Abstract
To improve diagnosis of asthma, we tend to confirm potential biomarkers by comparing sputum metabolome profiles between asthma patients and healthy controls, using ultra-high-performance liquid chromatography coupled to quadruple time-of-flight mass spectrometry (UHPLC-QTOF/MS). Thirty endogenous metabolites contributing to the separation of asthma patients and healthy controls were tentatively identified in positive mode, such as 1-hexadecanoyl-sn-glycerol, glycerol 1-stearate, sphingosine, Phe-Ser, Tyr-Ala and Phe-Gln, and 12 endogenous metabolites were identified in negative mode, such as cytidine 2’,3’-cyclic phosphate, 1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phospho-(1’-rac-glycerol), 1-octadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphoserine, thymidine, gamma-L-glutamyl-L-valine and adenine. Those differential metabolites were mainly participatedin glycerophospholipid metabolism, retrograde endocannabinoid signaling and metabolic pathways in positive mode and 2-oxocarboxylic acid metabolism, biosynthesis of amino acids, phenylalanine, tyrosine and tryptophan biosynthesis, valine, leucine and isoleucine degradation and metabolic pathways in negative mode. Importantly, several metabolic pathways including glycerophospholipid metabolism, inositol phosphate metabolism, and glycolysis or gluconeogenesis were found most important. These findings suggest sputum metabolomics can be used for the early diagnosis and risk prediction of asthma.
Keywords: Asthma, UHPLC-QTOF/MS, metabolomics, sputum
Introduction
Asthma is a complicated chronic airway allergic disease involving many cells and cytokines characterized by airway inflammation, hyperresponsiveness and remodeling [1]. Asthma is caused by a variety of inflammatory cell infiltrates, with symptoms of episodic dyspnea and reversible airway obstruction [2,3]. Recently, more than 300 million people suffer from asthma worldwide, with the prevalence rate in children and adult were 3%-38% and 2%-12%, respectively, and their morbidity and mortality are increased annually [4,5]. Recurrent attacks of asthma may cause a variety of complications, such as chronic obstructive emphysema, chronic pulmonary heart disease, pulmonary fibrosis, respiratory arrest, respiratory failure, pneumothorax and mediastinal emphysema [6]. Genetic variation and environmental changes have shown to trigger asthma, which genetic susceptibility and environmental exposure has significant influence on the development of asthma, and the interaction of genes and the environment can also lead to asthma and allergies [7].
Metabolomics are not only goal of qualitative and quantitative analysis of all metabolic components in particular biological samples, but also explain all the information about the metabolism of organisms from the point of view of systematic biology [8]. It is a science that analyzes the changes in the concentration and density of small molecular metabolites in biological cells, fluids (e.g. blood and urine), and tissue or tissue extracts [9]. Small molecule metabolites are the end product of the body’s metabolic activity, and the concentration changes of metabolites can therefore reaction of biochemical functions due to disease. Although metabolomics technology mainly includes the determination of nuclear magnetic resonance (NMR), mass spectrometry (MS), gas (GC) and high performance liquid chromatography (HPLC) and ultra HPLC (UHPLC). The ultra high pressure system can improve the separation and sensitivity of chromatographic peaks significantly, and UHPLC coupled to quadruple time-of-flight mass spectrometry (UHPLC-QTOF-MS) is more efficient and rapid in the analysis of complex samples [10,11]. Mayr et al. [12] combined proteomic with metabolomics techniques and found increased acyl coenzyme A dehydrogenase and decreased alanine and cytosolic malate dehydrogenase in blood vessel of mouse with coronary heart disease, suggesting that the metabolism of vascular fat cells may replace glucose metabolism, and the decrease in effective energy synthesis and glucose as well as oxidative stress is of great importance in the pathogenesis of coronary heart disease. Beger et al. [13] analyzed 40 patients with acute kidney injury in children after cardiac surgery, in which the vanillic acid was significantly increased in urine, suggesting that vanillic acid can be used as an early and sensitive diagnostic marker for acute kidney injury after cardiac surgery.
