Abstract
Galectin-4 is a member of multifunctional galactoside-binding lectin family with various biological functions, including tumor cells proliferation, cancer progression, cell adhesion, and tumor metastasis. In this study, we aimed to investigate the putative function of galectin-4 in tumor progression of hepatocellular carcinoma (HCC), and explore the possible mechanisms of galectin-4 related biological pathway. The result showed that the galectin-4 was significantly up-regulated in HCC tissues/cell lines compared to matched peritumor tissues/a normal liver cell line, respectively. Furthermore, overexpressing galectin-4 in HCCLM3 cells led to promotion of cell proliferation and inhibition of cell apoptosis via up-regulation of Cyclin D1, down-regulation of p21 and decreased pro-apoptotic Bax/Bcl-2 ratio, But knockdown of galectin-4 reversed these properties. Furthermore, galectin-4 could promote cell-cell adhesion via stabilizing the adherens junction complexes. These observations was further validated in the xenograft animal model. Taken together, galectin-4 plays an important role in HCC progression.
Keywords: Galectin-4, cell proliferation, cell apoptosis, cell adhesion, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC), one of most common cancers worldwide, ranks the third cancer-related deaths in China [1]. Although there are several advanced curative interventions (such as surgical resection, liver transplantation and target therapy) for diagnosis and management of HCC, the prognosis of patients remains poor due to cancer relapse and metastases [2-4]. Only 10 to 20% of patients with HCC tumors can receive resection at the time of diagnosis. This grim outlook has stimulated the investigation of cancer-related oncogenes and tumor suppressors, aiming to elucidate the biological basis of HCC and improve the diagnosis and management for HCC patients.
Recently, galectins, a family of glycan-binding proteins, have been emerged as pivotal regulators in the pathogenesis of HCC. Members of this family, including galectin-1, galectin-4, galectin-8 and galectin-9, are commonly aberrantly expressed in HCC and implicated in a variety of biological functions, including roles in cell adhesion, migration, cytokine synthesis, and survival [5-9]. Galectin-4, a tandem-repeat galectin, is predominantly expressed in healthy epithelium of the gastrointestinal tract, and serves as a lipid raft and adherens junction stabilizer with its glycan cross-linking capacity [10]. Recent studies have shown that aberrant level of galectin-4 was associated with several cancers [11]. According to previous studies, galectin-4 was down-regulated in colon cancer, absent in invasive carcinoma, and up-regulated in the pancreatic adenocarcinoma, hepatocellular carcinoma, lung cancer and so on [12-16]. These results suggested that galectin-4 played an important role in cancer progression and metastasis and thus could be used as a diagnosis/prognosis marker or drug target. Our pervious study reported that in HCC, galectin-4 expression was significantly down-regulated in tissues from early recurrent/metastatic HCC patients, when compared to non-recurrent/metastatic HCC patients [7]. However, the aberrant expression of galectin-4 in HCC tissue and the precise function of galectin-4 in HCC have not been fully characterized.
In this study, we aimed to evaluate the expression difference of galectin-4 between the HCC tissue and matched peritumor, and systematically investigated the function of galectin-4 by assessing the effect of knockdown and overexpression of galectin-4 on proliferation, cell cycle, cell apoptosis and cell adhesions in HCC cells in vitro and vivo.
Materials and methods
Clinical samples
A total of 20 paired HCC and matched peritumor tissues obtained from HCC patients receiving surgical resection, were evaluated for both mRNA and protein expression of galectin-4. Peritumor tissues were ≥2 cm away from the edge of the tumors. All tissues were obtained from patients that had undergone partial hepatectomy at Mengchao Hepatobiliary Hospital of Fujian Medical University between 2013 and 2014. Patients that had undergone treatment prior to surgery were excluded from the study. HCC and matched peritumor tissues were confirmed by pathological examination, and immediately stored in liquid nitrogen post-surgery. Written informed consent was obtained from either the patients or the families of the patients.
Cell culture
Human HCC cells (HepG2, Huh-7, and sk-hep-1) were purchased from the American Type Culture Collection. Additional human HCC cells (HL7702, MHCC97L, and HCCLM3) and Immortalized liver cell line (HL7702) were kind gifts from the Second Military Medical University of China (Shanghai, China). All of the cells were cultured in Dulbecco’s modified Eagle medium (DMEM) at 37°C in a 5% CO2 incubator. The medium was supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin.
