Abstract
This study was aimed to investigate the correlation of lnc-ITSN1-2, lnc-APOC3-2 and lnc-AL355149.1 expressions in plasma by qPCR with rheumatoid arthritis (RA) risk and disease activity. 30 RA patients and 30 health controls (HC) were enrolled in this study. Plasma sample were collected from RA patients before any treatment carried out and HCs. Top 3 RA related long non-coding RNAs (lncRNAs) (lnc-ITSN1-2, lnc-APOC3-2 and lnc-AL355149.1) were selected by a computational framework prediction. The expression of lnc-ITSN1-2, lnc-APOC3-2 and lnc-AL355149.1 were determined by qPCR method. Age (P=0.350) and gender (P=0.542) were similar between RA patients and HCs. lnc-ITSN1-2 level was extremely increased in RA patients compared with HCs (P<0.001), while both lnc-APOC3-2 and lnc-AL355149.1 expressions were numerically higher in RA patients but with no statistical significance (P=0.152 and P=0.139 respectively). Receiver Operating Characteristic (ROC) curves were performed and we found lnc-ITSN1-2 disclosed a great diagnostic value for RA with area under curve (AUC) 0.898, 95% CI 0.813-0.983, and sensitivity was 90.0% and specificity was 80.0% respectively at the best cut-off point. In addition, plasma lnc-ITSN1-2 level was illuminated to be positively associated with erythrocyte sedimentation rate (ESR) (P=0.049), C-reactive protein (CRP) (P<0.001) and disease activity score in 28 joints (DAS28) (P=0.007). Circulating lnc-ITSN1-2 expression was observed to be a novel and convincing biomarker for RA diagnosis as well as disease management.
Keywords: Long non-coding RNA (lncRNA), lnc-ITSN1-2, plasma, rheumatoid arthritis
Introduction
Rheumatoid arthritis (RA), as one of the most common inflammatory disease, affects more than 1% of the general population worldwide, which characterized by inflammation, synovitis and damage of articular cartilage as well as bone, and if not treated well 40%-70% patients would eventually progress to disability [1,2]. Despite of joint involved, patients with longer disease duration could have various extra-articular manifestations such as interstitial lung disease and cardiovascular disease [3,4]. These increasemuch disease and social burden in RA patients which pinnacles early diagnosis and sufficient treatment in RA management [5].
Long non-coding RNA (lncRNA), as an important part of non-coding RNA (ncRNA), consists of longer than 200 nucleotides, which involved in lots of important biological phenomena including imprinting genomic loci, shaping chromosome conformation, regulating enzymatic activity and so on [6]. Accumulating evidences have revealed that lncRNA is implicated in numerous diseases, such as cancers, diabetes as well as inflammatory diseases [7-9].
However, less researches on the role of lncRNA in RA were reported, especially the circulating lncRNAs. This study was aimed to predict potential lncRNAs associated with RA development through computational framework, and subsequently validate the correlation of top 3 related candidate lncRNAsin plasma by qPCR with RA risk and disease activity.
Patients and methods
Participants
30 RA patients from July 2016 to August 2016 in Guanghua Hospitalwere recruited in this study. All patients were diagnosed according to 1987 American College of Rheumatology (ACR) classification of RA with age above 18 years, and no treatments were performed before entering this study. Patients with the following conditions were excluded: history of severe infection, malignant tumor orjoint operations; abnormal hepatic and renal function; infection or tuberculosis (TB). 30 health controls (HC) were also enrolled during the same period with age and gender matched. This research was approved by the Ethics Committee of Guanghua Hospital. All participants provided written informed consents.
Disease assessments
In order to explore the association of studied lncRNAs with disease severity, several disease indexes were determined. Rheumatoid factor (RF) and anti-cyclic citrullinated peptide (CCP) antibody were determined in all RA patients.Inflammatory index including erythrocyte sedimentation rate (ESR) and C-reactive protein (CRP) was measure in RA patients as well. In addition, disease activity score in 28 joints (DAS28) and health assessment questionnaire (HAQ) score were assessed at the time blood sample were obtained from RA patients.