The pathogenesis of asthma is very complex, varies or overlaps in different types of asthma and has so far not been fully understood. Metabonomics can detect differences in small molecule metabolites in asthma patients and normal controls and find out the metabolic markers associated with pathogenesis of asthma, thus explaining the pathogenesis of asthma and providing a suitable treatment method [14,15]. NMR-based metabonomics applied to exhaled breath condensate can clearly identify biochemical metabolism in asthma patients with different severity [16]. LC-MS (liquid chromatography mass spectrometry) based metabolomics was also applied to the detection of urine samples from asthmatic patients, and representative markers were identified as the basis for the diagnosis of asthma [17]. Ried et al. [18] collected the serum samples from asthmatic patients and found that changes in lecithin and phosphatidylcholine concentrations may be used to identify and diagnose asthma.
As a direct secretion in the airways, sputum can reflect airway inflammation and is therefore used to identify and diagnose asthma and chronic obstructive pulmonary disease [19]. In the present study, UHPLC-QTOF/MS based metabolomics was performed to analyze the differential metabolites in sputum between patients with asthma in childhood and healthy controls. Biological correlates of metabolic pathways and potential biomarkers have been studied in depth to understand the metabolic disturbance of asthma in childhood. Insight obtained from these studies will be useful for developing novel diagnostic biomarkers and aiding in the prevention and control of asthma.
Materials and methods
Patient recruitment
35 clinical samples including 15 healthy controls and 20 asthma patients were collected from patients hospitalized in Child’s Hospital of Nanjing Medical University, stored in -80°C and prepared for UHPLC-QTOF-MS analysis. This prospective study was approved by the Ethics Committee of Affiliated Nanjing Children’s Hospital, Nanjing Medical University. Informed consent was obtained from all participants.
Sample collection and preparation
Sputum samples were collected from the 35 clinical samples and added into 900 μL extraction liquid and 10 μL L-2-chlorophenylalanineas (1 mg/mL stock in dH2O) as internal standard to the sample in 1.5 mL EP tubes. After vortex mixing for 30 s, samples were ultrasound treated for 10 min (incubated in ice water), incubated 1 h at -20°C and centrifuged for 15 min at 12000 rpm at 4°C. The supernatant (0.7 mL) fresh were transferred into a 2 mL LC/MS glass vial, dried in a vacuum concentrator without heating and added 100 μL extraction liquid. After vortex for 30 s and sonicate for 10 min (4°C water bath), samples were centrifuged for 15 min at 12000 rpm at 4°C, and then transfer the supernatant (60 μL) into a fresh 2 mL LC/MS glass vial, take 10 μL from each sample and pooling as QC samples and take 60 μL supernatant for the UHPLC-QTOF-MS analysis.
UHPLC-QTOF-MS analysis
LC-MS/MS analyses were performed using an UHPLC system (1290, Agilent Technologies) with a UPLC BEH Amide column (1.7 μm, 2.1×100 mm, Waters) coupled to TripleTOF 6600 (Q-TOF, AB Sciex). The mobile phase consisted of 25 mM NH4OAc and 25 mM NH4OH in water (pH=9.75) (A) and acetonitrile (B) was carried with elution gradient as follows: 0 min, 85% B; 2 min, 75% B; 9 min, 0% B; 14 min, 0% B; 15 min, 85% B; 20 min, 85% B, which was delivered at 0.3 mL min-1. The injection volume was 2 μL. The TripleTOF mass spectrometer was used during an LC/MS experiment as previously described [20].
Statistical analysis
The resulted three-dimensional data involving the peak number, sample name, and normalized peak area were fed to SIMCA14.1 software package (V14.1, MKS Data Analytics Solutions, Umea, Sweden) for PCA and OPLS-DA. The variable importance in the projection (VIP) value exceeding 1.0 was first filtered out as the changed metabolites. The remaining variables were then assessed by Student’s t-test (P-value < 0.05). Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) and MetaboAnalyst3.0 (http://www.metaboanalyst.ca) databases were utilized to analyze for the pathways of metabolites.
