclosed in this study, only 318 (38.18%) were overlapped with circBase (http://www.circbase.org/) and 515 (61.82%) were newly identified. Hierarchical clustering showed the 10 most upregulated and downregulated circRNAs between PTX-resistant and PTX-sensitive tissues of ovarian cancer (Figure 1A). Then, the five most upregulated circRNAs (hsa_circ_0063809, has_circ_0001946, has_circ_0026134, hsa_circ_0025033, and hsa_circ_0014130) were selected and validated by quantitative real-time PCR using PTX-resistant and PTX-sensitive tissue samples. As shown in Figures 1B–1F, except for hsa_circ_0014130, the circRNAs displayed a consistent expression level between the microarray and quantitative real-time PCR analyses.
Figure 1.
Profile of circRNAs in PTX-Resistant Ovarian Cancer Tissues
(A) circRNA microarray data of five pairs of PTX-sensitive ovarian cancer tissues and PTX-resistant ovarian cancer tissues are presented in a heatmap. (B) Relative expression of hsa_circ_0063809 in PTX-resistant ovarian cancer. (C) Relative expression of has_circ_0001946 in PTX-resistant ovarian cancer. (D) Relative expression of has_circ_0026134 in PTX-resistant ovarian cancer. (E) Relative expression of hsa_circ_0025033 in PTX-resistant ovarian cancer. (F) Relative expression of hsa_circ_0014130 in PTX-resistant ovarian cancer. (G) Determination of IC50 values of PTX for both resistant cell lines after inhibition of various circRNAs. (H) Relative expression of circCELSR1 in PTX-resistant ovarian cancer and PTX-sensitive ovarian cancer. (I) Relative expression of circCELSR1 in a panel of ovarian cancer cell lines. All tests were at least performed three times. Data are expressed as mean ± SD. **p < 0.01, ***p < 0.001.
Among them, we found that inhibition of circCELSR1 (hsa_circ_0063809) reversed PTX resistance in both SKOV3/PTX and HeyA-8/PTX cell lines, whereas the other four circRNAs showed little effect (Figure 1G). circCELSR1 is back-spliced of circularizing three exons of CELSR1 gene (chr22:46780439-46785396), located at 22q13.31 amplicon. We first verified its existence in many circRNA databases. According to the circBase database, circCELSR1 is detected in many types of cell lines, including H1hesc and Helas3 (http://www.circbase.org/cgi-bin/singlerecord.cgi?id1/4hsa_circ_0063809). To investigate the clinical significance of circCELSR1 expression in PTX sensitivity of ovarian cancer patients, the expression of circCELSR1 expression in normal ovarian tissues and ovarian cancer tissues from PTX-resistant or PTX-sensitive patients were analyzed. As shown in Figure 1H, there was an increasing trend in circCELSR1 levels from normal ovarian tissues to PTX-sensitive ovarian cancer tissues and then to PTX-resistant ovarian cancer tissues, and the differences among the three groups were significant (p < 0.001). Experiments were then performed at the cellular level. circCELSR1 expression was distinctively higher in PTX-resistant ovarian cancer cells SKOV3/PTX and HeyA-8/PTX cells, and clearly lower in normal ovarian surface IOSE-80 epithelial cells (Figure 1I; p < 0.01).
circCELSR1 Enhances the PTX Chemosensitivity of Ovarian Cancer In Vitro
To further validate the expression level of circCELSR1 on PTX resistance, circCELSR1 short hairpin RNA (shRNA) and circCELSR1 overexpression vector were constructed, and we performed loss-of-function and gain-of-function studies by knocking down or overexpressing circCELSR1 in ovarian cancer cells. First, we knocked down the expression of both circCELSR1 and CELSR1 mRNA. SKOV3/PTX and HeyA-8/PTX cells were transfected with three kinds of circCELSR1 shRNA (sh-circCELSR1#1, sh-circCELSR1#2, or sh-circCELSR1#3, respectively) or GFP lentivirus (sh-control [CTL]), as well as the sequence only in the linear transcript (si-CELSR1). As expected, shRNA directed against the back-spliced sequence knocked down only the circular transcript and did not affect the expression of linear species, and siRNA targeting the sequence in the linear transcript knocked down only the linear transcript and did not affect the expression of the circular transcript in SKOV3/PTX and HeyA-8/PTX cells (Figures S1A–S1H; p < 0.01). Due to the highest efficiency of interference, sh-circCELSR1#1 was chosen for the subsequent experiments. Meanwhile, we infected SKOV-3 and HeyA cells with the circCELSR1 overexpression adenovirus (circCELSR1 OE) or control GFP adenovirus (circCELSR1 CTL). The quantitative real-time PCR assay indicated the relative abundance of circCELSR1 in HeyA and SKOV-3 cells infected with adenovirus (Figures S1I and S1J; p < 0.01).
Next, we investigated the stability and localization of circCELSR1 in SKOV3/PTX and HeyA-8/PTX cells. Total RNAs from SKOV3/PTX and HeyA-8/PTX cells were isolated at the indicated time points after treatment with actinomycin D, an inhibitor of transcription. Then, quantitative real-time PCR was performed to measure the levels of circCELSR1 and CELSR1 mRNA. The results showed that the half-life of circCELSR1 exceeded 24 h, whereas that of CELSR1 mRNA was about 3 h in both SKOV3/PTX and HeyA-8/PTX cells (Figures S2A and S2B). Furthermore, we found that circCELSR1 was resistant to RNase R digestion (Figures S2C–S2D; p < 0.01). We then investigated the localization of circCELSR1. Quantitative real-time PCR of RNAs from nuclear and cytoplasmic fractions indicated that circCELSR1 was predominantly localized in the cytoplasm of SKOV3/PTX and HeyA-8/PTX cells (Figures S2E–S2F; p < 0.01). Collectively, the above data suggested that circCELSR1 harbored a loop structure and was predominantly localized in the cytoplasm.