Prediction of RA related lncRNAs
A computational framework to predict lncRNAs associated with RA was analyzed by combining human lncRNA expression profiles, gene expression profiles, and human disease-associated gene data as described in previous study [10]. And Top 3 related lncRNA (lnc-ITSN1-2, lnc-APOC3-2 and lnc-AL355149.1) were selected as candidates to be validated in this study.
Samples
Blood samples were collected in RA patients before any treatment initiation and HCs into sodium citrate tubes. The whole blood was stand for 3 h at -4°C before centrifuging at 1,500 g for 10 min at room temperature. The resultant plasma was then collectedandtotal RNA was subsequently extracted from plasma using TRIzol Reagent (TaKaRa, Japan).
qPCR analysis
Total RNA was reversely transcribed using PrimerScript Real-time reagent kit (TaKaRa, Japan) according to the manufacturer’s instructions. Quantitative analysis of studied lncRNAs expression was performed using SYBR Premix Ex TaqTM II (TaKaRa, Japan) with the primers presented in Table 1. Expression levels of candidate lncRNAs were calculated utilizing the 2-ΔΔt method with U6 as the internal reference.
Table 1.
Primers of candidate LncRNAs and U6
| Gene name | Primer sequence (5’ to 3’) | Amplicon size |
|---|---|---|
| Lnc-ITSN1-2-F | GCCTTTGGACACTTCTTCGGAA | 127 bp |
| Lnc-ITSN1-2-R | GGAGTGTCATCCATCAGCTGAA | |
| Lnc-APOC3-2-F | AGAACTGAAGCGTGGCAAGAT | 233 bp |
| Lnc-APOC3-2-R | TCCAGTCATCACATTGTACACCT | |
| Lnc-AL355149.1-F | TTCCTCCGTTCCGTCTATTCT | 221 bp |
| Lnc-AL355149.1-R | ATGATGATGCTACTCCTCCAAG | |
| U6-F | CTCGCTTCGGCAGCACA | 94 bp |
| U6-R | AACGCTTCACGAATTTGCGT |
Statistics
Statistical analysis was performed using SPSS V19.0 (SPSS, USA). Data was presented as mean values ± standard deviation, median and 25th-75th or counts (percentage). Significance of the comparison was determined by the Student test, Chi-square test or Wilcoxon rank sum test. Significance of the correlations was determined by Spearman test. Receiver Operating Characteristic (ROC) curve was drawn to evaluate the diagnostic value of candidate lncRNAs for RA. P Value <0.05 was considered statistically significant.
Results
Participants
30 RA patients with age 50.35±11.21 years and 24 (80%) female were included in the study. There were 25 cases (83.3%) with RF positive and 26 (86.7%) with anti-CCP positive. There were no difference found between RA patients and HCs in age and gender (P=0.350 and P=0.542 respectively). The other detailed clinical characteristics of RA patients were presented in Table 2.
Table 2.
Characteristics of participants
| Characteristics | RA patients (n=30) | Health controls (n=30) | P Value |
|---|---|---|---|
| Age (years) | 50.35±11.21 | 47.82±9.54 | 0.350 |
| Gender-Female (n/%) | 24 (80%) | 22 (73.3%) | 0.542 |
| RF positive (n/%) | 25 (83.3%) | - | - |
| Anti-CCP (n/%) | 26 (86.7%) | - | - |
| ESR (mm/h) | 25.54 (12.28-40.37) | - | - |
| CRP (mg/l) | 18.7 (7.99-33.26) | - | - |
| DAS28 | 4.85±1.12 | - | - |
| HAQ | 1.83±1.35 | - | - |
Data was presented as mean values ± standard deviation, median and 25th-75th or counts (percentage). Significance of the comparison was determined by the Student test or Chi-square test. P Value <0.05 was considered significant.RF, rheumatoid factor; CCP, cyclic citrullinated peptide; ESR, erythrocyte sedimentation rate; CRP, C-reactive protein; DAS28, disease activity score in 28 joints; HAQ, health assessment questionnaire.