Quantitative real-time RT-PCR
Total RNA were isolated from fresh-frozen HCC and matched peritumor tissues using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription was carried out using the Goscript Reverse Transcription System Kit (A5001; Promega, Madison, WI, USA) according to the manufacturer’s instructions. The primers for galectin-4 and the internal control gene (β-actin) were designed using the Primer-Blast Database (galectin-4 forward primer, CCCTTCTATGAGTACGGGCAC; galectin-4 reverse primer, TGGCCTCCGATGAAGTTGATT; β-actin forward primer, ATAGCACAGCCTGGATAGCAACGTAC; β-actin reverse primer, CACCTTCTACAATGAGCTGCGTGTG). For q-PCR analysis, aliquots of double-stranded cDNA were amplified using the GoTaq q-PCR Master Mix (A6002; Promega, Madison, WI, USA). The cycling parameters were 35 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 30 s. The relative gene expression was normalized to the geometric mean of the housekeeping gene β-actin, and calculated according to the Livak method (2-ΔΔCt). The experiments were independently repeated three times. Student’s t test was used to determine the significance of differences in comparisons by GragphPad Prism6.
Western blotting
After grinding, the fresh-frozen HCC tissues, matched peritumor tissues or HCC cell lines were lysed in ice-cold RIPA buffer (0.5 M Tris-HCl, 1.5 M NaCl (pH 7.4), 2.5% deoxycholic acid, 10% NP-40, 10 mM EDTA) containing protease inhibitor cocktail (Roche, Indianapolis, IN, USA). Tissue lysates were centrifuged at 17000 g for 30 min at 4°C. The extracts were quantified by BCA assay. Proteins of each sample (40 µg) were separated by 12% SDS-PAGE and transferred onto nitrocellulose membranes. Afterwards, the membranes were blocked for 2 h in the TBST buffer with 5% BSA and then probed with the galectin-4 primary antibody (1:500; Santa Cruz Biotechnology, Santa Cruz, CA, USA), the Cyclin D1, p21, Bax, Bcl-2, E-cadherin and β-catenin antibodies (1:1000, Cell Signaling Technology, Beverly, MA, USA), and β-actin antibody (1:2000; Transgen, Beijing, China) overnight at 4°C. The membranes were washed three times with TBST buffer for 10 min each time, then incubated with appropriate HRP conjugated secondary antibody (1:5000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h at room temperature. After washed again in the TBST buffer, the membranes were utilized for detection of protein expression levels by enhanced chemiluminescence and the results were visualized by autoradiography.
Lentivirus construction and the establishment of stable overexpression/knockdown galectin-4 cells
Complementary DNA of galectin-4 gene (GenBank, U82953.1) was cloned into PCDH-cmv-ef1-puro-eGFP vector for overexpression (designated as LV-Gal4); the empty vector was used as negative control (designated as LV-control). shRNA targeted for galectin-4 sequence was designed by Rosetta using a proprietary algorithm (shGal4, 5’-GGAAAGTCCCCGTTTATGAAA-3’). Lentiviral vectors encoding shGa4 and shNon were generated with pGreen-Puro (designated as LV-shGal4 and LV-shNon). Lentiviral particles were produced using the third-generation ViraPower (Invitrogen Corporation, Grand Island, NY, USA) according to the manufacturer’s instructions.The obtained particles were transfected into HCCLM3 cells with a multiplicity of infection (MOI) of 40 to 60 in the presence of polybrene (10 μg/ml). Transfected cells were selected for 2 weeks (start from 48 hours after transfection) with 4 μg/ml puromycin (Origene, Rockville, MD, USA). Four pooled populations of overexpression cells and knockdown cells (LV-control vs LV-Gal4, LV-shNon vs LV-shGal4), which were obtained 2 weeks after drug selection without subcloning, were both used in vitro and vivo experiments.
Cell viability assays
HCCLM3 cells (2×103) from above four groups were seeded onto 96-well plates. After 24 h, 10 µl of Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Santa Clara, CA, USA) reagent was added into each plate. 2 hours later, the optical densities (ODs) were determined using a microplate reader (MD) at 450 nm. Samples were then measured after 5 days of culture, likewise. The experiments were repeated independently five times. The cell viability curve of each group was drawn based on the absorbance at 450 nm. Student’s t test was used to determine the significance of differences in comparisons by GragphPad Prism6.
Cell cycle assays
The cell cycle of HCCLM3 cells from above four groups was detected by flow cytometry using propidium iodide (PI) staining according to the manufacturer’s instructions. The cells were harvested and adjusted to a concentration of 1×106 cells/ml, then fixed in 70% ethanol at 4°C overnight. The fixed cells were washed twice with cold PBS and then incubated for 30 min with RNase (8 µg/ml) and PI (10 µg/ml). The fluorescent signal was detected through the FL2 channel and the proportion of DNA in various phases was analyzed using ModfitLT Version 3.0 (Verity Software House, Topsham, ME, USA). The experiments were independently repeated three times. Student’s t test was used to determine the significance of differences in comparisons by GragphPad Prism6.