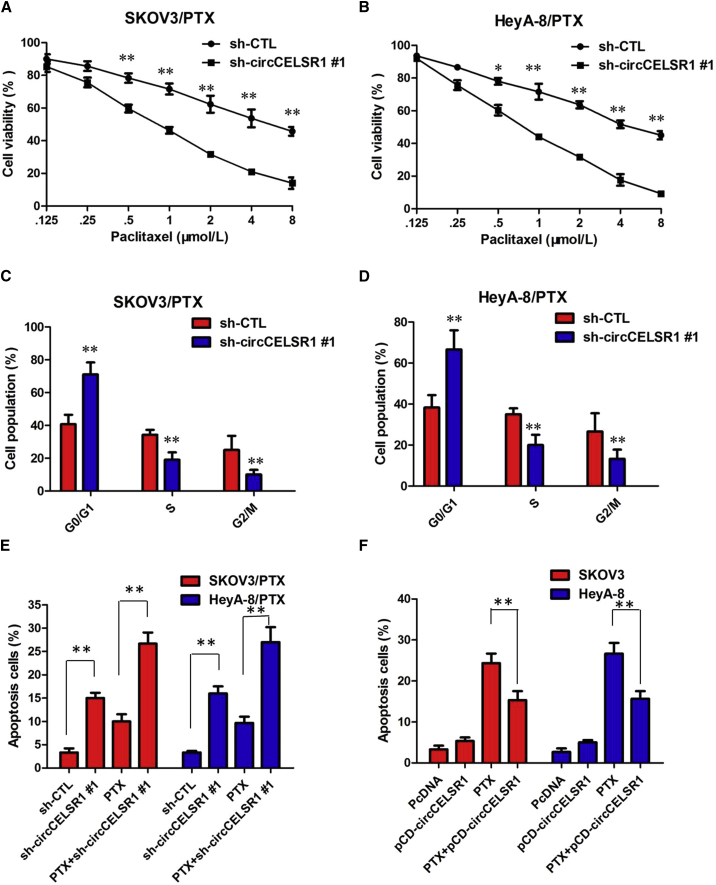
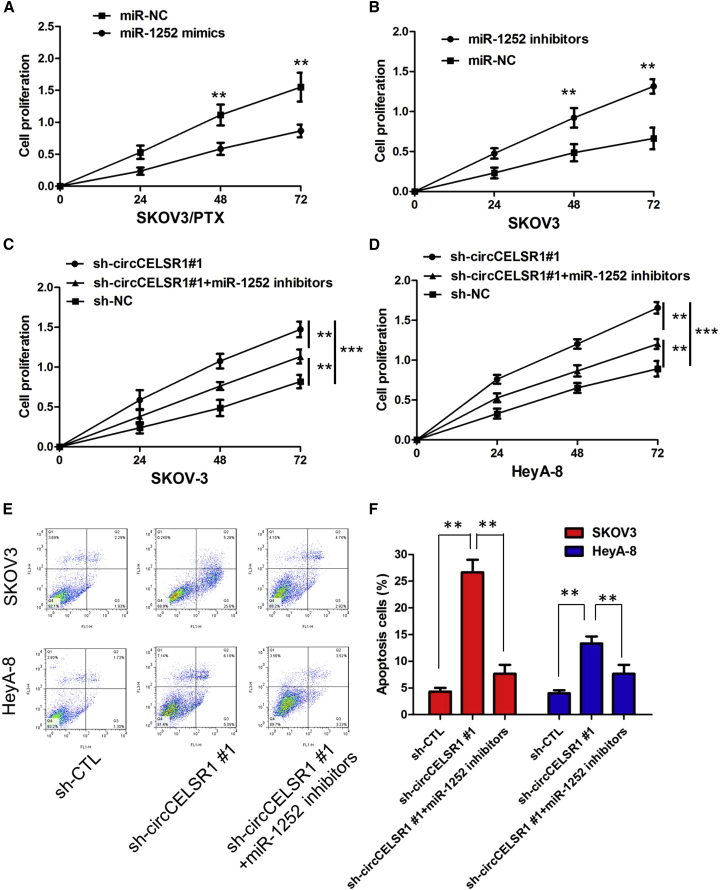
The Cell Counting Kit-8 (CCK-8) assay showed that sh-circCELSR1#1 transfection in SKOV3/PTX and HeyA-8/PTX cells rendered both cell lines more sensitive to PTX-mediated cytotoxicity compared with the control group, as demonstrated by the decreased 50% inhibitory concentration (IC50) value of PTX following circCELSR1 downregulation (Figures 2A and 2B; p < 0.01). To further evaluate whether the effect of circCELSR1 on PTX resistance was associated with cell cycle or apoptosis, we analyzed cell cycle and apoptosis using annexin V-allophycocyanin (APC)/DAPI double staining and flow cytometry. The knockdown of circCELSR1 significantly increased the percent of cells in the G0/G1 phase and decreased the percent of cells in G2 and S phase of SKOV3/PTX and HeyA-8/PTX cells in the presence of PTX (100 nM) (Figures 2C and 2D; p < 0.01). Flow cytometry analysis results demonstrated that inhibition of circCELSR1 promoted the PTX-induced cell apoptosis of resistant ovarian cancer cells in the presence of PTX (100 nM) (Figure 2E; p < 0.01). However, cell apoptosis assays revealed that following overexpression of circCELSR1, the PTX-induced apoptosis of HeyA and SKOV-3 cells was significantly decreased compared to the control group (Figure 2F; p < 0.01). Collectively, these results indicated that circCELSR1 silencing enhanced PTX-induced cytotoxicity in ovarian cancer cells.
Figure 2.
circCELSR1 Enhances the PTX Chemosensitivity of Ovarian Cancer In Vitro
(A) CCK-8 assay showed that inhibition of circCELSR1 rendered SKOV3/PTX cells sensitive to PTX. (B) CCK-8 assay showed that inhibition of circCELSR1 rendered HeyA-8/PTX cells sensitive to PTX. (C) Flow cytometry analysis showed that inhibition of circCELSR1 significantly increased the percentage of cells in the G0/G1 phase and decreased the percentage of cells in the G2 and S phase of SKOV3/PTX cells in the presence of PTX (100 nM). (D) Flow cytometry analysis showed that inhibition of circCELSR1 significantly increased the percentage of cells in the G0/G1 phase and decreased the percentage of cells in the G2 and S phase of HeyA-8/PTX cells in the presence of PTX (100 nM). (E) Flow cytometry analysis showed that inhibition of circCELSR1 promoted cell apoptosis of resistant ovarian cancer cells in the presence of PTX (100 nM). (F) Flow cytometry analysis showed that overexpression of circCELSR1 decreased cell apoptosis of HeyA and SKOV-3 cells in the presence of PTX (100 nM). All tests were at least performed three times. Data are expressed as mean ± SD. *p < 0.05, **p < 0.01.
