Abstract
Intervertebral degenerative disc disease (IDDD) is a common disease in clinic that causes pain and heavy financial burden on patients with poor prognosis. However, the pathogenesis of IDDD is not clear. Long non-coding RNA (LncRNA) is involved in regulating various body growth and pathological processes by affecting cell proliferation, differentiation, and apoptosis. However, the role of lncRNAs in IDDD is rarely reported. This study aims to investigate the role and mechanism of lncRNA MALAT1 in the development of IDDD. The nucleus pulposus of the intervertebral disc were collected and the primary nucleus pulposus cells were isolated and cultured. The cells were divided into three groups, including IDDD group, empty plasmid group transfected by pcDNA3.1, or MALAT1 group transfected by pcDNA3.1-MALAT1. MALAT1 expression was detected by real-time PCR. Cell proliferation was assessed by MTT assay. Caspase 3 activity was tested by the activity detection kit. IL-1 and IL-6 levels were analyzed by ELISA. The expression of MALAT1 in IDDD nucleus pulposus cells was significantly lower than that in control group (P < 0.05). The expression of MALAT1 was significantly increased after transfection with pcDNA3.1-MALAT1 plasmid in IDDD nucleus pulposus cells, which obviously inhibited cell proliferation, enhanced Caspase 3 activity, and promoted the secretion of IL-1 and IL-6 compared with IDDD group (P < 0.05). MALAT1 level decreased in IDDD nucleus pulposus cells. Upregulation of MALAT1 expression restrained IDDD through suppressing inflammation; inhibiting nucleus pulposus cell apoptosis, and promoting cell proliferation.
Keywords: MALAT1, disc degeneration disease, apoptosis, nucleus pulposus cells, inflammation
Introduction
Intervertebral disc degeneration disease (IDDD) is a common clinical disease. As a common bone disease, it can lead to neck and shoulder pain which brings severe problem to the global health [1,2]. Chronic low back pain is the most common clinical symptom, which can be induced by IDDD, lumbar disc herniation, lumbar spondylolisthesis, and lumbar spinal stenosis. IDDD is the main cause of chronic low back pain and has gradually become one of the global public health problems [3,4]. The incidence of IDDD in China increases year by year and exhibits the younger trend [5]. IDDD is induced by a variety of physical and chemical factors, as well as molecular biology and mechanical factors. Intervertebral disc cell structure and function changes, thereby affecting the nucleus pulposus cells and destroying the boundaries between the fiber ring and nucleus. It further causes the loss of water in the nucleus pulposus cells and decreases the load capacity of the intervertebral disc [6,7]. Following the lifestyle changes, movement reduction, aging, pace of human life acceleration, and long seated work, IDDD demonstrates an elevating incidence [8,9]. IDDD may lead to loss of labor capacity and even disability [10]. So far, however, the pathogenesis of IDDD is unclear. The pathogenic factors of IDDD include genetic factors, aging, nucleus pulposus immunodeficiency reduction, and nucleus pulposus cell death [11,12].
There are a large number of noncoding RNA transcripts in eukaryotes. Noncoding transcripts account for the majority in the human genome [13], and can be divided into long noncoding RNA (lncRNA) and small RNA (siRNA, miRNA) according to the length [14]. LncRNA refers to transcripts that are more than 200 nt in length and are not involved in protein coding. They are initially defined as the “noise” of transcripts. However, with the study in-depth, they are found to participate in the regulation of gene expression at epigenetic, transcriptional, and post-transcriptional level [15,16]. LncRNA MALAT1 participates in the occurrence and development of tumor and autoimmune diseases [17,18]. However, there is still lack of reports about the role of lncRNAs MALAT1 in IDDD. This study intends to explore the role and mechanism of lncRNA MALAT1 in the occurrence of IDDD.
Materials and methods
General information
Five IDDD patients received operation in Gansu Provincial people’s Hospital between Jun 2015 and Jun 2016 were enrolled, including 3 males and 2 females with mean age at 36.2 ± 3.2 (35-49) years old. Inclusion criteria: diagnosed as IDDD by lumbar nuclear magnetic resonance imaging (MRI); degenerative grade 3 according to Christian MRI classification criteria; younger than 45 years old; underwent removal of nucleus pulposus or resection of the intervertebral disc. Exclusion criteria [9]: accompanied by other lumbar disc lesions, the presence of infectious diseases, malignancies, severe diabetes, severe liver and kidney disease, pulmonary fibrosis, bone metabolic diseases, systemic immune diseases, and malignant tumor complications. Another 6 patients with idiopathic scoliosis were selected, including 3 males and 3 females with average age at 36.1 ± 3.3 (28-45) years, excluding IDDD or other lumbar disc disease. All the patients and their families had signed informed consent. The study was approved by the medical ethics committee in Gansu Provincial people’s Hospital.