Candidate lncRNAs selection by computational framework prediction
Three top RA related lncRNAs were selected by computational framework prediction as presented as follows: lnc-ITSN1-2 (TCONS_00029004, NONCODE id: NONHSAT081856), lnc-APOC3-2 (TCONS_00019155, NONCODE id: NONHSAT024352) and lnc-AL355149.1-1 (TCONS_00000848, NONCODE id: NONHSAT001134). The other detailed information about the three lncRNAs was listed in Table 3.
Table 3.
Top 3 RA-related lncRNAs by computational framework prediction
| TCONS_00029004 | Gene_id | Transcript_id | NONCODE_id | Disease | P-value | Corrected P-value (Bonferroni) | FDR | Chr |
|---|---|---|---|---|---|---|---|---|
| TCONS_00000848 | lnc-ITSN1-2 | lnc-ITSN1-2:6 | NONHSAT081856 | Arthritis, Rheumatoid | 1.56351E-13 | 8.05206E-10 | 2.92988E-09 | chr21 |
| TCONS_00019155 | lnc-AL355149.1-1 | lnc-AL355149.1-1:2 | NONHSAT001134 | Arthritis, Rheumatoid | 2.37216E-10 | 1.22166E-06 | 3.1873E-06 | chr1 |
| TCONS_00029004 | lnc-APOC3-2 | lnc-APOC3-2:1 | NONHSAT024352 | Arthritis, Rheumatoid | 1.2405E-09 | 6.38857E-06 | 2.20968E-05 | chr11 |
FDR, false discovery rate.
Expressions of lnc-ITSN1-2, lnc-APOC3-2 and lnc-AL355149.1 in plasma of RA patients and HCs
As presented in Figure 1, lnc-ITSN1-2 level was extremely increased in RA patients (8.433 (6.186-12.341)) compared with HCs (3.999 (3.338-4.896)), P<0.001. Both lnc-APOC3-2 and lnc-AL355149.1 expressions were numerically higher in RA patients than in HCs, but with no statistical significance (P=0.152 and P=0.139 respectively).
Figure 1.
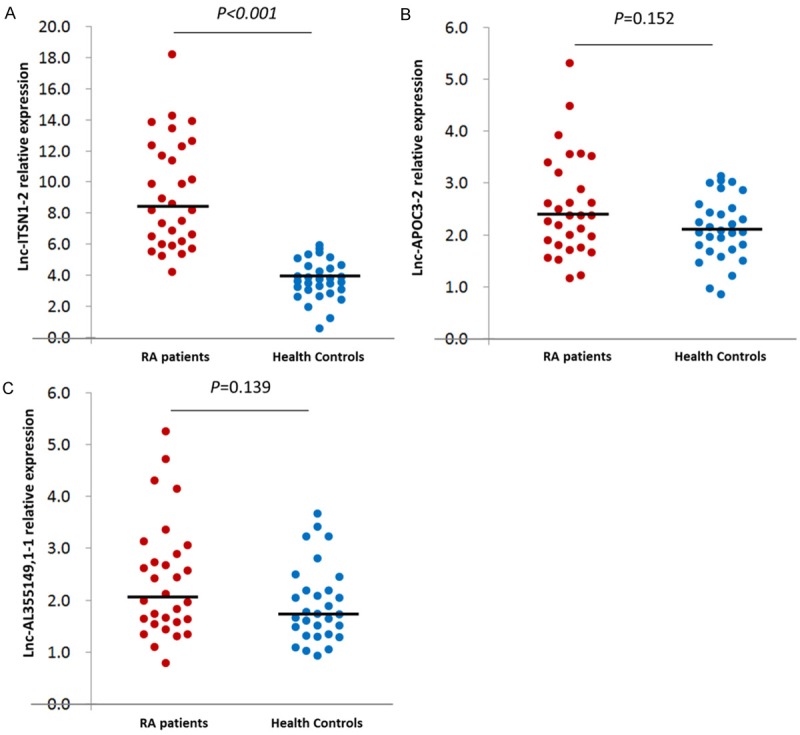
Expression of candidate lncRNAs in RA patients and HCs. Lnc-ITSN1-2 level was increased in RA patients compared to HCs (A), while no difference were found in lnc-APOC3-2 (B) and lnc-AL355149, 1-1 (C) between two groups.