Results
Multivariate data analysis
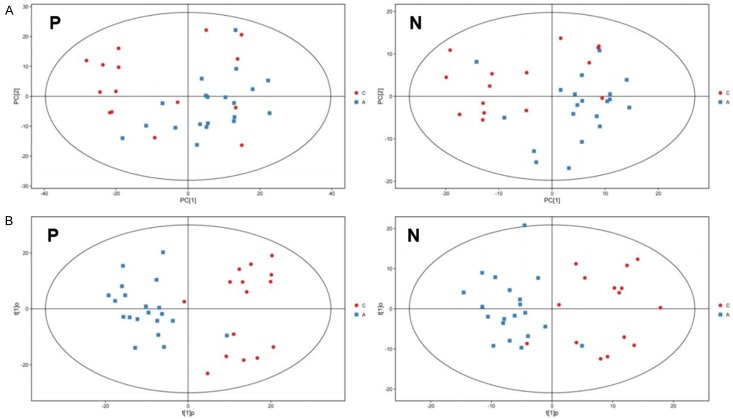
First of all, matrices showed 1514 and 651 features for UHPLC-QTOF-MS in positive and negative mode, respectively. For multivariate analysis, PCA that showed the distribution of origin data was performed to prediction of control and asthma groups for UHPLC-QTOF-MS in both positive and negative mode. As shown in Figure 1A, control and asthma groups had no differences in metabolic profiles and no strong outliers were observed.
Figure 1.
The sputum PCA (A) and OPLS-DA score plot (B) between healthy controls and asthma patients in childhood in both positive and negative mode. P: positive mode; N: negative mode. C: healthy control; A: asthma patients.
In order to gain higher levels of population separation and obtain better understandings of variables that are responsible for classification, supervised orthogonal projections to latent structures-discriminate analysis (OPLS-DA) was presented. As shown in Figure 1B, clear separation of groups in UHPLC-QTOF-MS in both positive and negative mode was found obviously. Afterwards, the parameters for the classification from the software were R2Y=0.54 and Q2=-0.86 in positive modes and R2Y=0.52 and Q2=-1.07 in negative modes. 7-fold cross validation is used to estimate the robustness and predictive power of our model; such permutation experiments are carried out for further verification of the model, which is stable and fitted and predicted well. These results suggest that the method can identify potential biomarkers for distinguishing between asthmatics and healthy people.
Identification and quantization of potential metabolites
Volcano plot analysis using Student’s t-test (P-value < 0.05) with VIP score >1 when compared between asthma patients and healthy control (Figure 2A). Combined with retention time, accurate molecular mass and mass spectrometric analysis of the data provided by the method, the preliminary screening of endogenous metabolites can be made. Our data revealed 30 potential biomarkers in positive mode, among which 5 metabolites were increased and 25 metabolites were decreased in asthma patients compared with healthy controls with Log2fold-change ≥ 1.5 and P < 0.05 by Student’s t-test (Table 1). Meanwhile, there were 12 potential biomarkers in negative mode, among which 4 metabolites were increased and 8 metabolites were decreased in asthma patients compared with healthy controls (Table 2). Top 3 increased and decreased metabolites in healthy controls compared with asthma patients in positive mode, including 1-Hexadecanoyl-sn-glycerol, Glycerol 1-stearate, Sphingosine, Phe-Ser, Tyr-Ala and Phe-Gln, were shown in Figure 2B and 2C, respectively. Top 3 increased and decreased metabolites in healthy controls compared with asthma patients in negative mode, including Cytidine 2’,3’-cyclic phosphate, 1-Hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phospho-(1’-rac-glycerol), 1-Octadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphoserine, Thymidine, Gamma-L-Glutamyl-L-valine and Adenine, were shown in Figure 2D and 2E, respectively.
Figure 2.