Main reagents and instruments
Type II collagenase, Trizol reagent, and IL-10 were purchased from Sigma (USA). RNA extraction kit, RT-PCR primer, reverse transcription (RT) kit, and real-time PCR reagent were purchased from Axygen (USA). DMEM/F12 medium, fetal bovine serum (FBS), and penicillin-streptomycin were purchased from Hyclone (USA). Dimethyl sulfoxide (DMSO) and MTT powder were purchased from Gibco (USA). Trypsin was purchased from Sigma (USA). IL-1 and IL-6 ELISA kits were purchased from R&D (USA). PCR amplification kit and PCR product purification kit were purchased from Promega (USA). Plasmid extraction kit and restriction enzyme were purchased from Roche (USA). Surgical microscopes were purchased from Suzhou medical equipment factory. Caspase 3 activity detection kit was purchased from Cell Signaling (USA). CO2 incubator was purchased from Sanyo (Japan). ABI7900 HT Real-time PCR was purchased from ABI (USA). Labsystem Version1.3.1 microplate reader was purchased from Bio-Rad (USA).
Primary nucleus pulposus cells isolation and cultivation
The nucleus pulposus or resected intervertebral disc obtained during surgery was placed in a sterile environment and rinsed repeatedly with 0.85% saline. The annulus fibrosus around the intervertebral disc and other non-nucleus connective tissue were removed in a sterile petri dish. Next, the specimens were rinsed with D-Hank solution to remove the blood. The nucleus pulposus was cut into pieces at l mm3 size. The tissue was then digested by 0.1% type II collagenase at 37°C for 45 min. After removing the supernatant, the tissue was centrifuged at 1500 rpm for 5 min and seeded in flask supplemented with 4 ml fresh DMEM medium. The LECs cells were seeded at 1 × 105 cells/cm2 in 6-well plate containing 10% FBS, 90% high glucose DMEM/F12 medium, 100 U/ml penicillin, and 100 μg/ml streptomycin. The cells were washed by D-Hanks solution and digested with 0.25% trypsin-0.02% EDTA for 5 to 10 min for subculture at 1:2. The cells in 2nd-5th generation were used for experiment. The cells were divided into three groups, including control group, empty plasmid group, and MALAT1 group transfected by pcDNA3.1 or pcDNA3.1-MALAT1 plasmid, respectively.
MALAT1 plasmid construction and transfection
The primers of MALAT1 sequence were designed by Primer6.0 software and inserted into pcDNA3.1 vector. The cDNA was constructed using cloned cDNA library as template and pcDNA3.1-MALAT1 containing the cleavage site. The PCR product was purified and digested to connect to pcDNA3.1 plasmid. Next, the recombinant plasmid pcDNA3.1-MALAT1 was identified by restriction enzyme digestion and sequencing. pcDNA3.1-MALAT1 and pcDNA3.1 empty plasmid were added into 200 μl serum-free medium and incubated at room temperature for 15 min, respectively. The mixed lipo2000 was added with pcDNA3.1-MALAT1 plasmid or empty pcDNA3.1 plasmid and incubated at room temperature for 30 min. The cells were washed by PBS and added with the mixture together with 1.6 ml FBS free DMEM medium. After 6 h incubation, the medium was changed and the cells were used for experiment after another 48 h incubation.
MTT assay
The cells in logarithmic phase were added with 20 μl MTT for 4 h. Then, the plate was added with 150 μl DMSO for 10 min and tested at 570 nm to obtain the absorbance value. Each experiment was repeated for three times.
ELISA
ELISA was used to test IL-1 and IL-6 contents in the serum. A total of 50 μl diluted standard substance were added to each well to establish standard curve. Next, the plate was added with 50 μl sample and washed for five times. Then the plate was incubated in 50 μl conjugate reagent at 37°C for 30 min. After washed for five times, the plate was treated by 50 μl color agent A and B at 37°C avoid of light for 30 min. At last, the plate was added with 50 μl stop buffer to stop the reaction and tested at 450 nm to obtain the OD value. The OD value of standard substance was used to prepare the linear regression equation, which was adopted to calculate the concentration of samples.