Diagnostic value of lnc-ITSN1-2 level for RA
In order to further investigate the prediction of candidate lncRNAs for RA risk, ROC curve was performed (Figure 2). We found lnc-ITSN1-2 disclosed a great diagnostic value for RA with area under curve (AUC) 0.898, 95% CI 0.813-0.983, and sensitivity was 90.0% and specificity was 80.0% respectively at the best cut-off point (defined as the point at which the value was highest by adding sensitivity to specificity). However, lnc-APOC3-2 and lnc-AL355149.1 lacked predictive value for RA risk with AUC 0.608, 95% CI 0.464-0.752 and AUC 0.611, 95% CI 0.468-0.754 respectively.
Figure 2.
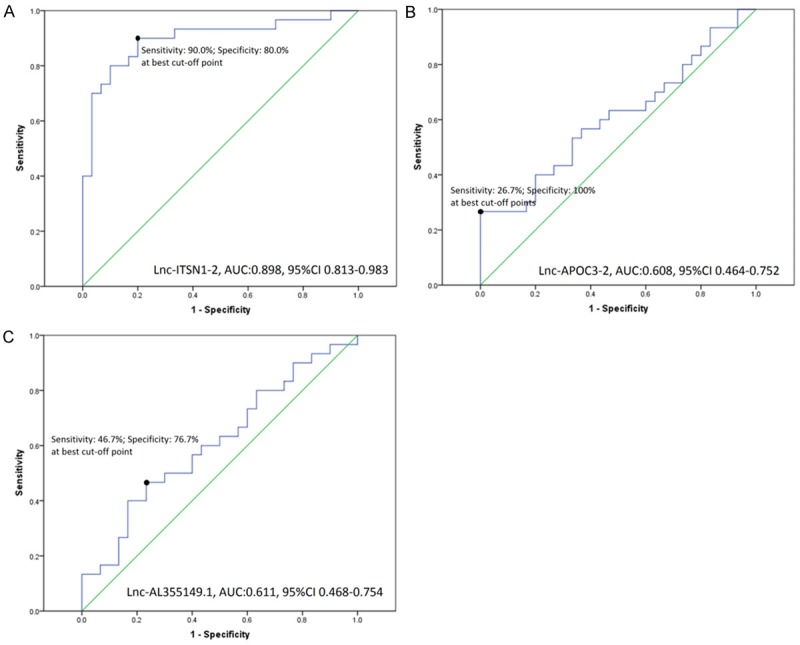
ROC curves of candidate lncRNAs for RA diagnosis. Lnc-ITSN1-2 presented a good value in diagnosis of RA with AUC 0.898, 95% CI 0.813-0.983 (A), but lnc-APOC3-2 (B) and lnc-AL355149.1 (C) lacked predictive value for RA risk.
The correlation of lnc-ITSN1-2, lnc-APOC3-2 and lnc-AL355149.1 level with clinical features in RA patients
Correlation between candidate lncRNAs and clinical features was subsequently determined by Spearman test. As shown in Table 4, plasma lnc-ITSN1-2 level was illuminated to be positively associated with ESR (P=0.049), CRP (P<0.001) and DAS 28 (P=0.007), while no difference was found to be correlated with RF positive (P=0.398), anti-CCP positive (P=0.338) and HAQ (P=0.109). Expect for that lnc-AL355149.1 level was positively associated with DAS 28 score (P=0.026), no other correlation of lnc-APOC3-2 and lnc-AL355149.1 with clinical features were found in our study.
Table 4.
Correlation of Lnc-ITSN1-2, Lnc-APOC3-2 and Lnc-AL355149.1 level with clinical features in RA patients
| Items (N=30) | Parameters | RF positive | Anti-CCP positive | ESR | CRP | DAS28 | HAQ |
|---|---|---|---|---|---|---|---|
| Lnc-ITSN1-2 | r | -0.160 | -0.181 | 0.363 | 0.636 | 0.480 | 0.299 |
| P value | 0.398 | 0.338 | 0.049 | <0.001 | 0.007 | 0.109 | |
| Lnc-APOC3-2 | r | -0.191 | -0.227 | -0.025 | 0.274 | -0.051 | 0.005 |
| P value | 0.312 | 0.229 | 0.894 | 0.143 | 0.787 | 0.978 | |
| Lnc-AL355149.1 | r | -0.202 | -0.351 | 0.174 | 0.212 | 0.406 | 0.194 |
| P value | 0.286 | 0.057 | 0.357 | 0.260 | 0.026 | 0.305 |
Significance of the correlation was determined by Spearman test. P Value <0.05 was considered significant. RF, rheumatoid factor; CCP, cyclic citrullinated peptide; ESR, erythrocyte sedimentation rate; CRP, C-reactive protein; DAS28, disease activity score in 28 joints; HAQ, health assessment questionnaire.