Cell apoptosis assays
Apoptosis of HCCLM3 cells from above four groups was determined by flow cytometric analysis using Annexin V-PE/7AAD kit (Becton-Dickinson, San Jose, CA, USA) according to the manufacturer’s instructions. Annexin V-positivity and 7AAD-negativity or Annexin V/7AAD double-positivity indicated the presence of early or late apoptotic cells, respectively. Each experiment was performed in triplicate and repeated a minimum of three times. Student’s t test was used to determine the significance of differences in comparisons by GragphPad Prism6.
Immunofluorescence
Cells (5×104) were seed in the confocal laser dishes (diameter = 35 mm) and cultured for 24 hrs to allow cell attachment. Afterward, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 and then incubated with E-cadherin antibody and β-catenin antibody (1:100; Cell Signaling Technology, Beverly, MA, USA) at 4°C overnight, respectively. Then, the cells were washed twice with cold PBS and incubated with PE-conjugated goat anti-rabbit IgG in the dark for 60 min at room temperature. After wash with twice with cold PBS, the cell nuclei were stained with Dapi (Molecular Probes). Subsequently, the cells were imaged by a confocal microscope (LSM 780, Germany).
Tumor cell xenograft
Animal experimental protocols were approved by the Institutional Animal Care and Use Committee of Fujian medical University (Fuzhou, China). In total, 20 male BALB/c nude mice (6 weeks, 18-20 g) were purchased from Wushi Experimental Animal Center. The animals were raised in pathogen-free experimental animal rooms with free access to water and food. The mice were randomly divided into four groups, each group including 5 mice. For in vivo growth assays, 4×106 HCCLM3 cells of above four groups suspended in 100 μl of phosphate-buffered saline were subcutaneously injected into the dorsal flank of the nude mice, respectively (Day 0). Once palpable tumors (about 10 days) were formed, tumor growth was monitored every three day using a linear caliper, and the volume (V) was estimated using the formula Volume = (a×b2)/2 mm3 (a, longer diameter of the tumor; b, shorter diameter of the tumor). After 30 days, these mice were killed by Cervical Dislocation (Day 30). Tumors were excised, measured, weighed and photographed. Student’s t test was used to determine the significance of differences in comparisons by GragphPad Prism6.
Ki-67 staining
The paraffin-embedded isolated tumor tissues were cut into 4-μm-thick serial sections. Slides were deparaffinized in xylene, rehydrated in graded alcohol. The slides were incubated in 1% hydrogen peroxide for 30 min to block endogenous peroxidase activity and then rehydrated in distilled water followed by PBS. After heat-induced epitope retrieval in 0.1 mol/L EDTA (pH 9.0) for 3 min, the slides were incubated with a primary antibody ki67 (monoclonal mouse; 1:100; Santa Cruz, CA, USA) for 1 hour at room temperature. Subsequently, slides were incubated with HRP-labeled secondary antibody (PV9002; ZSGB-BIO, Beijing, China) at room temperature for another 1 h. DAB (3, 3-diaminobenzidine) was used as a chromagen, and sections were counterstained with hematoxylin, dehydrated and mounted. The results were independently assessed by two pathologists double-blindly. Quantification of hepatocyte proliferation was determined by immunostaining for the proliferation marker Ki67 in tumor sections.
Statistical analysis
Each assay was performed at least three times, and data were presented as the mean ± standard deviation. Student’s t test and analysis of variance were used to determine the significance of differences in comparisons. All data were analyzed with GragphPad Prism6. P<0.05 was considered threhold for statistically significant difference.
Results
Galectin-4 was up-regulated in HCC tissues and HCC cell lines
To investigate the expression of galectin-4 in HCC tissue, we assessed its expression in 20 paired HCC patients at both mRNA and protein levels. Results of q-PCR and western blotting analysis revealed that galectin-4 was significantly up-regulated (3.33-fold in mRNA level and 3.27-fold in protein level, respectively) in tumor tissues, compared to their matched peritumor tissues (Figure 1A and 1B).
Figure 1.