circCELSR1 Knockdown Enhanced the Anti-Tumor Effect of PTX in Ovarian Cancer In Vivo
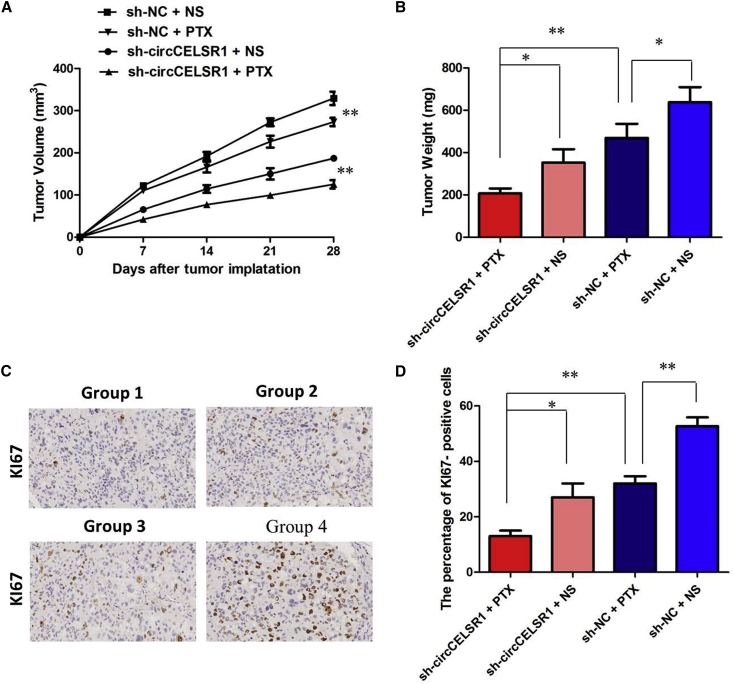
To confirm the effects of circCELSR1 on the chemosensitivity of ovarian cancer cells to PTX in vivo, SKOV3/PTX cells stably infected with sh-circCELSR1 or sh-CTL were subcutaneously injected into each mouse, followed by administration with PTX. According to the treatment, the tumors on the mice were actually assigned to the following groups: group 1, sh-circCELSR1-transfected cells + PTX; group 2, sh-circCELSR1-transfected cells + normal saline (NS); group 3, sh-negative control (NC)-transfected cells + PTX; and group 4, sh-NC-transfected cells + NS. The results showed that PTX treatment significantly inhibited the growth of tumor cells when compared with control groups (group 1 versus group 2, p < 0.01; group 3 versus group 4, p < 0.01). More importantly, with PTX treatment, tumor cells infected with sh-circCELSR1 grew at lower levels than did controls (group 1 versus group 3, p < 0.01), suggesting that circCELSR1 knockdown enhances the PTX chemosensitivity in vivo (Figure 3A). Tendencies in tumor weight were consistent with those in tumor volume (Figure 3B, group 1 versus group 2, p < 0.05; group 3 versus group 4, p < 0.05; group 1 versus group 3, p < 0.01). Moreover, an immunohistochemistry assay showed that the tumors treated with sh-circCELSR1 plus PTX displayed an increased proliferation percentage of Ki-67-positive tumor cells compared with the control group (Figures 3C and 3D; group 1 versus group 3, p < 0.01). Collectively, these results implicated that circCELSR1 knockdown displayed a synergic effect with PTX in suppressing ovarian cancer cell growth in vivo.
Figure 3.
circCELSR1 Knockdown Enhanced the Anti-Tumor Effect of PTX in Ovarian Cancer In Vivo
(A) Volume of tumors that developed in xenografts from different groups. (B) Weights of tumors that developed in xenografts from different groups. (C) The immunohistochemistry assay showed that the tumors treated with sh-circCELSR1 plus PTX displayed an increased proliferation percentage of Ki-67 positive tumor cells compared with the control group. (D) The percentage of KI67 positive cells in xenografts from different groups. All tests were at least performed three times. Data are expressed as mean ± SD. *p < 0.05, **p < 0.01.
circCELSR1 Functioned as a Molecular Sponge of miR-1252 in Ovarian Cancer Cells
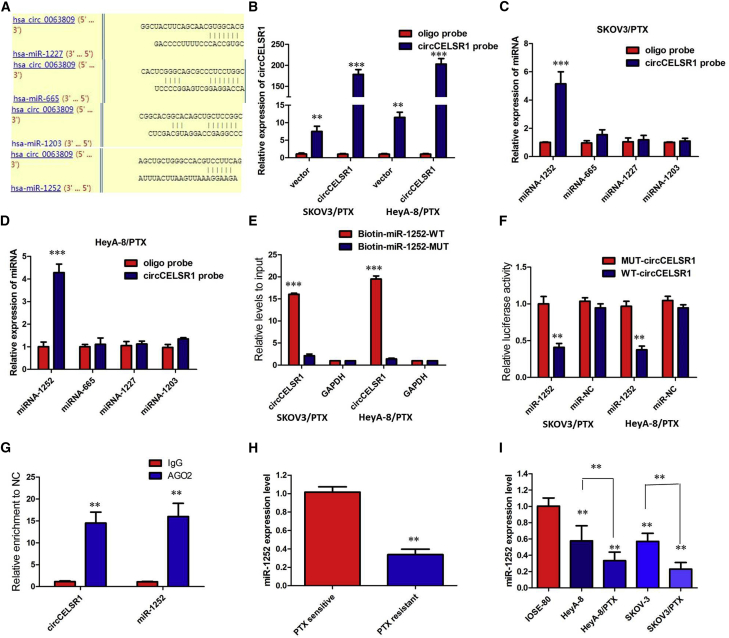
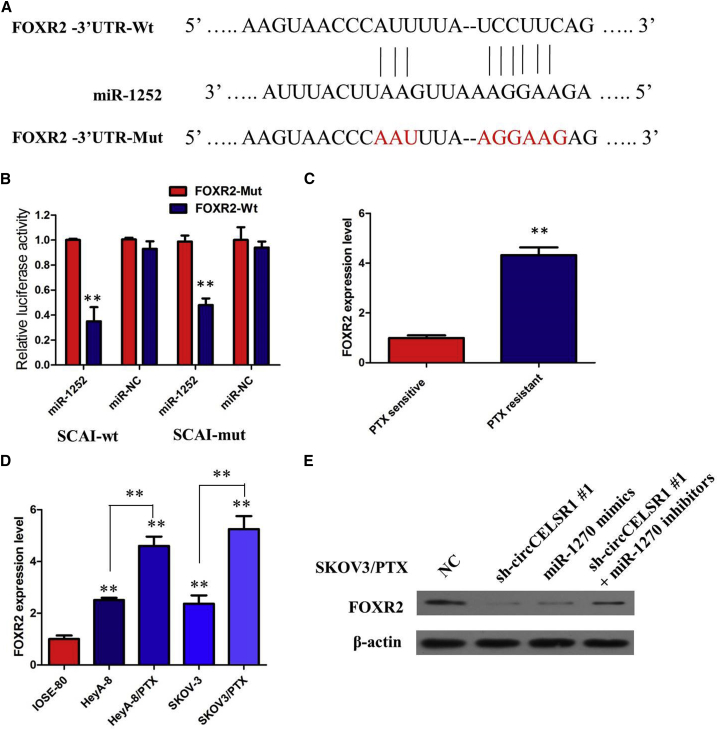
Up to now, accumulating evidence indicated that circRNAs exerted their functions by interacting with microRNAs (miRNAs). Therefore, to investigate the effect of circCELSR1 on the expression of miRNAs, the bioinformatics prediction analysis was performed with TargetScan and miRanda databases. Based on competing endogenous RNA (ceRNA) analysis, circCELSR1 was able to directly bind to 27 miRNAs (Table S1); however, 4 miRNAs (miRNA-665, miRNA-1227, miRNA-1252, and miRNA-1203) were finally selected because they ranked highly in correspondence with the positions of the putative binding sites in the 3′ untranslated region (3′ UTR) of circCELSR1 (Figure 4A). To investigate these potential target miRNAs, we designed a 3′-terminal-biotinylated circCELSR1 probe that was verified to pull down circCELSR1 in ovarian cancer cells, and overexpression of circCELSR1 enhanced the pull-down efficiency (Figure 4B). The quantitative real-time PCR analysis of the levels of the four candidate miRNAs revealed that only miR-1252 was abundantly pulled down by the circCELSR1 probe in both SKOV3/PTX and HeyA-8/PTX cells (Figures 4C and 4D). To validate the direct binding of miR-1252 and circCELSR1, we designed biotin-labeled miR-1252 and its mutant mimics to pull down circCELSR1 in SKOV3/PTX and HeyA-8/PTX cells overexpressing circCELSR1. Quantitative real-time PCR analysis revealed that wild-type miR-1252 captured more circCELSR1 compared with the mutant (Figure 4E). In order to further validate the interaction, a circCELSR1 sequence containing the putative or mutated miR-1252 binding site was cloned into the downstream area of the luciferase reporter gene, generating wild-type (WT)-circCELSR1 or mutant (MUT)-circCELSR1 luciferase reporter plasmids. Then, the effect of miR-1252 on WT-circCELSR1 or MUT-circCELSR1 luciferase reporter systems was determined. The results showed that miR-1252 mimic considerably reduced the luciferase activity of the WT-circCELSR1 luciferase reporter vector compared with negative control, while miR-1252 mimic did not pose any impact on the luciferase activity of MUT-circCELSR1-transfected SKOV3/PTX and HeyA-8/PTX cells (p < 0.01, Figure 4F). In a further RNA immunoprecipitation (RIP) experiment, circCELSR1 and miR-1252 simultaneously existed in the production precipitated by anti-AGO2 (p < 0.01, Figure 4G), suggesting that miR-1252 is a circCELSR1-targeting miRNA.