Discussion
In the present study, we found plasma lnc-ITSN1-2 was dramatically elevated in RA patients compared with HCs, and it had a good diagnostic value for RA with AUC 0.898. In addition, lnc-ITSN1-2 was positively associated with disease activity (ESR, CRP and DAS 28), while lnc-AL355149.1 was found to be associated with DAS 28 as well.
lncRNAs, which are a diverse class of molecules lackpotential of coding protein composed of longer than 200 nucleotides, are characterized by unique regulatory mechanisms, alternative forms of biogenesis, cis-regulatory activities and functional structured RNA domains [6,11]. Although The deregulation of lncRNAs was reported to be associated with various diseases including cancers, nutrition metabolism and endocrine disorder, cardiovascular disease as well as autoimmune disease [8,9,12,13].
260 lncRNAs are illustrated to be differentially expressedin the synovium between adjuvant-induced arthritis (AA) and normal rats by lncRNA/mRNA microarray, and six lncRNAs consisting of XR_008357, U75927, MRAK046251, XR_006457, DQ266363 and MRAK003448 are further validated to be increased in AA rats by qPCR [14]. Furthermore, 135 differentially expressed lncRNAs were observed in fibroblast-like synoviocytes (FLSs) from RA patients compared to normal FLSs, and lnc-ENST00000483588 was up regulated while lnc-ENST00000438399, lnc-uc004afb.1, and lnc-ENST00000452247 was down regulated by qPCR in RA FLSs [9]. These indicates lncRNAs deregulations play important role in RA pathogenesis, however, comprehensive understanding about how lncRNAs function in RA etiology was seldom investigated.
Hotair, as one of the most key imbalanced lncRNA which involved in pathogenesis of various diseases, is found to be notably elevated in blood mononuclear cells and serum exosome of RA patients and leads active macrophagemigration, while Hotair level was remarkably reduced in differentiated osteoclasts and synoviocytes with down-regulating MMP-2 as well MMP-3 expressions [15]. Oncofetal H19 RNA is revealed to be increased in synovial tissue of RA patients than controls, and isolated low expression of H19 in FLSs was raised markedly on starvation despite of addition of interleukin-1beta (IL-1beta), tumor necrosis factor-alpha (TNF-alpha) or platelet-derived growth factor [16]. What’s more, C5T1 lncRNA which starts in the 3’ untranslated region (UTR) of C5 influences mRNA expression of RA candidate gene C5 [17]. These studies provided some but insufficient information about lncRNA functions in RA etiology.
Lnc-ITSN1-2, located in chromosome 21, with NONCODE gene ID NONHSAG032726.2 and NONCODE transcript ID NONHSAT081856.2, start from 33976355 to end site 33976982 and length was 451 bp (http://www.noncode.org/show_rna.php?id=NONHSAT081856.2). No previous report has been described about the function of lnc-ITSN1-2 or any biological information. In our study, we found lnc-ITSN1-2 was extremely increased in plasma of RA patients compared with HCs, and was positively correlated with inflammatory indexes (ESR and CRP) and disease activity (DAS 28). ESR and CRP are regarded as critical parameters for RA diagnosis and disease assessments for inflammation, ACR as well as European League against Rheumatism (EULAR) recommended ESR and CRP as routine examination for RA patients, while DAS 28 is also a gold standard for disease activity assessing score and goal of treating-to-target therapy [18-20]. These indicated lnc-ITSN1-2 could be served as a novel circulating biomarker for RA diagnosis and disease management.