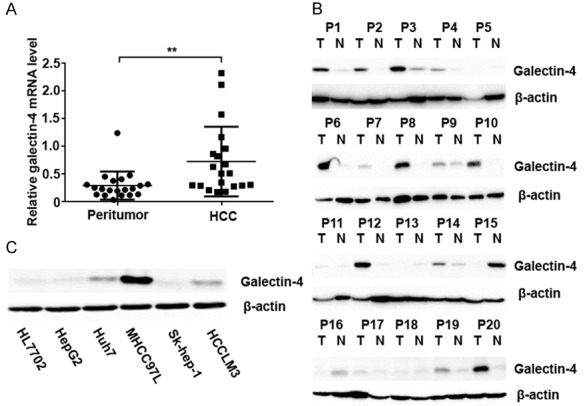
Galectin-4 was significantly up-regulated in HCC samples. A. Quantitative PCR analysis of galectin-4 expression in HCC and matched peritumor tissues. B. Western blotting analysis of galectin-4 expression in HCC and matched peritumor tissues. C. Western blotting analysis of galectin-4 expression in different HCC cell lines. *P<0.05. T: tumor; N: peritumor.
Furthermore, the expression of galectin-4 was also evaluated in six HCC cell lines (HL7702, HepG2, Huh7, MHCC97L, Sk-hep-1, and HCCLM3). As shown in Figure 1C, HCC cell lines (Huh7, MHCC97L, and HCCLM3) expressed higher level of galectin-4 compared with the normal hepatic epithelium cell line (HL7702). Overall, these results clearly indicated that galectin-4 expression was significantly increased at both mRNA and protein levels in HCC tumors.
Galectin-4 overexpression promoted cell proliferation in vitro
To explore the functional role of galectin-4 in HCC progression, we selected HCCLM3 cells with the exhibition of moderate level of galectin-4 expression to establish stable galectin-4 overexpression and knockdown cell model by lentivirus transduction. Furthermore, to investigate the effect of galectin-4 expression on cell proliferation, cell viability curves were measured using CCK-8 assays. HCCLM3 cells with overexpression of galectin-4 showed higher cell viability compared to the negative control cells, while knockdown of galectin-4 led to significant decrease of cell viability in HCCLM3 cells (Figure 2A). As cell viability directly linked to cell proliferation, we explored the effect of galectin-4 on cell cycle progression using flow cytometry. As shown in Figure 2B, galectin-4 overexpression cells showed increased percentage of G2/M phase cells and decreased percentage of G1 phase cells. This reversed tendency was found in the HCCLM3 cells with galectin-4 knockdown. These results suggest that galectin-4 may play an important role in proliferation promotion effect of HCC. To confirm the mechanism of galectin-4 on cell cycle, we detected the expression of cell cycle associated proteins TP53, p21, p27, cyclin D1. As shown in Figure 2C, galectin-4 overexpression markedly increased the expression of P21 and decreased the expression of cyclin D1. Meanwhile, galectin-4 knockdown significantly suppressed the expression of P21 and increased the expression of cyclin D1. However, it did not significantly change the TP53 and P27 levels in HCC cells (data no shown). These results suggested that galectin-4 may promote the cell cycle progression in HCC cells via regulating the expression of cyclin D1 and P21.
Figure 2.
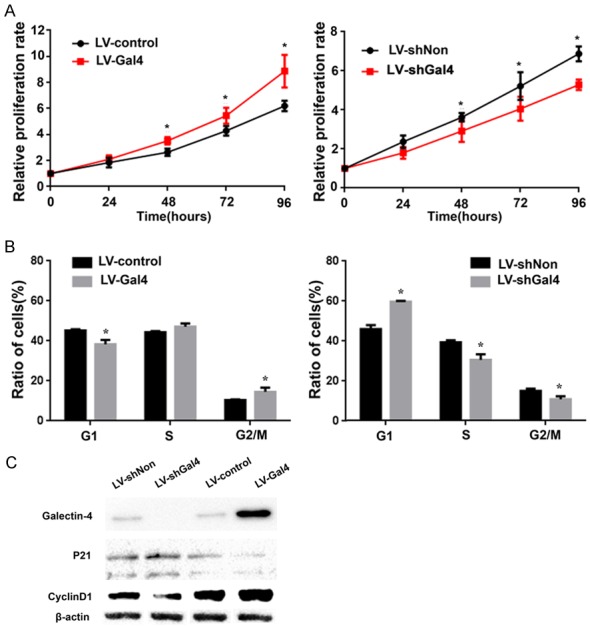
Galectin-4 promoted HCC cell proliferation by regulation of p21 and Cyclin D1 Bcl-2 in vitro. A. CCK-8 assay was performed to measure cell viability in HCCLM3 cells with stable galectin-4 overexpression/knockdown. B. Flow cytometry was used to analyze the cell cycle in HCCLM3 cells with stable galectin-4 overexpression/knockdown. C. Western blotting analysis of p21 and cyclin D1 in HCCLM3 cells with stable galectin-4 overexpression/knockdown. *P<0.05.