Figure 4.
circCELSR1 Functioned as a Molecular Sponge of miR-1252 in Ovarian Cancer Cells
(A) StarBase v2.0 results showing the sequence of circCELSR1 with highly conserved putative four miRNAs (miRNA-665, miRNA-1227, miRNA-1252, and miRNA-1203) binding sites. (B) Quantitative real-time RT-PCR analysis of circCELSR1 expression in lysates of SKOV3/PTX and HeyA-8/PTX cells with circCELSR1 overexpression following a biotinylated circMAT2B pull-down assay. (C) Expression of four candidate miRNAs was quantified by quantitative real-time PCR after the biotinylated circCELSR1 pull-down assay in SKOV3/PTX cells. miR-1252 was pulled down by circCELSR1 in SKOV3/PTX cells. (D) Expression of four candidate miRNAs was quantified by quantitative real-time PCR after the biotinylated circCELSR1 pull-down assay in HeyA-8/PTX cells. miR-1252 was pulled down by circCELSR1 in HeyA-8/PTX cells. (E) Biotinylated wild-type/mutant miR-1252 was transfected into SKOV3/PTX and HeyA-8/PTX cells with circCELSR1 overexpression. After streptavidin capture, circCELSR1 expression was detected by quantitative real-time PCR. (F) Luciferase activity in SKOV3/PTX and HeyA-8/PTX cells co-transfected with luciferase reporters containing circCELSR1 sequences with wild-type or mutated miR-1252 binding sites and mimics of miR-1252 or controls. (G) circCELSR1 and miR-1252 simultaneously existed in the production precipitated by anti-AGO2. (H) Relative expression of miR-1252 in PTX-resistant ovarian cancer and PTX-sensitive ovarian cancer. (I) Relative expression of miR-1252 in a panel of ovarian cancer cell lines. All tests were at least performed three times. Data are expressed as mean ± SD. **p < 0.01, ***p < 0.001.
The quantitative real-time PCR analysis indicated that there was a decreasing trend in miR-1252 levels from normal ovarian tissues to PTX-sensitive ovarian cancer tissues and then to PTX-resistant ovarian cancer tissues, and the differences among the three groups were significant (p < 0.01, Figure 4H). We also confirmed that the expression of miR-1252 was obviously decreased in PTX-resistant cells than that in PTX-sensitive cells, indicating the opposite result to circCELSR1 expression (p < 0.01, Figure 4I). Subsequently, the effect of circCELSR1 on miR-1252 expression was also observed in SKOV3/PTX and HeyA-8/PTX cells. The results manifested that miR-1252 expression was specifically inhibited after overexpression of circCELSR1 in PTX-sensitive ovarian cancer cells and evidently promoted following knockdown of circCELSR1 in PTX-resistant ovarian cancer cells (p < 0.01, Figures S3A and S3B). Therefore, we concluded that circCELSR1 could directly bind to miR-1252 to suppress its expression in PTX-resistant ovarian cancer cells.
circCELSR1 Knockdown Inhibited PTX Resistance by Upregulating miR-1252 in PTX-Resistant Ovarian Cancer Cells
To gain insight into whether circCELSR1 affected PTX resistance of ovarian cancer cells via modulation of miR-1252, we further performed rescue assays to confirm how miR-1252 modulated PTX resistance. We transfected miR-1252 mimics or inhibitors into ovarian cancer cell lines, and the proliferation curves were performed. Our results showed that miR-1252 mimics markedly inhibit the cell growth in PTX-resistant cells when compared with cells transfected with miR-NC (Figure 5A; Figure S3C; p < 0.01), whereas PTX-sensitive cells transfected with miR-1252 inhibitors grew at a dramatically higher rate as compared with controls (Figure 5B; Figure S3D; p < 0.01). Moreover, CCK-8 assay proved that downregulation of circCELSR1 markedly inhibits the cell growth in PTX-resistant cells, whereas the sh-circCELSR1#1-induced decrease of cell growth was partially restored by miR-1252 inhibition (Figures 5C–5D; p < 0.01). Furthermore, flow cytometry analysis indicated that miR-1252 overexpression or sh-circCELSR1#1 knockdown dramatically aggravated PTX-induced apoptosis of SKOV3/PTX and HeyA-8/PTX cells; however, sh-circCELSR1#1-triggered apoptosis was attenuated after co-transfection with miR-1252 inhibitor (Figures 5E–5F; p < 0.01). Taken together, these data hinted that inhibition of miR-1252 could significantly reverse circCELSR1-mediated PTX toxicity in ovarian cancer cells.