Caspase 3 activity detection
Caspase 3 activity was assessed according to the kit instruction. The cells were digested by trypsin and centrifuged at 600 g and 4°C for 5 min. After removing the supernatant, the cells were lysed on ice for 15 min and centrifuged at 20000 g and 4°C for 5 min. Next, the cells were treated by 2 mM Ac-DEVD-pNA and tested at 405 nm.
Real-time PCR
Total RNA was extracted from the cells by Trizol and reverse transcribed to cDNA. The primers were designed using Primer 6.0 software and synthetized by Invitrogen (Shanghai, China) (Table 1). Real-time PCR was performed at 35 cycles of 92°C for 30 s, 58°C for 45 s, and 72°C for 35 s. GAPDH was selected as internal reference. The relative expression of mRNA was calculated by 2-ΔCt method.
Table 1.
Primer sequences
| Gene | Forward 5’-3’ | Reverse 5’-3’ |
|---|---|---|
| GAPDH | ACCAGGTATCTTGGTTG | TAACCATGTCAGCGTGGT |
| MALAT1 | CAGGTCGTGTATCTCCGCCA | TCATACTACCAGTCTCACAC |
Statistical analysis
All data analyses were performed on SPSS19.0 software. All data were presented as mean ± standard deviation and compared by one-way ANOVA. P < 0.05 was depicted as statistical significance.
Results
LncRNA MALAT1 expression in nucleus pulposus cells from IDDD
The expression of lncRNA MALAT1 in IDDD nucleus pulposus cells was detected by Real-time PCR (original data in Supplementary Figure 1). The results showed that the expression of lncRNA MALAT1 in IDDD nucleus pulposus cells was significantly lower than that in control group (P < 0.05) (Figure 1), suggesting that lncRNA MALAT1 expression was downregulated in nucleus pulposus cells during IDDD occurrence.
Figure 1.
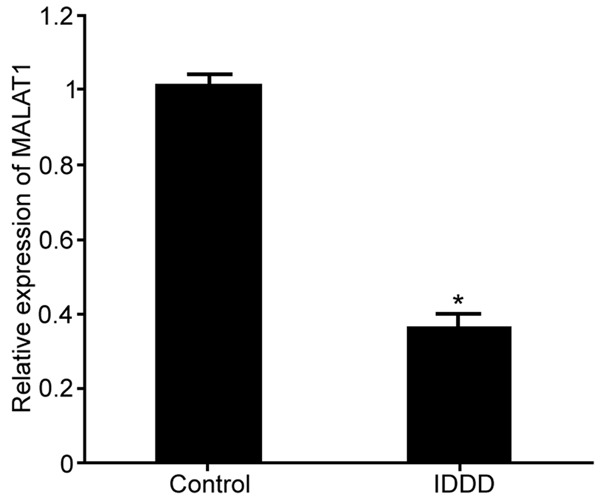
LncRNA MALAT1 expression in nucleus pulposus cells from IDDD. *P < 0.05, compared with control.
Regulation of lncRNA MALAT1 on MALAT1 expression in IDDD nucleus pulposus cells
The effect of MALAT1 plasmid on the expression of MALAT1 in IDDD nucleus pulposus cells was further detected by real-time PCR (original data in Supplementary Figure 2). The results demonstrated that MALAT1 transfection obviously promoted the expression of MALAT1 in IDDD nucleus pulposus cells compared with IDDD group (P < 0.05) (Figure 2).
Figure 2.
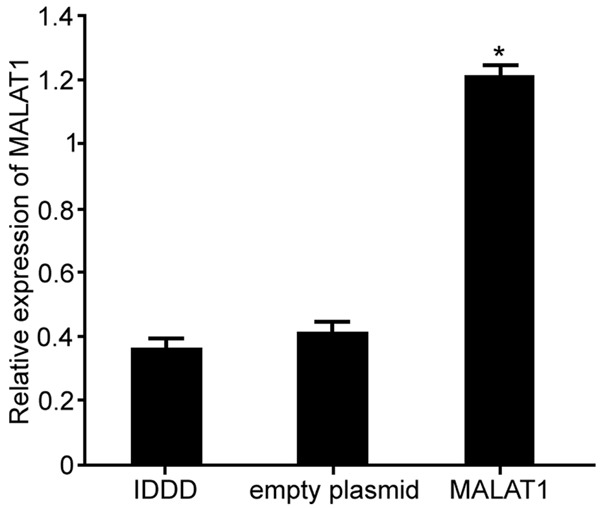
Regulation of lncRNA MALAT1 on MALAT1 expression in IDDD nucleus pulposus cells. *P < 0.05, compared with IDDD group.