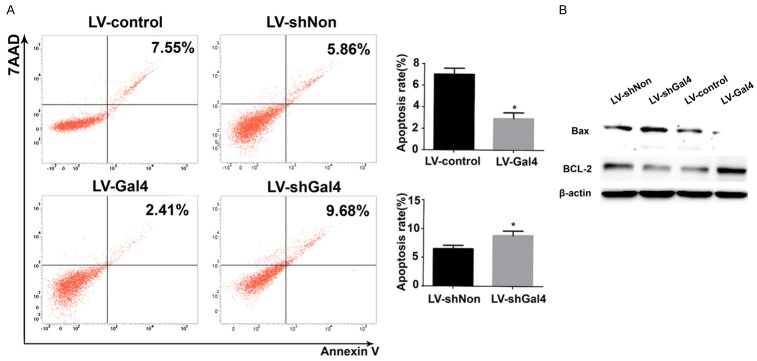
Galectin-4 overexpression promoted cell apoptosis in vitro
To investigate whether the effect of galectin-4 overexpression on the promotion of cell growth was associated with an induction of apoptosis, the Annexin V-7AAD binding assay was performed. After 72 hours, the cells were collected for apoptosis analysis by flow cytometry. Analysis of the proportion of apoptotic cells revealed that the percentage of galectin-4 overexpressing cells with undergoing apoptosis significantly decreased. Meanwhile, the opposite trend was observed in galectin-4 silence cells (Figure 3A and 3B). Moreover, we further determined potential target proteins involved in cell apoptosis. The results suggested that high expression of galectin-4 led to decreasing the expression of pro-apoptotic Bax, while increasing the expression of anti-apoptotic Bcl-2, which resulted in inhibiting cell apoptosis (Figure 3C).
Figure 3.
Galectin-4 inhibited HCC cell apoptosis by regulation of Bax and Bcl-2 in vitro. A. Flow cytometry was used to analyze the cell apoptosis in HCCLM3 cells with stable galectin-4 overexpression/knockdown. B. Western blotting analysis of Bax and Bcl-2 in HCCLM3 cells with stable galectin-4 overexpression/knockdown. *P<0.05.
Galectin-4 overexpression promoted cell adhesions in vitro
Galectin-4 is located at apical membrane, suggesting a role in cell adhesions and cell migration [17,18]. During cell culture, we noticed that HCCLM3 cells overexpressing galectin-4 exhibited an overcrowded morphology, whereas HCCLM3 cells with galectin-4 knockdown showed a loose morphology compared with its corresponding control cells (Figure 4A). These morphological changes indicated that galectin-4 might be involved in the cell adhesions. To get insight into the signaling pathways mediating the adhesion effect of galectin-4, we examined the expression of adherence junction components by immunofluorescence. E-cadherin and β-catenin were presented diffusely in the cytoplasm and expressed at cell-cell contacts of control HCCLM3 cells. In galectin-4 HCCLM3 cells, signals of E-cadherin and β-catenin were positive along sites of cell-cell adhesion, whereas HCCLM3 cells with galectin-4 knockdown showed a decreased expression compared with its corresponding control cells. Furthermore, the Western blotting results also showed that E-cadherin and β-catenin were significantly increased after galectin-4 overexpression in HCCLM3 cells, while they were significantly depressed by galectin-4 knockdown. These result suggested that galectin-4 may act as a regulator of adherens junction stability.
Figure 4.
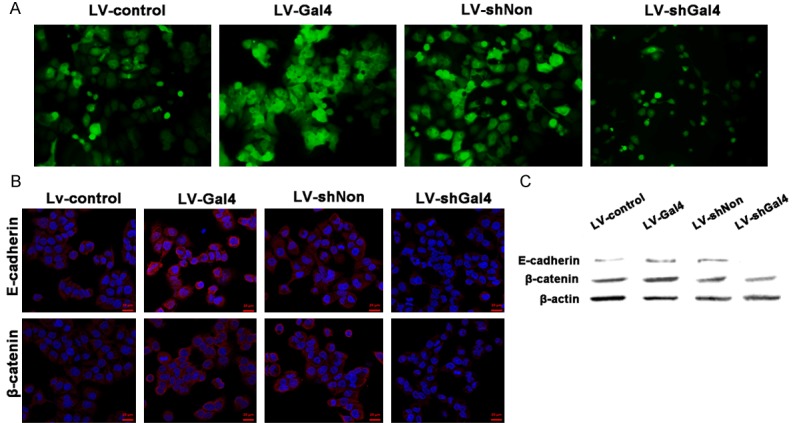
Galectin-4 promoted cell adhesion by stabilizing adherens junction complexes in vitro. A. Morphology of HCCLM3 cells with stable galectin-4 overexpression/knockdown. B. Immunofluorescence analysis of E-cadherin and β-catenin in HCCLM3 cells with stable galectin-4 overexpression/knockdown. C. Western blotting analysis of E-cadherin and β-catenin in HCCLM3 cells with stable galectin-4 overexpression/knockdown.