Figure 5.
circCELSR1 Knockdown Inhibited PTX Resistance by Upregulating miR-1252 in PTX-Resistant Ovarian Cancer Cells
(A) CCK-8 assay showed that miR-1252 mimics markedly inhibited the cell growth of SKOV3/PTX cells when compared with cells transfected with miR-NC. (B) CCK-8 assay showed that miR-1252 inhibitor markedly promoted the cell growth of SKOV3 cells when compared with cells transfected with miR-NC. (C) Downregulation of circCELSR1 markedly inhibits the cell growth in SKOV3/PTX cells, whereas the sh-circCELSR1#1-induced decrease of cell growth was partially restored by miR-1252 inhibition. (D) Downregulation of circCELSR1 markedly inhibits the cell growth in HeyA-8/PTX cells, whereas the sh-circCELSR1#1-induced decrease of cell growth was partially restored by miR-1252 inhibition. (E) The images of cell apoptosis of SKOV3/PTX and HeyA-8/PTX cells. (F) The flow cytometry analysis indicated that miR-1252 overexpression or sh-circCELSR1#1 knockdown dramatically aggravated PTX-induced apoptosis of SKOV3/PTX and HeyA-8/PTX cells; however, sh-circCELSR1#1-triggered apoptosis was attenuated after co-transfection with miR-1252 inhibitor. All tests were at least performed three times. Data are expressed as mean ± SD. **p < 0.01.
circCELSR1 Positively Regulated FOXR2 Expression by Interacting with miR-1252 in PTX-Resistant Ovarian Cancer Cells
To further investigate the role and downstream mechanism with which miR-1252 affects PTX resistance of ovarian cancer cells, bioinformatics tools (microRNA.org and miRBase) were used to search for the potential targets of miR-1252 (Table S1). As displayed in Figure 6A, binding sites of miR-1252 were identified in the 3′ UTR of FOXR2 mRNA. In order to verify whether miR-1252 may directly bind the FOXR2 gene, miR-1252 wild-type and mutant sequences were constructed into the luciferase reporter gene and co-transfected with FOXR2, and luciferase assay confirmed that miR-1252 targeted FOXR2 and negatively regulated FOXR2 expression (Figure 6B; p < 0.01). We examined FOXR2 expression in ovarian cancer tissues and cell lines. The results of real-time PCR analysis demonstrated that the FOXR2 was higher in PTX-resistant ovarian cancer tissues compared with those in PTX-sensitive ovarian cancer tissues (Figure 6C; p < 0.01). The expression of FOXR2 was obviously increased in PTX-resistant ovarian cancer cell lines than that in their parental ovarian cancer cell lines (Figure 6D; p < 0.01). Furthermore, we found that circCELSR1 knockdown or miR-1252 overexpression triggered a substantial decline of FOXR2 protein level in SKOV3/PTX cells. Moreover, inhibition of the circCELSR1-mediated decrease of FOXR2 protein expression was significantly recuperated following miR-1252 inhibitors (Figure 6E). All of these data made us draw a conclusion that circCELSR1 positively regulated FOXR2 expression by interacting with miR-1252 in PTX-resistant ovarian cancer cells.
Figure 6.
circCELSR1 Positively Regulated FOXR2 Expression by Interacting with miR-1252 in PTX-Resistant Ovarian Cancer Cells
(A) Bioinformatics analysis revealed that the binding sites of miR-1252 were identified in the 3′ UTR of FOXR2 mRNA. (B) Luciferase reporter assay demonstrated that miR-1252 mimics significantly decreased the luciferase activity of FOXR2-WT in ovarian cancer cells. (C) Relative expression of FOXR2 in PTX-resistant ovarian cancer and PTX-sensitive ovarian cancer. (D) Relative expression of FOXR2 in a panel of ovarian cancer cell lines. (E) Inhibition of the circCELSR1-mediated decrease of FOXR2 protein expression was significantly recuperated following miR-1252 inhibitors. All tests were at least performed three times. Data are expressed as mean ± SD. **p < 0.01, ***p < 0.001.
Discussion
circRNAs are a brand-new kind of non-coding RNAs, becoming a hotspot research field.14 Recently, a handful of studies demonstrated that several circRNAs possess miRNA binding sites, and, accordingly, serve as ceRNAs to arrest the activity of miRNAs and ultimately regulate their downstream targets.15 Previous studies have found that miRNAs and long noncoding RNAs (lncRNAs) play an important role in the development, progression, and drug resistance of ovarian cancer.16,17 However, biological functions of circRNAs in ovarian cancer, especially in drug resistance, are the tip of the iceberg.
Therefore, in the present study, we screened the differentially expressed circRNAs between PTX-resistant ovarian cancer tissues and PTX-sensitive ovarian cancer tissues using microarrays. The data show that that the expression of circCELSR1 was significantly increased in both PTX-resistant ovarian cancer tissues and cell lines. Moreover, inhibition of circCELSR1 could increase the PTX sensitivity of ovarian cancer cells. Mechanistically, circCELSR1 functions as a ceRNA by sponging miR-1252 to abolish the suppressive effect of this miRNA on its target gene FOXR2. These results suggested that circCELSR1 may have the potential to regulate the sensitivity of ovarian cancer cells to PTX, in turn promoting the progression of ovarian cancer.
Emerging evidence shows that dysregulation of circRNAs plays important roles in chemoresistance.18 In this study, we determined that circRNA expression is associated with PTX resistance in ovarian cancer using circRNAs microarray. We found a novel circRNA termed circCELSR1 that was upregulated in tissue samples from patients with PTX-resistant ovarian cancer and in PTX-resistant cell lines. Moreover, circCELSR1 was expressed at higher levels than other candidate circRNAs in PTX-resistant ovarian cancer patients, which meant that it may play a more important role than other circRNAs in ovarian cancer. Based on the abovementioned studies, we conducted a series of experiments and demonstrated that upregulated circCELSR1 could decrease the PTX sensitivity of ovarian cancer cells. We downregulated circCELSR1 in two PDX-resistant ovarian cancer cell lines, SKOV3/PTX and HeyA-8/PTX. Loss-of-function experiments revealed that circCELSR1 silencing inhibited ovarian cancer cell proliferation, induced apoptosis, and sensitized ovarian cancer cells to PTX in vitro and in vivo.
Further investigations showed that circCELSR1 interacted with miR-1252, and miR-1252 mimics reversed circCELSR1-mediated effects. We verified that circCELSR1 had an endogenous sponge-like effect on miR-1252 in ovarian cancer. First, bioinformatics prediction and a luciferase reporter assay showed that circCELSR1 3ʹ UTR shares identical miR-1252 response elements and might therefore bind competitively to miR-1252. Second, circCELSR1 and miR-1252 simultaneously existed in the production precipitated by anti-AGO2. Third, knockdown or overexpression of circCELSR1 significantly affected miR-1252 expression. Noncoding RNAs, including miRNAs, lncRNAs, and circRNAs, play an important role in the evolution and progression of drug resistance in cancers. We found that the miR-1252 was significantly lower in PTX-resistant ovarian cancer tissue and cell lines.