The influence of lncRNA MALAT1 on the proliferation of IDDD nucleus pulposus cells
MTT assay was used to detect the effect of MALAT1 on the proliferation of IDDD nucleus pulposus cells (original data in Supplementary Figure 3). The results revealed that the expression of MALAT1 was up-regulated by MALAT1, which markedly facilitated the proliferation of IDDD nucleus pulposus cells compared with IDDD group (P < 0.05) (Figure 3). It indicated that regulation of lncRNA MALAT1 can affect the proliferation of IDDD nucleus pulposus cells.
Figure 3.
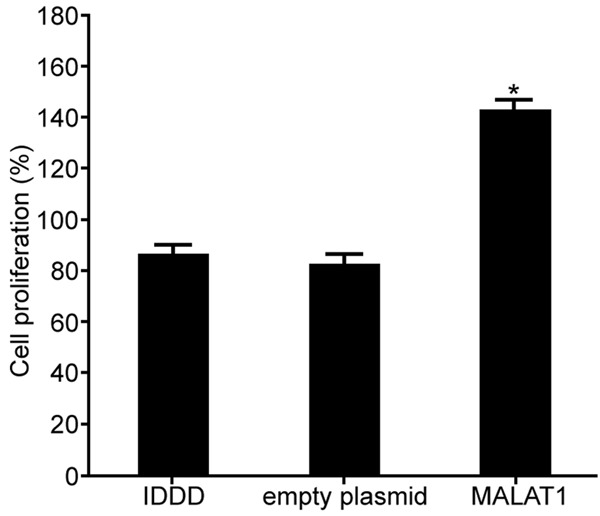
The influence of lncRNA MALAT1 on the proliferation of IDDD nucleus pulposus cells. *P < 0.05, compared with IDDD group.
The impact of lncRNA MALAT1 on the activity of Caspase 3 in IDDD nucleus pulposus cells
The effect of lncRNA MALAT1 plasmid transfection on the activity of Caspase 3 in nucleus pulposus cells was assessed by Caspase 3 activity kit (original data in Supplementary Figure 4). The results exhibited that the expression of MALAT1 was upregulated after lncRNA MALAT1 transfection, which significantly inhibited the activity of Caspase 3 in nucleus pulposus cells compared with IDDD group (P < 0.05) (Figure 4). The results showed that regulation of lncRNA MALAT1 expression affected IDDD nucleus pulposus cell apoptosis, thereby influencing cell proliferation.
Figure 4.
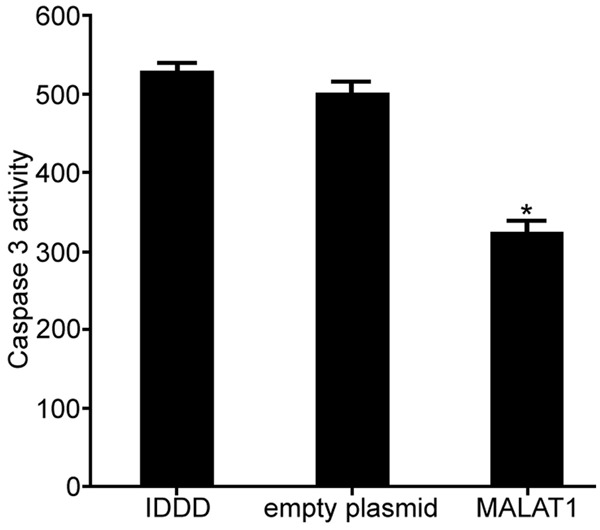
The impact of lncRNA MALAT1 on the activity of Caspase 3 in IDDD nucleus pulposus cells. *P < 0.05, compared with IDDD group.
The influence of lncRNA MALAT1 on the expression of inflammatory factors in nucleus pulposus cells
The expression of IL-1 and IL-6 in the supernatant of nucleus pulposus cells was detected by ELISA (original data in Supplementary Figure 5). The results showed that upregulation of MALAT1 obviously suppressed the expression of IL-1 and IL-6 in the nucleus pulposus cells compared with IDDD group (P < 0.05) (Figure 5). The above results suggested that regulation of lncRNA MALAT1 restrained the secretion of inflammatory factors in IDDD nucleus pulposus cells, thereby regulating the progression of IDDD.