Galectin-4 overexpression promoted tumor growth in vivo
To determine the efficacy of galectin-4 in tumor growth in vivo, we performed experiments with a HCC xenograft animal model. As shown in Figure 5A and 5B, mice injected with galectin-4 overexpressing cells showed a significant increase in tumor growth, compared with those injected with negative control cells (P = 0.028). Oppositely, knockdown of galectin-4 exhibited a decreased capacity for tumorigenesis in the xenograft animal model (P = 0.002). Furthermore, since the Ki-67 protein is a well-established biomarker for cell proliferation, the isolated tumor tissues were subjected to IHC staining for Ki-67. The results showed that the Ki-67 expression significantly increased in galectin-4 overexpression-group compared to control-group, and the tumors with knockdown of galectin-4 reversed the effect (Figure 5C). These data indicated that galectin-4 positively correlated with tumor growth.
Figure 5.
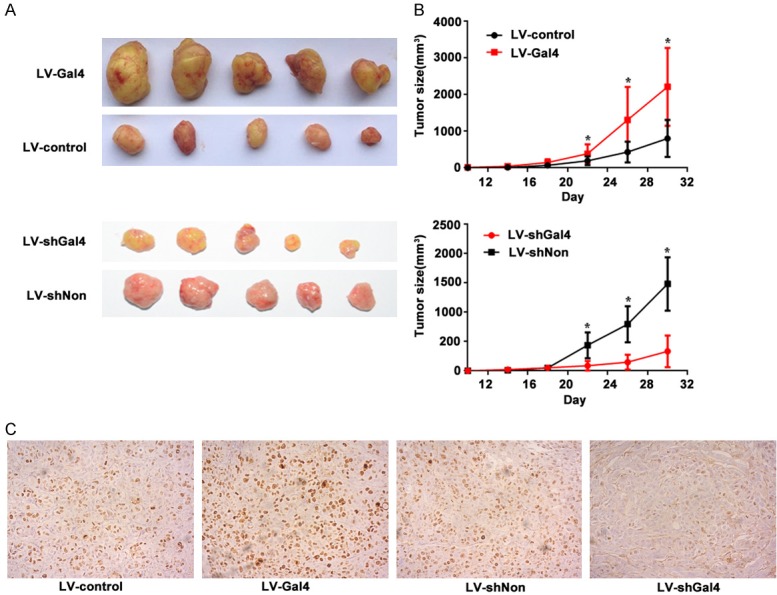
Galectin-4 promoted HCC tumor growth in vivo. A. Macroscopic appearance of representative tumor specimens from HCC xenograft model that was injected with LV-control, LV-Gal4, LV-shNon and LV-Gal4 cells, respectively. B. Tumor growth curves for the xenograft model that was injected with LV-control, LV-Gal4, LV-shNon and LV-Gal4 cells. Data were mean ± SEM of 5 animals per group. C. Representative Ki67 stained tumor sections from xenograft model that was injected with LV-control, LV-Gal4, LV-shNon and LV-Gal4 cells, respectively. *P<0.05.
Discussion
Galectin-4, defined as a multifunctional member of galectin family, is of great importance in the biological behavior of various tumors [19]. It has been reported that reduced level of galectin-4 could promote the invasive behavior in colon adenocarcinoma and pancreatic cancer [20,21]. And our previous report have shown an association between galectin-4 expression and HCC [7]. Down-regulation of galectin-4 could promote recurrence/metastasis of HCC. In contrast, the possible promoting metastasis role of galectin-4 is identified in lung adenocarcinoma [12]. Taken together, these results suggest that dysregulation of galectin-4 has an important influence on tumor development and progression in a context-dependent mean. However, the precise role of galectin-4 in HCC development is rarely studied in the above literature.
Galectin-4 is not found in healthy liver tissues, while shows aberrant high expression in HCC tissues, suggesting that tumors may benefit from high expression of galctin-4. In our study, we detected mRNA and protein expression of galectin-4 in 20 paired HCC and matched peritumor tissues as well as six HCC cell lines. Compared to normal tissues, galectin-4 expression in most of HCC tissues was at a high level. This tendency was also verified in HCC cell lines.