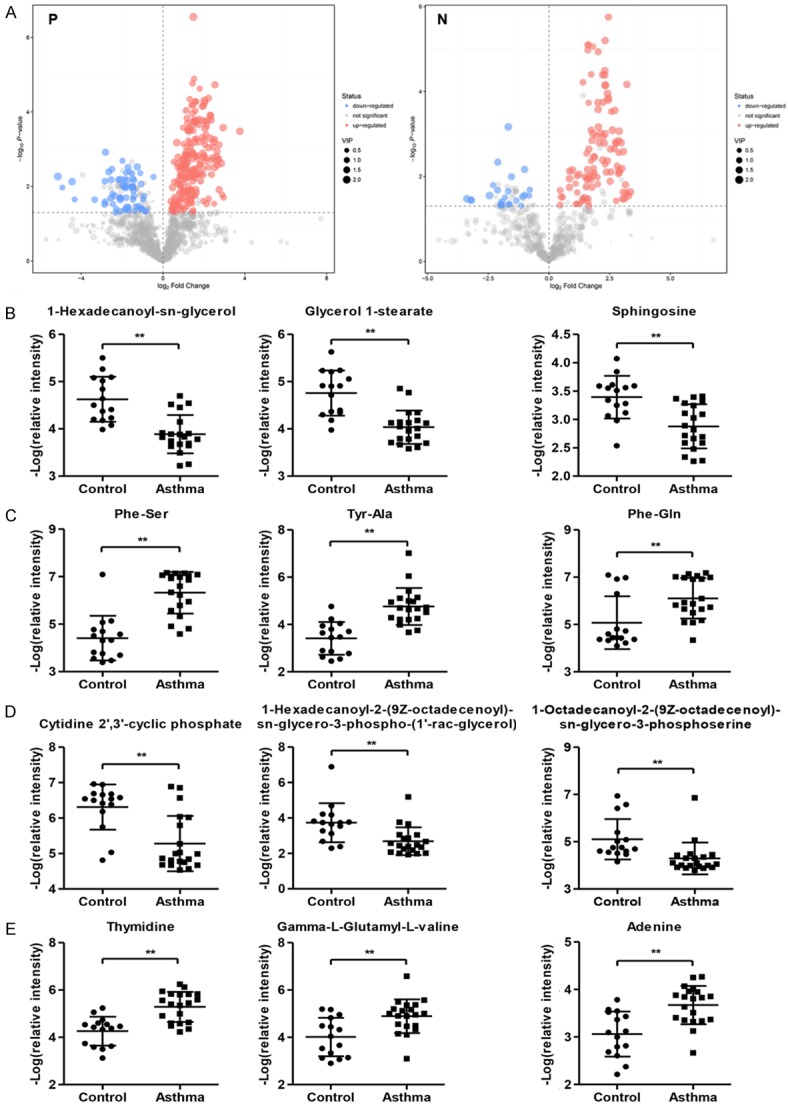
Different metabolites between asthma patients in childhood and healthy controls. (A) Volcano plot between asthma patients and healthy controls in positive and negative mode. Top 3 increased (B) and decreased metabolites (C) in healthy controls compared with asthma patients in positive mode. Top 3 increased (D) and decreased metabolites (E) in healthy controls compared with asthma patients in negative mode. P: positive mode; N: negative mode.
Table 1.
Different metabolites in asthma patients compared with healthy control using UHPLC-QTOF-MS in positive ion mode
| MS2 name | MS2 score | Type | P-value | Log2Fold change |
|---|---|---|---|---|
| 1-Hexadecanoyl-sn-glycerol_1 | 0.817 | MS2 forward | 0.001 | 2.3 |
| Glycerol 1-stearate_1 | 0.623 | MS2 reverse | 0.000 | 2.0 |
| Sphingosine_2 | 0.677 | MS2 forward | 0.003 | 1.7 |
| PC (16:0/16:0) | 16.000 | MS2 reverse | 0.005 | 1.6 |
| 1-Hexadecanoyl-sn-glycerol_2 | 0.961 | MS2 reverse | 0.048 | 1.5 |
| Glu-Pro_1 | 0.729 | MS2 reverse | 0.010 | -1.5 |
| Pro-Arg_2 | 0.624 | MS2 forward | 0.011 | -1.5 |
| L-Arginine_4 | 0.987 | MS2 reverse | 0.010 | -1.5 |
| Phe-His_1 | 0.937 | MS2 forward | 0.004 | -1.6 |
| N-Acetyl-D-glucosamine_2 | 0.643 | MS2 forward | 0.043 | -1.7 |
| Gamma-L-Glutamyl-L-valine_1 | 0.942 | MS2 forward | 0.003 | -1.7 |
| Glu-Pro_2 | 0.652 | MS2 forward | 0.007 | -1.8 |
| Lys-Pro_3 | 0.739 | MS2 forward | 0.010 | -1.8 |
| Phe-Ile | 0.742 | MS2 forward | 0.035 | -1.8 |
| Phe-His_3 | 0.937 | MS2 forward | 0.004 | -1.8 |
| Pro-Val_2 | 0.608 | MS2 reverse | 0.009 | -1.9 |
| Urocanic acid_1 | 0.933 | MS2 forward | 0.002 | -2.0 |
| Tyr-Pro_2 | 0.935 | MS2 reverse | 0.021 | -2.1 |
| His-Pro | 0.907 | MS2 forward | 0.007 | -2.1 |
| Tyr-Pro_1 | 0.954 | MS2 reverse | 0.022 | -2.1 |
| N-Acetyl-D-glucosamine_1 | 0.908 | MS2 reverse | 0.047 | -2.1 |
| Lys-Phe_1 | 0.774 | MS2 forward | 0.020 | -2.1 |
| Thr-Phe_1 | 0.791 | MS2 forward | 0.007 | -2.2 |
| L-Citrulline_1 | 0.935 | MS2 reverse | 0.049 | -2.3 |
| Arg-Phe_1 | 0.807 | MS2 forward | 0.003 | -2.5 |
| Adenine_1 | 0.834 | MS2 reverse | 0.007 | -2.5 |
| Phe-Tyr_1 | 0.802 | MS2 forward | 0.004 | -2.6 |
| Phe-Gln_1 | 0.888 | MS2 forward | 0.001 | -2.8 |
| Tyr-Ala_2 | 0.935 | MS2 forward | 0.007 | -4.4 |