Furthermore, circRNAs and miRNAs can regulate different signaling pathways through downstream mRNA. In this study, circCELSR1 binds with miR-1252 to promote the expression of FOXR2. Human Fox genes belong to the family of winged/forkhead transcription factors, including at least 43 members from FoxA1 to FoxQ1.19 FoxR2 is a more recently discovered member of the Fox transcription factor family, first identified in 2004.20 An increasing number of studies have found that FoxR2 was involved in the development and progression of many human tumors.21,22 Our findings were similar to the above results. FOXR2 was higher in PTX-resistant ovarian cancer tissues and cell lines. Furthermore, we found that circCELSR1 knockdown or miR-1252 overexpression triggered a substantial decline of FOXR2 protein level in SKOV3/PTX cells. Moreover, inhibition of the circCELSR1-mediated decrease of FOXR2 protein expression was significantly recuperated following miR-1252 inhibitors. Taken together, the study revealed that a circCELSR1/miR-1252/FOXR2 axis exists in ovarian cancer.
Conclusions
In summary, our study first revealed that circCELSR1 is frequently activated in PTX-resistant ovarian cancer tissues and cell lines and is associated with aggressive ovarian cancer phenotypes. Additionally, circCELSR1 silencing suppressed ovarian cancer proliferation in both in vitro and in vivo experiments. Mechanistically, circCELSR1 functions as a molecular sponge to downregulate miR-1252, thereby resulting in partial abolition of the translational repression of its target gene FOXR2 in ovarian cancer cells. In conclusion, we identified that the circCELSR1/miR-1252/FOXR2 axis may provide a foundation for developing novel potential therapeutic strategies for ovarian cancer.
Materials and Methods
Patients and Tissue Samples
Thirty-six ovarian carcinoma specimens were collected from ovarian cancer patients receiving oophorectomies between July 2018 and January 2019 at the Department of Gynecological Oncology, Fudan University Shanghai Cancer Center. In all of the cases, the diagnoses were confirmed by two experienced pathologists, which were done in accordance with the principles laid down in the latest World Health Organization classification. Samples were promptly frozen in liquid nitrogen and maintained at −80°C until use. Patient samples were divided into two groups based on the response to the first-line chemotherapy: treatment-sensitive patients (S, n = 20) and treatment-resistant patients (R, n = 16). According to the National Comprehensive Cancer Network (NCCN) guidelines, intrinsically treatment-resistant tumors were regarded as those with persistent or recurrent disease within 6 months after the initiation of first-line taxol-platinum-based combination chemotherapy. Treatment-sensitive tumors were classified as those with a complete response to chemotherapy and a platinum-free interval of 6 months. This study was approved by the Ethics Committee of Fudan University Shanghai Cancer Center, and written informed consent was provided by every participant prior to surgery.
Cell Lines and Culture
Human ovarian carcinoma cell lines (SKOV3 and HeyA-8) and a normal ovarian epithelial cell line (IOSE-80) were purchased from ATCC (Manassas, VA, USA) and BNCC (Beijing, China), respectively. The corresponding PTX-resistant ovarian cancer cells SKOV3/PTX and HeyA-8/PTX cells were established from the parental cell lines by stepwise exposure to escalating concentrations of PTX, as previously described.23 Cells were cultured in RPMI 1640 medium (HyClone, Logan, UT, USA) with 10% (v/v) fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA) and antibiotics (100 U/mL penicillin, 100 μg/mL streptomycin) (Sigma-Aldrich, St. Louis, MO, USA) in a 95% air/5% CO2 atmosphere at 37°C. To maintain the PTX-resistant phenotype of SKOV3/PTX and HeyA-8/PTX cells, 5 nM PTX was additionally added into the culture medium.
circRNA Microarrays
Five pairs of PTX-sensitive ovarian cancer tissues and PTX-resistant ovarian cancer tissues were used for circRNA microarrays. Total RNAs were digested with RNase R (20 U/μL, Epicenter, USA) to remove linear RNAs and enrich circRNAs. The enriched circRNAs were amplified and transcribed into fluorescent cRNA utilizing a random priming method (Super RNA labeling kit; Arraystar, Rockville, MD, USA). The labeled cRNAs were hybridized onto the human circRNA array (8 × 15K, Arraystar). The slides were incubated for 17 h at 65°C in a hybridization oven (Agilent, Santa Clara, CA, USA). circRNAs differentially expressed with statistical significance between HCC and paired normal tissues (fold change [FC] ≥ 2 and p ≤ 0.05) were identified through volcano plot filtering. Hierarchical clustering was performed to show the distinguishable expression pattern of circRNAs among samples.
Actinomycin D and RNase R Treatment
To block transcription, 2 mg/mL actinomycin D or dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO, USA) as a negative control was added into the cell culture medium. For RNase R treatment, total RNA (2 μg) was incubated for 30 min at 37°C with or without 3 U/μg RNase R (Epicenter Technologies, Madison, WI, USA). After treatment with actinomycin D and RNase R, quantitative real-time PCR was performed to determine the expression levels of circCELSR1 and CELSR1 mRNA.
Isolating RNAs from Nucleus and Cytoplasmic Fractions
The nuclear and cytoplasmic fractions were isolated using a PARIS kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Briefly, cells were collected and lysed with a cell fractionation buffer, followed by centrifugation to separate the nuclear and cytoplasmic fractions. The supernatant containing the cytoplasmic fraction was collected and then transferred to a fresh RNase-free tube. The nuclear pellet was lysed with cell disruption buffer. The cytoplasmic fraction and nuclear lysate were mixed with 2× lysis/binding solution and then added with 100% ethanol. The sample mixture was drawn through a filter cartridge, followed by washing with wash solution. The RNAs of nuclear and cytoplasmic fractions were eluted with elution solution. U6 small nuclear RNA (snRNA) and 18S rRNA were employed as positive controls for nuclear and cytoplasmic fractions, respectively.
Transfection
For transfections, cells at the confluence of 50%–80% were infected with 1 × 106 recombinant lentivirus-transducing units and 6 μg/mL Polybrene (Sigma, Shanghai, China). Stably transfected cells were selected via treatment with 2 μg/mL puromycin for 2 weeks. Stably transfected cells were picked via flow cytometry for subsequent assays. Plasmid, lentivirus, miRNA inhibitor, and miRNA mimics used in this study were purchased from GenePharma (Shanghai, China), and pHBV1.3 copy was purchased from Miaolingbio (Wuhan, China). Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) was utilized for transfection.
RNA Preparation and Quantitative Real-Time PCR
Total RNA extraction and quantification, RNA purification, and cDNA synthesis were conducted as described previously.21 For RNase R treatment, 2 μg of total RNA was incubated for 15 min at 37°C with or without 3 U/μg RNase R (Epicenter Technologies, Madison, WI, USA). To detect RNA expression, quantitative real-time PCR was performed with PowerUp SYBR Green Master Mix (Thermo Fisher, Waltham, MA, USA) and the Applied Biosystems StepOnePlus real-time PCR detection system (Life Technologies, CA, USA). The relative gene expression was calculated using the 2−ΔCT method normalized to GAPDH, and the fold change of gene expression was calculated by the 2−ΔΔCT method. Bulge-loop miRNA quantitative real-time PCR primer sets (one reverse transcription [RT] primer and a pair of qPCR primers for each set) specific for miR-1252 was designed by RiboBio (Guangzhou, China). The relative expression of miR-1252 was normalized to human U6 snRNA.