Some limitations existed in this study. Firstly, sample size of this study was relatively small with 30 RA patients and 30 HCs; however, it has been the largest sample for circulating lncRNA determination study in RA patients. Secondly, the patients and HCs were enrolled only in one center, so some confounding factors might exist. Thirdly, we identified lnc-ITSN1-2 deregulation in RA patients and its correlation with clinical features, but how lnc-ITSN1-2 function in the pathogenesis of RA was not investigated, which we would explore in the future study.
In conclusion, circulating lnc-ITSN1-2 expression was observed to be a novel and convincing biomarker for RA diagnosis as well as disease management.
Acknowledgements
This research was supported by Shanghai New Three-year Action Plan-Innovation Connotation Construction of Traditional Chinese Medicine (ZY3-CCCX-3-3021). This research was supported by Psoriasis Key Specialty (20161003) and Project of XELJANZ (tofacitinib CP-690550)in the immunoregulatory effects of rheumatoid arthritis (CNKW2013207).
Disclosure of conflict of interest
None.
References
- 1.Catrina AI, Svensson CI, Malmstrom V, Schett G, Klareskog L. Mechanisms leading from systemic autoimmunity to joint-specific disease in rheumatoid arthritis. Nat Rev Rheumatol. 2017;13:79–86. doi: 10.1038/nrrheum.2016.200. [DOI] [PubMed] [Google Scholar]
- 2.Smolen JS, Aletaha D, Mclnnes IB. Rheumatoid arthritis. Lancet. 2016;4:427–442. doi: 10.1016/S0140-6736(16)30173-8. [DOI] [PubMed] [Google Scholar]
- 3.Radner H, Lesperance T, Accortt NA, Solomon DH. Incidence and prevalence of cardiovascular risk factors among patients with rheumatoid arthritis, psoriasis, or psoriatic arthritis. Arthritis Care Res (Hoboken) 2016 doi: 10.1002/acr.23171. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 4.Zamora-Legoff JA, Krause ML, Crowson CS, Ryu JH, Matteson EL. Patterns of interstitial lung disease and mortality in rheumatoid arthritis. Rheumatology (Oxford) 2016 doi: 10.1093/rheumatology/kew391. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor PC, Moore A, Vasilescu R, Alvir J, Tarallo M. A structured literature review of the burden of illness and unmet needs in patients with rheumatoid arthritis: a current perspective. Rheumatol Int. 2016;36:685–695. doi: 10.1007/s00296-015-3415-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 7.Redis RS, Vela LE, Lu W, Ferreira de Oliveira J, Ivan C, Rodriguez-Aguayo C, Adamoski D, Pasculli B, Taguchi A, Chen Y, Fernandez AF, Valledor L, Van Roosbroeck K, Chang S, Shah M, Kinnebrew G, Han L, Atlasi Y, Cheung LH, Huang GY, Monroig P, Ramirez MS, Catela Ivkovic T, Van L, Ling H, Gafa R, Kapitanovic S, Lanza G, Bankson JA, Huang P, Lai SY, Bast RC, Rosenblum MG, Radovich M, Ivan M, Bartholomeusz G, Liang H, Fraga MF, Widger WR, Hanash S, Berindan-Neagoe I, Lopez-Berestein G, Ambrosio AL, Gomes Dias SM, Calin GA. Allele-specific reprogramming of cancer metabolism by the long non-coding RNA CCAT2. Mol Cell. 2016;61:520–534. doi: 10.1016/j.molcel.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X, Wong D. Long non-coding RNA-mediated regulation of glucose homeostasis and diabetes. Am J Cardiovasc Dis. 2016;6:17–25. [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Xu YZ, Sun N, Liu JH, Chen FF, Guan XL, Li A, Wang F, Zhao QF, Wang HY, Song SS, Yu W, Zhao JN, Li XJ. Long noncoding RNA expression profile in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Res Ther. 2016;18:227. doi: 10.1186/s13075-016-1129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu MX, Chen X, Chen G, Cui QH, Yan GY. A computational framework to infer human disease-associated long noncoding RNAs. PLoS One. 2014;9:e84408. doi: 10.1371/journal.pone.0084408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Gesualdo F, Capaccioli S, Lulli M. A pathophysiological view of the long non-coding RNA world. Oncotarget. 