| Phe-Ser_1 | 0.856 | MS2 forward | 0.005 | -5.1 |
Table 2.
Different metabolites in asthma patients compared with healthy control using UHPLC-QTOF-MS in negative ion mode
| MS2 name | MS2 score | Type | P-value | Log2Fold change |
|---|---|---|---|---|
| Cytidine 2’,3’-cyclic phosphate | 0.994 | MS2 forward | 0.000 | 2.6 |
| 1-Hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phospho-(1’-rac-glycerol) | 0.983 | MS2 forward | 0.001 | 2.2 |
| 1-Octadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphoserine | 0.839 | MS2 reverse | 0.000 | 2.0 |
| 1-Stearoyl-sn-glycerol 3-phosphocholine | 0.622 | MS2 reverse | 0.010 | 1.5 |
| Thymine_2 | 0.999 | MS2 forward | 0.010 | -1.5 |
| p-Chlorophenylalanine_3 | 0.623 | MS2 reverse | 0.046 | -1.7 |
| Phenylpyruvate_2 | 0.887 | MS2 forward | 0.001 | -1.7 |
| Phosphoenolpyruvate | 0.996 | MS2 forward | 0.045 | -2.0 |
| Urocanic acid | 0.938 | MS2 forward | 0.005 | -2.1 |
| Adenine_1 | 0.810 | MS2 forward | 0.021 | -2.1 |
| Gamma-L-Glutamyl-L-valine_1 | 0.707 | MS2 forward | 0.028 | -2.4 |
| Thymidine | 0.927 | MS2 forward | 0.034 | -3.4 |
Metabolic pathway analysis
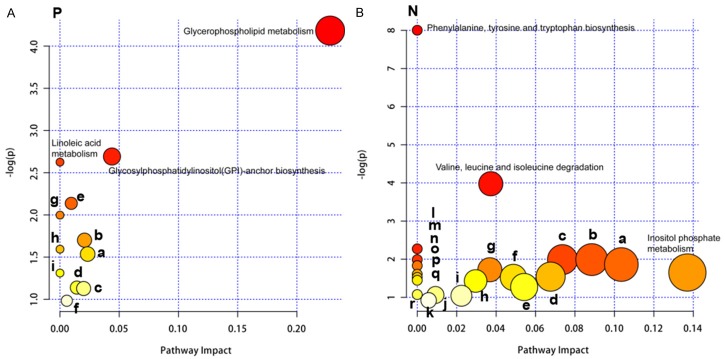
KEGG pathways representing the differentially expressed metabolites in asthma patients compared with healthy control using UHPLC-QTOF-MS in positive and negative mode were shown in Tables 3 and 4, respectively. KEGG analysis identified only the pathways in which all differential metabolites were involved, but further metabolic pathway analysis was needed to determine whether these pathways were closely related to experimental conditions. Using the comprehensive analysis of the pathway of differential metabolites (including enrichment and topological analysis), the pathway can be further screened to find the most critical pathway associated with the different metabolites. We used the KEGG metabolic pathway as a background repository. Our data showed that several metabolic pathways, including glycerophospholipid metabolism (positive mode), inositol phosphate metabolism (negative mode) and glycolysis or gluconeogenesis (negative mode) with the impact-value 0.23, 0.14 and 0.10, respectively, were found most important (Figure 3A and 3B), with the impact-value ≥ 0.10 as potential target pathway [21].