Some reports showed that galectin-4 could enhance the epithelial cells’ ability to counteract apoptosis, which is likely to be advantageous in hyperplastic tissues of pre-malignant and malignant tumors [11,17]. In the present study, we established the stable galectin-4 knockdown and overexpression in HCCLM3 cells to investigate its effects on the biological behavior. Galectin-4 overexpression led to promotion of cell proliferation, while also resulted in decreased cell apoptosis. Meanwhile, reduced galectin-4 expression in the HCC cells exhibited the opposite effect. This finding was consistent with observations in the HCC xenograft animal model. These results indicated that galectin-4 was also associated with growth events in HCC cells. Therefore, galectin-4 may implicate in the modulation of cell growth and thus affect the biological behavior of HCC, although it was reported to induce opposite role in cell proliferation of colon cancer.
Furthermore, the molecular mechanism of galectin-4 in HCC is still incompletely understood. It has been reported that galectin-4 could inhibit tumorigenesis of CRC cells through Wnt/β-catenin and IL-6/NF-κB/STAT3 signaling pathways [13,14]. However, in this research, we found that the galectin-4 involved in cell proliferation and apoptosis on HCCLM3 cells were mediated by Cyclin D1, p21, Bax and Bcl-2. Our results showed that overexpression of galectin-4 increased the expression of Cyclin D1, Bcl-2, whereas p21 and Bax expression was markedly decreased. Furthermore, we also demonstrated that galectin-4 silencing significantly deceased the expression of adherens junction complexes (such as E-cadherin and β-catenin) and thus could facilitate tumor escape by loss of cell-cell interaction, enhancing migratory properties. Those results implied that galectin-4 may act as an “adhesin” participating in the development of HCC.
In summary, we here reported the identification of galectin-4 as a protein up-regulated in HCC tissues. Together with the previously published functional result for galectin-4, we supposed a dual active role during development of HCC. Overexpression of galectin-4 may enhance cell proliferation and inhibit cell apoptosis in primary HCC, while further down-regulated in the advanced stage could promote the recurrence/metastasis of HCC. This similar galectin-4 expression pattern were also observed in pancreatic cancer and ileal carcinoid [21,22]. Accordingly, these results identified the role of galectin-4 in tumorigenesis and indicated that galectin-4 may be a candidate biomarker and a target for new therapies of HCC.
Acknowledgements
This work is supported by the Specialized Science and Technology Key Project of Fujian Province (Grant No. 2013YZ0002-3); the Science and Technology Infrastructure Construction Program of Fujian Province (Grant No. 2014Y2005); the Science Foundation of Fujian provinces (Grant No. 2015Y0056 and 2017J05144), the Scientific Foundation of Fuzhou (Grant No. 2015-S-143-16, 2016-S-124-11).
Disclosure of conflict of interest
None.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.De Lope CR, Tremosini S, Forner A, Reig M, Bruix J. Management of HCC. J Hepatol. 2012;56(Suppl 1):S75–87. doi: 10.1016/S0168-8278(12)60009-9. [DOI] [PubMed] [Google Scholar]
- 3.Vilarinho S, Calvisi DF. New advances in precision medicine for hepatocellular carcinoma recurrence prediction and treatment. Hepatology. 2014;60:1812–1814. doi: 10.1002/hep.27311. [DOI] [PubMed] [Google Scholar]
- 4.Lubezky N, Goykhman Y, Nakache R, Nachmany I. Resection of hepatocellular carcinoma. Springer; 2016. pp. 467–475. [Google Scholar]
- 5.Bacigalupo ML, Manzi M, Espelt MV, Gentilini LD, Compagno D, Laderach DJ, Wolfensteintodel C, Rabinovich GA, Troncoso MF. Galectin-1 triggers epithelial-mesenchymal transition in human hepatocellular carcinoma cells. J Cell Physiol. 2015;230:1298–1309. doi: 10.1002/jcp.24865. [DOI] [PubMed] [Google Scholar]