CCK-8 Assay
Cell proliferation was assessed using the CCK-8 assay (Beyotime Biotechnology, Nantong, China). Cells (2 × 103) were seeded into each well of 96-well plates. 10 μL of CCK-8 solution was added to each well at six time points. After 1.5 h of incubation at 37°C, the absorbance at 450 nM was measured using a SpectraMax 250 spectrophotometer (Molecular Devices, Sunnyvale, CA, USA). Experiments were independently performed in triplicate.
Cell Cycle and Apoptosis Assay
SKOV3/PTX and HeyA-8/PTX cells were seeded into six-well plates and treated with PTX for 48 h. To assess the cell cycle and apoptosis, 3 × 105 treated cells were seeded into six-well plates and cultured for 48 h at 37°C. The cells for cell cycle analysis were digested using trypsin (HyClone), washed twice with phosphate-buffered saline (PBS), and fixed in 70% ethanol overnight at 4°C. The cells were centrifuged at 500 × g for 5 min, washed twice with cold PBS, and centrifuged. After treating with RNase A (0.1 mg/mL) and propidium iodide (PI, 0.05 mg/mL) purchased from 4A Biotech (Beijing, China) for 30 min at 37°C, cell cycle analysis was performed through fluorescence-activated cell sorting flow cytometry (Beckman Coulter, Palo Alto, CA, USA). For the analysis of apoptosis, cells were trypsinized followed by two PBS washing steps. The cells were stained using the annexin V/PI detection kit (4A Biotech, Beijing, China) for 5 min at room temperature. The apoptotic cells were measured using flow cytometry (Beckman Coulter). All experiments were repeated at least three times.
Tumor Xenograft Model
Six-week-old female BALB/c athymic nude mice (n = 5 per group) were purchased from Shanghai Experimental Animal Center (Shanghai, China). Mice were subcutaneously injected into the back with 1 × 106 SKOV3/PTX cells stably transfected with sh-circCELSR1 or sh-NC suspended in 100 μL of Hank’s balanced salt solution. After 6 days of injection, mice were intraperitoneally administrated with PBS or 3 mg/kg PTX (Sigma-Aldrich) every 3 days for five cycles. According to the treatment, the tumors on the mice were actually assigned to the following groups: group 1, sh-circCELSR1-transfected cells + PTX; group 2, sh-circCELSR1-transfected cells + NS; group 3, sh-NC-transfected cells + PTX; and group 4, sh-NC-transfected cells + NS. The tumor size was measured every 3 days with a caliper, and tumor volume was calculated according to the formula: volume = (length × width2)/2. All mice were sacrificed on day 21 after inoculation. The resected tumor masses were harvested for subsequent weight and quantitative real-time PCR analysis. The animal experiments were approved by the Animal Care and Use Committee of Huaihe Hospital and were performed in accordance with the Institutional Guide for the Care and Use of Laboratory Animals.
Immunohistochemistry
Other tissue sections were used for immunohistochemistry staining of Ki-67. The deparaffinized and dehydrated slides were heat-treated with citrate buffer (pH 6.0) to retrieval antigen and washed with 3% H2O2 in 1× Tris-buffered saline (TBS) to block endogenous peroxidase activity. After that, the sections were incubated with a Ki-67 antibody (dilution 1:200; Cell Signaling Technology) overnight at 4°C and a biotinylated secondary antibody for another 30 min. An avidin-biotin-peroxidase complex (Dako LSAB2 System; Dako, Produktionsvej, Denmark) was added, and the color was developed using diaminobenzidine (DAB). Counterstaining was performed with hematoxylin. The sections were viewed under a BX51 system microscope (Olympus, Tokyo, Japan). Images were recorded using a digital microscope camera (DP70; Olympus).
Biotinylated RNA Pull-Down Assay
The pull-down assay was performed with biotinylated RNA as previously described. Briefly, for circCELSR1 pulled-down miRNAs, the biotinylated-circCELSR1 probe was incubated with C-1 magnetic beads (Life Technologies, Carlsbad, CA, USA) at 25°C for 2 h to generate probe-coated beads. These were then incubated with sonicated cells at 4°C overnight, eluted with the wash buffer, and analyzed by quantitative real-time PCR. For miR-1252 pulled-down circCELSR1, cells with circCELSR1 overexpression were transfected with biotinylated miR-1252 mimics or mutant miR-1252 using Lipofectamine 3000. The cells were harvested 48 h after transfection, lysed, sonicated, and incubated with C-1 magnetic beads (Life Technologies, Carlsbad, CA, USA), eluted with the wash buffer, and analyzed by quantitative real-time PCR. The biotinylated circCELSR1 probe and biotinylated miR-1252 mimics or mutant were designed and synthesized by RiboBio (Guangzhou, China).
Luciferase Reporter Assay
The circCELSR1 full fragments or its mutant containing the putative miR-1252-binding sites in circCELSR1 was synthesized and cloned into downstream of firefly luciferase gene in pGL3 plasmids (Promega, Fitchburg, WI, USA), named as pGL3-circCELSR1-wild-type (WT) and pGL3-circCELSR1 mutant (MUT). Similarly, the 3′ UTR of FOXR2 containing the predicted miR-1252-binding sites or mutant sites was amplified by PCR and inserted into pGL3 plasmids, named as pGL3-FOXR2-3′ UTR-WT and pGL3-FOXR2-3′ UTR-MUT. SKOV3/PTX and HeyA-8/PTX cells were maintained in 96-well plates and co-transfected with circCELSR1-WT (or circCELSR1-MUT) or FOXR2-WT (or FOXR2-MUT) and miR-1252 mimic, or miR-NC using Lipofectamine 3000 (Invitrogen). Luciferase activity was measured in cell lysates 24 h after transfection using a Dual-Luciferase reporter system (Promega, Madison, WI, USA).
RIP Assay
The RIP assay was carried out using a Magna RIP RNA-binding protein immunoprecipitation kit (Bersinbio, Guangzhou, China) according to the manufacturer’s protocol. Huh7 cells (2 × 107) were lysed in complete RIP lysis buffer and the cell lysates were divided into two equal parts and incubated with either 5 μg of human anti-Argonaute2 (AGO2) antibody (Millipore, Billerica, MA, USA) or non-specific anti-immunoglobulin G (IgG) antibody (Millipore) with rotation at 4°C overnight. Magnetic beads were added to the cell lysates and incubation was continued at 4°C for 1 h. The samples were then incubated with proteinase K at 55°C for 1 h. The enriched RNA was obtained using RNA extraction reagent (enol/chloroform/isoamylol at 125:24:1, pH of <5.0; Solarbio). The purified RNA was used to detect the expression levels of the genes of interest by quantitative real-time PCR.