2014;5:10976–10996. doi: 10.18632/oncotarget.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Q, Zhou X, Xia Q, Shen J, Yan J, Zhu J, Li X, Shu M. Long non-coding RNA CCAT1 that can be activated by c-Myc promotes pancreatic cancer cell proliferation and migration. Am J Transl Res. 2016;8:5444–5454. [PMC free article] [PubMed] [Google Scholar]
- 13.Ruhle F, Stoll M. Long non-coding RNA Databases in Cardiovascular Research. Genomics Proteomics Bioinformatics. 2016;14:191–199. doi: 10.1016/j.gpb.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang H, Qin XJ, Li WP, Ma R, Wang T, Li ZQ. LncRNAs expression in adjuvant-induced arthritis rats reveals the potential role of LncRNAs contributing to rheumatoid arthritis pathogenesis. Gene. 2016;593:131–142. doi: 10.1016/j.gene.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Song J, Kim D, Han J, Kim Y, Lee M, Jin EJ. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin Exp Med. 2015;15:121–126. doi: 10.1007/s10238-013-0271-4. [DOI] [PubMed] [Google Scholar]
- 16.Stuhlmuller B, Kunisch E, Franz J, Martinez-Gamboa L, Hernandez MM, Pruss A, Ulbrich N, Erdmann VA, Burmester GR, Kinne RW. Detection of oncofetal h19 RNA in rheumatoid arthritis synovial tissue. Am J Pathol. 2003;163:901–911. doi: 10.1016/S0002-9440(10)63450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Messemaker TC, Frank-Bertoncelj M, Marques RB, Adriaans A, Bakker AM, Daha N, Gay S, Huizinga TW, Toes RE, Mikkers HM, Kurreeman F. A novel long non-coding RNA in the rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels. Genes Immun. 2016;17:85–92. doi: 10.1038/gene.2015.54. [DOI] [PubMed] [Google Scholar]
- 18.Calabrese LH, Calabrese C, Kirchner E. The 2015 American College of Rheumatology Guideline for the treatment of rheumatoid arthritis should include new standards for hepatitis B screening: comment on the article by singh et al. Arthritis Care Res (Hoboken) 2016;68:723–724. doi: 10.1002/acr.22865. [DOI] [PubMed] [Google Scholar]
- 19.Smolen JS, Landewe R, Breedveld FC, Buch M, Burmester G, Dougados M, Emery P, Gaujoux-Viala C, Gossec L, Nam J, Ramiro S, Winthrop K, de Wit M, Aletaha D, Betteridge N, Bijlsma JW, Boers M, Buttgereit F, Combe B, Cutolo M, Damjanov N, Hazes JM, Kouloumas M, Kvien TK, Mariette X, Pavelka K, van Riel PL, Rubbert-Roth A, Scholte-Voshaar M, Scott DL, Sokka-Isler T, Wong JB, van der Heijde D. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2013 update. Ann Rheum Dis. 2014;73:492–509. doi: 10.1136/annrheumdis-2013-204573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smolen JS, Breedveld FC, Burmester GR, Bykerk V, Dougados M, Emery P, Kvien TK, Navarro-Compan MV, Oliver S, Schoels M, Scholte-Voshaar M, Stamm T, Stoffer M, Takeuchi T, Aletaha D, Andreu JL, Aringer M, Bergman M, Betteridge N, Bijlsma H, Burkhardt H, Cardiel M, Combe B, Durez P, Fonseca JE, Gibofsky A, Gomez-Reino JJ, Graninger W, Hannonen P, Haraoui B, Kouloumas M, Landewe R, Martin-Mola E, Nash P, Ostergaard M, Ostor A, Richards P, Sokka-Isler T, Thorne C, Tzioufas AG, van Vollenhoven R, de Wit M, van der Heijde D. Treating rheumatoid arthritis to target: 2014 update of the recommendations of an international task force. Ann Rheum Dis. 2016;75:3–15. doi: 10.1136/annrheumdis-2015-207524. [DOI] [PMC free article] [PubMed] [Google Scholar]