Figure 5.
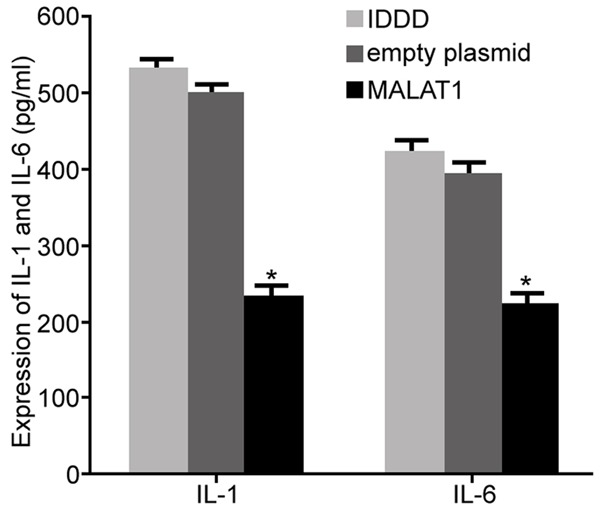
The influence of lncRNA MALAT1 on the expression of inflammatory factors in nucleus pulposus cells. *P < 0.05, compared with IDDD group.
Discussion
LncRNA is the most transcribed non-protein coding sequence in the genome [19]. It was found that lncRNA can be regulated at gene level and participates in physiological activities in a variety of ways, including chromatin modification, genomic blotting, intranuclear transport, chromosomal gene silencing, transcriptional activation, etc. [20]. LncRNA regulates the normal and pathological states of cells, such as growth, proliferation, cell cycle, and apoptosis. Therefore, lncRNA is an important regulatory factor in the development and progression of human disease [20,21]. LncRNA MALAT1 has been shown to be expressed in many normal tissues, but significantly reduced in a wide range of diseases, including tumors and diabetes [18]. However, the role and mechanism of lncRNA MALAT1 in IDDD has not been reported. This study confirmed that lncRNA MALAT1 obviously declined in nucleus pulposus cells of IDDD patients, suggesting that lncRNA MALAT1 involved in the occurrence and development of IDDD.
In this study, we further observed that after transfecting pcDNA3.1-MALAT1 plasmid to IDDD nucleus pulposus cells markedly upregulated MALAT1 expression, inhibited cell proliferation, increase the activity of Caspase 3, facilitated inflammatory factors IL-1 and IL-6 secretion. Most studies revealed that nucleus pulposus cell as one type of the chondrocytes play a significant role in the intervertebral disc lesions. Nucleus pulposus cells account for more than half of the intervertebral discs [22]. During the IDDD lesions, inflammatory factors participate in the metabolism of extracellular matrix and cell proliferation. Increased secretion of inflammatory factors cause the cell matrix cannot supply nutrition, leading to nucleus pulposus damage, nucleus pulposus cells reduce and structural injury, decreased nucleus pulposus elasticity, and abnormal disc function [23,24]. The proteoglycan (GAG) and type II collagen in the nucleus pulposus of the intervertebral discs can maintain the elasticity of the intervertebral disc nucleus and ensure the intervertebral disc tolerance [25,26]. Enhanced secretion of inflammatory factors leads to inflammation, induces nucleus pulposus cells apoptosis, and promotes nucleus pulposus damage [27,28]. This study investigated the expression and role of lncRNA MALAT1 in nucleus pulposus cells of IDDD patients, while its specific mechanism needs further exploration.
Conclusion
MALAT1 level decreased in IDDD nucleus pulposus cells. Upregulation of MALAT1 expression restrained IDDD through suppressing inflammation, inhibiting nucleus pulposus cell apoptosis, and promoting cell proliferation.