Western Blot Assay
The lysates from cells were collected by radio immunoprecipitation assay (RIPA) buffers (Beyotime Biotechnology, Shanghai, China) and then boiled for 5 min at 100°C. Protein concentration was determined using the bicinchoninic acid (BCA) protein assay kit (Promega, Madison, WI, USA). The protein samples (20 μg) in loading buffer were separated on 10% SDS polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Bio-Rad Laboratories, Hercules, CA, USA). The membranes were blocked with 5% (w/v) non-fat milk in TBS with Tween 20 (TBST) for 1 h and incubated with antibodies against FOXR2 (1:1,000 dilution; ab244513; Abcam, Cambridge, UK) overnight at 4°C. After incubation with appropriately diluted horseradish peroxidase (HRP)-conjugated secondary antibodies (1:2,000 dilution), the immunoreactive protein bands were visualized using a chemiluminescent substrate (ECL kit; Amersham Biosciences, Piscataway, NJ, USA). The density of each protein band was normalized to β-actin.
Statistical Analysis
Results are presented expressed as mean ± SD (standard deviation). Student’s t test was performed to measure the difference between two groups, and differences between more than two groups were assessed using one-way ANOVA. p < 0.05 was considered significant.
Author Contributions
J.C. and C.Q. contributed flow cytometry assay. H.W. contributed animal experiments. Z.F. collected and classified the human tissue samples. Q.H. and J.Z. analyzed the data. Y.H. and S.Z. wrote the paper. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the Academician Li Jieshou Gut Research Fund 2017.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.12.005.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Vaughan S., Coward J.I., Bast R.C., Jr., Berchuck A., Berek J.S., Brenton J.D., Coukos G., Crum C.C., Drapkin R., Etemadmoghadam D. Rethinking ovarian cancer: recommendations for improving outcomes. Nat. Rev. Cancer. 2011;11:719–725. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinato D.J., Graham J., Gabra H., Sharma R. Evolving concepts in the management of drug resistant ovarian cancer: dose dense chemotherapy and the reversal of clinical platinum resistance. Cancer Treat. Rev. 2013;39:153–160. doi: 10.1016/j.ctrv.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Tsujioka H., Yotsumoto F., Hikita S., Ueda T., Kuroki M., Miyamoto S. Targeting the heparin-binding epidermal growth factor-like growth factor in ovarian cancer therapy. Curr. Opin. Obstet. Gynecol. 2011;23:24–30. doi: 10.1097/GCO.0b013e3283409c91. [DOI] [PubMed] [Google Scholar]
- 5.Pokhriyal R., Hariprasad R., Kumar L., Hariprasad G. Chemotherapy resistance in advanced ovarian cancer patients. Biomark Cancer. 2019;11 doi: 10.1177/1179299X19860815. 1179299X19860815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stage T.B., Bergmann T.K., Kroetz D.L. Clinical pharmacokinetics of paclitaxel monotherapy: an updated literature review. Clin. Pharmacokinet. 2018;57:7–19. doi: 10.1007/s40262-017-0563-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei Y., Pu X., Zhao L. Preclinical studies for the combination of paclitaxel and curcumin in cancer therapy (Review) Oncol. Rep. 2017;37:3159–3166. doi: 10.3892/or.2017.5593. [DOI] [PubMed] [Google Scholar]
- 8.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 9.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Hansen T.B., Kjems J., Damgaard C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 11.Wei S., Zheng Y., Jiang Y., Li X., Geng J., Shen Y., Li Q., Wang X., Zhao C., Chen Y. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 2019;44:182–193. doi: 10.1016/j.ebiom.2019.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shang J., Chen W.M., Wang Z.H., Wei T.N., Chen Z.Z., Wu W.B. circPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. Exp. Hematol. 2019;70:42–54.e3. doi: 10.1016/j.exphem.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Kun-Peng Z., Xiao-Long M., Chun-Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int. J. Biol. Sci. 2018;14:321–330. doi: 10.7150/ijbs.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Z., Ji M., Wang Q., He N., Li Y. Circular RNA Cdr1as upregulates SCAI to suppress cisplatin resistance in ovarian cancer via miR-1270 suppression. Mol. Ther. Nucleic Acids. 2019;18:24–33. doi: 10.1016/j.omtn.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Xia X., Li X., Li F., Wu X., Zhang M., Zhou H., Huang N., Yang X., Xiao F., Liu D. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol. Cancer. 2019;18:131. doi: 10.1186/s12943-019-1056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang P., Zhu J., Zheng Y., Zhang H., Sun H., Gao S. miRNA-574-3p inhibits metastasis and chemoresistance of epithelial ovarian cancer (EOC) by negatively regulating epidermal growth factor receptor (EGFR) Am. J. Transl. Res. 2019;11:4151–4165. [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X., Mangala L.S., Mooberry L., Bayraktar E., Dasari S.K., Ma S., Ivan C., Court K.A., Rodriguez-Aguayo C., Bayraktar R. Identifying and targeting angiogenesis-related microRNAs in ovarian cancer. Oncogene. 2019;38:6095–6108. doi: 10.1038/s41388-019-0862-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao D., Qi X., Zhang X., Fang K., Guo Z., Li L. hsa_circRNA_0006528 as a competing endogenous RNA promotes human breast cancer progression by sponging miR-7-5p and activating the MAPK/ERK signaling pathway. Mol. Carcinog. 2019;58:554–564. doi: 10.1002/mc.22950. [DOI] [PubMed] [Google Scholar]
- 19.Katoh M., Igarashi M., Fukuda H., Nakagama H., Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013;328:198–206. doi: 10.1016/j.canlet.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 20.Katoh M., Katoh M. Identification and characterization of human FOXN6, mouse Foxn6, and rat Foxn6 genes in silico. Int. J. Oncol. 2004;25:219–223. [PubMed] [Google Scholar]
- 21.Xu W., Chang J., Liu G., Du X., Li X. Knockdown of FOXR2 suppresses the tumorigenesis, growth and metastasis of prostate cancer. Biomed. Pharmacother. 2017;87:471–475. doi: 10.1016/j.biopha.2016.12.120. [DOI] [PubMed] [Google Scholar]
- 22.Wang X.H., Cui Y.X., Wang Z.M., Liu J. Down-regulation of FOXR2 inhibits non-small cell lung cancer cell proliferation and invasion through the Wnt/β-catenin signaling pathway. Biochem. Biophys. Res. Commun. 2018;500:229–235. doi: 10.1016/j.bbrc.2018.04.046. [DOI] [PubMed] [Google Scholar]
- 23.Li Z., Hu S., Wang J., Cai J., Xiao L., Yu L., Wang Z. miR-27a modulates MDR1/P-glycoprotein expression by targeting HIPK2 in human ovarian cancer cells. Gynecol. Oncol. 2010;119:125–130. doi: 10.1016/j.ygyno.2010.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.