Table 3.
KEGG pathways representing the differentially expressed compounds in asthma patients compared with healthy control using UHPLC-QTOF-MS in positive ion mode
| Pathway | Compounds |
|---|---|
| Metabolic pathways-Homo sapiens (human) | Adenine, Phosphatidylcholine, L-Citrulline, Phosphatidylethanolamine, Sphingomyelin, Betaine, 2-Methylprop-2-enoyl-CoA |
| Retrograde endocannabinoid signaling-Homo sapiens (human) | Phosphatidylcholine, Phosphatidylethanolamine |
| Glycerophospholipid metabolism-Homo sapiens (human) | Phosphatidylcholine, Phosphatidylethanolamine |
Table 4.
KEGG pathways representing the differentially expressed compounds in asthma patients compared with healthy control using UHPLC-QTOF-MS in negative ion mode
| Pathway | Compounds |
|---|---|
| Metabolic pathways-Homo sapiens (human) | Phosphoenolpyruvate, myo-Inositol, 3-Methyl-2-oxobutanoic acid, Adenine, Phenylpyruvate, Thymidine, Hexadecanoic acid, Indole, N-Acetyl-L-glutamate, Urocanate, 2-Methylprop-2-enoyl-CoA |
| Biosynthesis of amino acids-Homo sapiens (human) | Phosphoenolpyruvate, 3-Methyl-2-oxobutanoic acid, Phenylpyruvate, N-Acetyl-L-glutamate |
| 2-Oxocarboxylic acid metabolism-Homo sapiens (human) | 3-Methyl-2-oxobutanoic acid, Phenylpyruvate, N-Acetyl-L-glutamate |
| Phenylalanine, tyrosine and tryptophan biosynthesis-Homo sapiens (human) | Phosphoenolpyruvate, Phenylpyruvate, Indole |
| Valine, leucine and isoleucine degradation-Homo sapiens (human) | 3-Methyl-2-oxobutanoic acid, 2-Methylprop-2-enoyl-CoA |
Figure 3.
Summary of pathway analysis of sputum specimens of asthma patients. A. Pathways based on positive mode. (a) Glycine, serine and threonine metabolism; (b) Valine, leucine and isoleucine degradation; (c) Arginine and proline metabolism; (d) Tyrosine metabolism; (e) Sphingolipid metabolism; (f) Purine metabolism; (g) alpha-Linolenic acid metabolism; (h) Phenylalanine metabolism; (i) Arachidonic acid metabolism. B. Pathways based on negative mode. (a) Glycolysis or Gluconeogenesis; (b) Valine, leucine and isoleucine biosynthesis; (c) Pantothenate and CoA biosynthesis; (d) Histidine metabolism; (e) Pyrimidine metabolism; (f) Phenylalanine metabolism; (g) Ubiquinone and other terpenoid-quinone biosynthesis; (h) Fatty acid metabolism; (i) Tryptophan metabolism; (j) Arginine and proline metabolism; (k) Purine metabolism; (l) Citrate cycle (TCA cycle); (m) Fatty acid elongation in mitochondria; (n) Pyruvate metabolism; (o) Galactose metabolism; (p) Ascorbate and aldarate metabolism; (q) Fatty acid biosynthesis; (r) Tyrosine metabolism. P: positive mode; N: negative mode.