Acknowledgements
This work was supported by Shaanxi Province Natural Science Basic Research Project, 2016JQ8052.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Wu X, Liu W, Duan Z, Gao Y, Li S, Wang K, Song Y, Shao Z, Yang S, Yang C. The involvement of protease nexin-1 (PN1) in the pathogenesis of intervertebral disc (IVD) degeneration. Sci Rep. 2016;6:30563. doi: 10.1038/srep30563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xue JB, Zhan XL, Wang WJ, Yan YG, Liu C. OPG rs2073617 polymorphism is associated with upregulated OPG protein expression and an increased risk of intervertebral disc degeneration. Exp Ther Med. 2016;12:702–710. doi: 10.3892/etm.2016.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shao J, Yu M, Jiang L, Wei F, Wu F, Liu Z, Liu X. Differences in calcification and osteogenic potential of herniated discs according to the severity of degeneration based on Pfirrmann grade: a cross-sectional study. BMC Musculoskelet Disord. 2016;17:191. doi: 10.1186/s12891-016-1015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson CA, Roffey DM, Chow D, Alkherayf F, Wai EK. A systematic review of preoperative predictors for postoperative clinical outcomes following lumbar discectomy. Spine J. 2016;16:1413–1422. doi: 10.1016/j.spinee.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Capoor MN, Ruzicka F, Machackova T, Jancalek R, Smrcka M, Schmitz JE, Hermanova M, Sana J, Michu E, Baird JC, Ahmed FS, Maca K, Lipina R, Alamin TF, Coscia MF, Stonemetz JL, Witham T, Ehrlich GD, Gokaslan ZL, Mavrommatis K, Birkenmaier C, Fischetti VA, Slaby O. Prevalence of propionibacterium acnes in intervertebral discs of patients undergoing lumbar microdiscectomy: a prospective cross-sectional study. PLoS One. 2016;11:e0161676. doi: 10.1371/journal.pone.0161676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lv X, Liu Y, Zhou S, Wang Q, Gu H, Fu X, Ding Y, Zhang B, Dai M. Correlations between the feature of sagittal spinopelvic alignment and facet joint degeneration: a retrospective study. BMC Musculoskelet Disord. 2016;17:341. doi: 10.1186/s12891-016-1193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang F, Jiang D. IL-1beta/HMGB1 signalling promotes the inflammatory cytokines release via TLR signalling in human intervertebral disc cells. Biosci Rep. 2016:36. doi: 10.1042/BSR20160118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nisolle JF, Bihin B, Kirschvink N, Neveu F, Clegg P, Dugdale A, Wang X, Vandeweerd JM. Prevalence of age-related changes in ovine lumbar intervertebral discs during computed tomography and magnetic resonance imaging. Comp Med. 2016;66:300–307. [PMC free article] [PubMed] [Google Scholar]
- 9.Miles DE, Mitchell EA, Kapur N, Beales PA, Wilcox RK. Peptide: glycosaminoglycan hybrid hydrogels as an injectable intervention for spinal disc degeneration. J Mater Chem B Mater Biol Med. 2016;4:3225–3231. doi: 10.1039/c6tb00121a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant MP, Epure LM, Bokhari R, Roughley P, Antoniou J, Mwale F. Human cartilaginous endplate degeneration is induced by calcium and the extracellular calcium-sensing receptor in the intervertebral disc. Eur Cell Mater. 2016;32:137–151. doi: 10.22203/ecm.v032a09. [DOI] [PubMed] [Google Scholar]
- 11.Walter BA, Purmessur D, Moon A, Occhiogrosso J, Laudier DM, Hecht AC, Iatridis JC. Reduced tissue osmolarity increases TRPV4 expression and pro-inflammatory cytokines in intervertebral disc cells. Eur Cell Mater. 2016;32:123–136. doi: 10.22203/ecm.v032a08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou PH, Wang ST, Ma HL, Liu CL, Chang MC, Lee OK. Development of a two-step protocol for culture expansion of human annulus fibrosus cells with TGF-beta1 and FGF-2. Stem Cell Res Ther. 2016;7:89. doi: 10.1186/s13287-016-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Shen Q, Zhang X, Yang C, Cui S, Sun Y, Wang L, Fan X, Xu S. The long non-coding RNA XIST controls non-small cell lung cancer proliferation and invasion by modulating miR-186-5p. Cell Physiol Biochem. 2017;41:2221–2229. doi: 10.1159/000475637. [DOI] [PubMed] [Google Scholar]