- 6.Jiang S, Weng D, Wang Q, Pan K, Zhang Y, Li Y, Li J, Zhao J, He J, Lv L. Galectin-3 is associated with a poor prognosis in primary hepatocellular carcinoma. J Transl Med. 2014;12:273–273. doi: 10.1186/s12967-014-0273-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai Z, Zeng Y, Xu B, Gao Y, Wang S, Zeng J, Chen L, Huang A, Liu X, Liu J. Galectin-4 serves as a prognostic biomarker for the early recurrence/metastasis of hepatocellular carcinoma. Cancer Sci. 2014;105:1510–1517. doi: 10.1111/cas.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bacigalupo ML, Manzi M, Rabinovich GA, Troncoso MF. Hierarchical and selective roles of galectins in hepatocarcinogenesis, liver fibrosis and inflammation of hepatocellular carcinoma. World J Gastroenterol. 2013;19:8831–8849. doi: 10.3748/wjg.v19.i47.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujita K, Iwama H, Sakamoto T, Okura R, Kobayashi K, Takano J, Katsura A, Tatsuta M, Maeda E, Mimura S. Galectin-9 suppresses the growth of hepatocellular carcinoma via apoptosis in vitro and in vivo. Int J Oncol. 2015;46:2419–2430. doi: 10.3892/ijo.2015.2941. [DOI] [PubMed] [Google Scholar]
- 10.Bumerdene K, Leffler H, Nilsson UJ, Blanchard H. Structural characterisation of human galectin-4 N-terminal carbohydrate recognition domain in complex with glycerol, lactose, 3’-sulfo-lactose, and 2’-fucosyllactose. Sci Rep. 2016;6:20289. doi: 10.1038/srep20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huflejt ME, Leffler H. Galectin-4 in normal tissues and cancer. Glycoconj J. 2004;20:247–255. doi: 10.1023/B:GLYC.0000025819.54723.a0. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi T, Saito T, Fujimura T, Hara K, Takamochi K, Mitani K, Mineki R, Kazuno S, Oh S, Ueno T. Galectin-4, a novel predictor for lymph node metastasis in lung adenocarcinoma. PLoS One. 2013;8:e81883. doi: 10.1371/journal.pone.0081883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SW, Park KC, Jeon SM, Ohn TB, Kim TI, Kim WH, Cheon JH. Abrogation of galectin-4 expression promotes tumorigenesis in colorectal cancer. Cell Oncol (Dordr) 2013;36:169–178. doi: 10.1007/s13402-013-0124-x. [DOI] [PubMed] [Google Scholar]
- 14.Maftouh M, Belo AI, Avan A, Funel N, Peters GJ, Giovannetti E, Van Die I. Galectin-4 expression is associated with reduced lymph node metastasis and modulation of Wnt/β-catenin signalling in pancreatic adenocarcinoma. Oncotarget. 2014;5:5335–5349. doi: 10.18632/oncotarget.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondoh N, Wakatsuki T, Ryo A, Hada A, Aihara T, Horiuchi S, Goseki N, Matsubara O, Takenaka K, Shichita M. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 1999;59:4990–4996. [PubMed] [Google Scholar]
- 16.Shi Y, Wang H, Yin Y, Sun W, Li Y, Zhang C, Wang Y, Wang S, Chen W. Identification and analysis of tumour-associated antigens in hepatocellular carcinoma. Br J Cancer. 2005;92:929–934. doi: 10.1038/sj.bjc.6602460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huflejt ME, Jordan ET, Gitt MA, Barondes SH, Leffler H. Strikingly different localization of Galectin-3 and Galectin-4 in human colon adenocarcinoma T84 Cells. Galectin-4 is localized at sites of cell adhesion. J Biol Chem. 1997;272:14294–14303. doi: 10.1074/jbc.272.22.14294. [DOI] [PubMed] [Google Scholar]
- 18.Delacour D, Gouyer V, Zanetta J, Drobecq H, Leteurtre E, Grard G, Moreauhannedouche O, Maes E, Pons A, Andre S. Galectin-4 and sulfatides in apical membrane trafficking in enterocyte-like cells. J Cell Biol. 2005;169:491–501. doi: 10.1083/jcb.200407073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thijssen VL, Heusschen R, Caers J, Griffioen AW. Galectin expression in cancer diagnosis and prognosis: a systematic review. Biochim Biophys Acta. 2015;1855:235–247. doi: 10.1016/j.bbcan.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Satelli A, Rao PS, Thirumala S, Rao US. Galectin-4 functions as a tumor suppressor of human colorectal cancer. Int J Cancer. 2011;129:799–809. doi: 10.1002/ijc.25750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belo AI, Der Sar AM, Tefsen B, Van Die I. Galectin-4 reduces migration and metastasis formation of pancreatic cancer cells. PLoS One. 2013;8:e65957. doi: 10.1371/journal.pone.0065957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rumilla KM, Erickson LA, Erickson AK, Lloyd RV. Galectin-4 expression in carcinoid tumors. Endocr Pathol. 2006;17:243–250. doi: 10.1385/ep:17:3:243. [DOI] [PubMed] [Google Scholar]