Discussion
In the present study, we found that 30 potential biomarkers in positive mode and 12 potential biomarkers in negative mode were different between healthy controls and asthma patients, based on UHPLC-QTOF-MS analysis. Metabolic pathways, retrograde endocannabinoid signaling and glycerophospholipid metabolism in positive mode and metabolic pathways, biosynthesis of amino acids, 2-oxocarboxylic acid metabolism, phenylalanine, tyrosine and tryptophan biosynthesis and valine, leucine and isoleucine degradation in negative mode represented the differentially expressed compounds in asthma patients compared with healthy control using UHPLC-QTOF-MS. Moreover, several metabolic pathways, including glycerophospholipid metabolism, inositol phosphate metabolism, and glycolysis or gluconeogenesis, were found most important.
Asthma and related respiratory disease are the most common chronic disease in industrialized countries, with doubled prevalence rate in recent decades, affecting 26 million children in the United States [22]. The development of new treatments for asthma, however, has not kept up with the rise in prevalence, which mainly due to the incomplete understanding of the pathological and physiological mechanism of asthma. Asthma is a heterogeneous syndrome, including many different subtypes and multiple phenotypes. Explaining its complex, underlying biological mechanisms requires new approaches. New biomarkers and therapeutic targets are needed to reflect new approaches to heterogeneity of asthma. Although the metabolomics of asthma has so far promoted studies of biological metabolites and metabolic pathways that are associated with asthma development and performance, the validation of these findings is lacking, which mainly due to the heterogeneity of asthma [23]. To better understand which metabolites are important in asthma, we performed metabolomic analysis of sputum from asthma patients and healthy controls.
The metabolomic is a biochemical phenotype of a cell or tissue, and metabolite, the component of the metabolomic, is the final product of gene expression and is produced under the action of metabolic enzymes. Many life activities in cells actually occur at the metabolite level, such as cell signaling, energy delivery and intercellular communication, are regulated by metabolites [24]. By analyzing the metabolic groups of different physiological states, we can fully understand the biochemical state of the organism or cell. The metabolomic analysis of phenotype from biological or physiological state of recently acquired information, so metabolomics analysis provided more information to reveal the relationship between gene and phenotype and to achieve the purpose of monitoring and inferring gene function [25]. By using NMR based metabolomics to study saliva in patients with intractable asthma, it was found that metabolomics successfully predicted 86% of the patients; however, the rate of success was 81% by using traditional exhaled NO content and forced expiratory volume in one second as the index [26]. Metabolomics study of hydrocortisone induced kidney deficiency in rats showed that the model group showed obvious metabolite difference compared with the normal group [27]. Kim et al. [28] collected urine samples from 50 kidney cancer patients and 13 normal controls and found that a profile analysis of low molecular weight metabolites in urine, including cluster analysis, PCA and linear discriminant analysis, can be used to screen for patients with kidney cancer, and it therefore provides a basis for the feasibility of metabonomics in the diagnosis of kidney cancer.
Mattarucchi et al. [17] applied the LC-MS technology to analyze the urine of 41 cases of children with asthma, which showed significant differences in urine metabonomics between asthmatic and normal children, with the discrimination rate of 98%, and the changes of metabolites are closely related to airway inflammation in asthma. Moreover, to assess the association between urinary romotyrosine level and children with asthma, the HPLC with online electro spray ionization tandem MS (HPLC-ESI-MS) was utilized and showed that increased romotyrosine levels contributed to the severe asthma, suggesting that urinary romotyrosine can be used to assess the risk of asthma in children and to predict the risk of an acute attack of asthma [29]. The HPLC-QTOF-MSF method was developed previously for urine metabolite profiling study and showed nineteen differential metabolites between the Shao-yao-Gancao decoction treatment group and the asthma group [30]. In the present study, 30 different metabolites in asthma patients compared with healthy control using UHPLC-QTOF-MS in positive ion mode were found, including 1-hexadecanoyl-sn-glycerol_1, glycerol 1-stearate_1, sphingosine_2, PC (16:0/16:0) and 1-hexadecanoyl-sn-glycerol_2 were increased and Phe-Ser_1, Tyr-Ala_2, Phe-Gln_1, Phe-Tyr_1, Adenine_1, etc. were decreased. 12 different metabolites in asthma patients compared with healthy control using UHPLC-QTOF-MS in negative ion mode were also found, including cytidine 2’,3’-cyclic phosphate, 1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phospho-(1’-rac-glycerol), 1-octadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphoserine, and 1-stearoyl-sn-glycerol 3-phosphocholine were increased and thymidine, gamma-L-glutamyl-L-valine_1, adenine_1, urocanic acid, phosphoenolpyruvate, phenylpyruvate_2, p-chlorophenylalanine_3, and thymine_2 were decreased. These data suggest that these different metabolites may associate with the development of asthma.