- 14.Lin J, Zhan Y, Liu Y, Chen Z, Liang J, Li W, He A, Zhou L, Mei H, Wang F, Huang W. Increased expression of ZEB1-AS1 correlates with higher histopathological grade and promotes tumorigenesis in bladder cancer. Oncotarget. 2017;8:24202–24212. doi: 10.18632/oncotarget.15527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z, Chen Z, Fan R, Jiang B, Chen X, Chen Q, Nie F, Lu K, Sun M. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Mol Cancer. 2017;16:82. doi: 10.1186/s12943-017-0651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu F, Zhang J. Long non-coding RNA HOTAIR functions as miRNA sponge to promote the epithelial to mesenchymal transition in esophageal cancer. Biomed Pharmacother. 2017;90:888–896. doi: 10.1016/j.biopha.2017.03.103. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Li J, Tang N. Long noncoding RNA Malat1 is a potent autophagy inducer protecting brain microvascular endothelial cells against oxygen-glucose deprivation/reoxygenation-induced injury by sponging miR-26b and upregulating ULK2 expression. Neuroscience. 2017;354:1–10. doi: 10.1016/j.neuroscience.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 18.Hu M, Wang R, Li X, Fan M, Lin J, Zhen J, Chen L, Lv Z. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with beta-catenin. J Cell Mol Med. 2017 doi: 10.1111/jcmm.13189. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xin JW, Jiang YG. Long noncoding RNA MALAT1 inhibits apoptosis induced by oxygen-glucose deprivation and reoxygenation in human brain microvascular endothelial cells. Exp Ther Med. 2017;13:1225–1234. doi: 10.3892/etm.2017.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Z, Rayile A, Zhang X, Li Y, Zhao Q. Ulinastatin protects against lipopolysaccharide-induced cardiac microvascular endothelial cell dysfunction via downregulation of lncRNA MALAT1 and EZH2 in sepsis. Int J Mol Med. 2017;39:1269–1276. doi: 10.3892/ijmm.2017.2920. [DOI] [PubMed] [Google Scholar]
- 21.Huo Y, Li Q, Wang X, Jiao X, Zheng J, Li Z, Pan X. MALAT1 predicts poor survival in osteosarcoma patients and promotes cell metastasis through associating with EZH2. Oncotarget. 2017;8:46993–47006. doi: 10.18632/oncotarget.16551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li D, Yue J, Jiang L, Huang Y, Sun J, Wu Y. Correlation between expression of high temperature requirement serine protease A1 (HtrA1) in nucleus pulposus and T2 value of magnetic resonance imaging. Med Sci Monit. 2017;23:1940–1946. doi: 10.12659/MSM.904018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun D, Liu P, Cheng J, Ma Z, Liu J, Qin T. Correlation between intervertebral disc degeneration, paraspinal muscle atrophy, and lumbar facet joints degeneration in patients with lumbar disc herniation. BMC Musculoskelet Disord. 2017;18:167. doi: 10.1186/s12891-017-1522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang L, Yang C, Song Y, Zhao K, Liu W, Hua W, Wang K, Tu J, Li S, Yin H, Zhang Y. MicroRNA-494 promotes apoptosis and extracellular matrix degradation in degenerative human nucleus pulposus cells. Oncotarget. 2017;8:27868–27881. doi: 10.18632/oncotarget.15838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruber HE, Marrero E, Ingram JA, Hoelscher GL, Hanley EN Jr. Constitutive expression of IL-22 in the human intervertebral disc and its reduction by exposure to pro-inflammatory cytokines in vitro. Biotech Histochem. 2017;92:222–229. doi: 10.1080/10520295.2017.1300834. [DOI] [PubMed] [Google Scholar]
- 26.Shan Z, Lin X, Wang S, Zhang X, Pang Y, Li S, Yu T, Fan S, Zhao F. An injectable nucleus pulposus cell-modified decellularized scaffold: biocompatible material for prevention of disc degeneration. Oncotarget. 2017;8:40276–40288. doi: 10.18632/oncotarget.16831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Liu W, Song Y, Wu X, Zhang Y, Li S, Gao Y, Tu J, Liu Y, Yang C. The role of angiopoietin-2 in nucleus pulposus cells during human intervertebral disc degeneration. Lab Invest. 2017;97:971–982. doi: 10.1038/labinvest.2017.35. [DOI] [PubMed] [Google Scholar]
- 28.Bian Q, Ma L, Jain A, Crane JL, Kebaish K, Wan M, Zhang Z, Edward Guo X, Sponseller PD, Seguin CA, Riley LH, Wang Y, Cao X. Mechanosignaling activation of TGFbeta maintains intervertebral disc homeostasis. Bone Res. 2017;5:17008. doi: 10.1038/boneres.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