In the present study, metabolic pathways, retrograde endocannabinoid signaling and glycerophospholipid metabolism in positive mode and metabolic pathways, biosynthesis of amino acids, 2-oxocarboxylic acid metabolism, phenylalanine, tyrosine and tryptophan biosynthesis and valine, leucine and isoleucine degradation in negative mode were represented the differentially expressed compounds in asthma patients compared with healthy control. Moreover, several metabolic pathways including glycerophospholipid metabolism, inositol phosphate metabolism, and glycolysis or gluconeogenesis were found most important. Glycerophospholipid metabolism was the most significantly perturbed pathways in experimental allergic asthma [31]. Inositol phosphates are important intracellular second messengers in eukaryotic cells and are of great importance in diverse cellular functions, such as Ca2+-signaling pathways, cell growth, cell differentiation, apoptosis, endocytosis, cell migration, mRNA exportation and maintenance of genomic stability [32,33]. These findings demonstrated obvious correlation between these metabolic pathways and the development of asthma. However, some of the different metabolites in asthma patients should be further validated in our further investigation by using Real-time PCR and western blot analysis.
Our study illustrates metabolic characteristics of sputum specimens in asthma patients and emphasizes the importance of metabolomics by using UHPLC-QTOF/MS. Several metabolites, signaling pathways and metabolic pathways were found difference between healthy controls and asthma patients. The observations presented here reveal that metabolomic analysis of sputum on basis of UHPLC-QTOF/MS may have clinical implications for the early diagnosis and risk prediction of asthma.
Acknowledgements
This work was funded by the National Natural Sciences Foundation of China (NSFC 81302552) and Nanjing Medical Science and technique Development Foundation.
Disclosure of conflict of interest
None.
References
- 1.Holgate ST, Holloway J, Wilson S, Howarth PH, Haitchi HM, Babu S, Davies DE. Understanding the pathophysiology of severe asthma to generate new therapeutic opportunities. J Allergy Clin Immunol. 2006;117:496–506. doi: 10.1016/j.jaci.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 2.Lazarus SC. Clinical practice. Emergency treatment of asthma. N Engl J Med. 2010;363:755–764. doi: 10.1056/NEJMcp1003469. [DOI] [PubMed] [Google Scholar]
- 3.Lemanske RF, Busse WW. Asthma: clinical expression and molecular mechanisms. J Allergy Clin Immunol. 2010;125:S95–102. doi: 10.1016/j.jaci.2009.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masoli M, Fabian D, Holt S, Beasley R Global Initiative for Asthma (GINA) Program. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy. 2004;59:469–478. doi: 10.1111/j.1398-9995.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- 5.Cavkaytar O, Sekerel BE. Baseline management of asthma control. Allergol Immunopathol. 2014;42:162–168. doi: 10.1016/j.aller.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Akinbami LJ, Schoendorf KC. Trends in childhood asthma: prevalence, health care utilization, and mortality. Pediatrics. 2002;110:315–322. doi: 10.1542/peds.110.2.315. [DOI] [PubMed] [Google Scholar]
- 7.Kabesch M. Gene by environment interactions and the development of asthma and allergy. Toxicol Lett. 2006;162:43–48. doi: 10.1016/j.toxlet.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Fens N, de Nijs SB, Peters S, Dekker T, Knobel HH, Vink TJ, Willard NP, Zwinderman AH, Krouwels FH, Janssen HG, Lutter R, Sterk PJ. Exhaled air molecular profiling in relation to inflammatory subtype and activity in COPD. Eur Respir J. 2011;38:1301–1309. doi: 10.1183/09031936.00032911. [DOI] [PubMed] [Google Scholar]
- 9.Saude EJ, Skappak CD, Regush S, Cook K, Ben-Zvi A, Becker A, Moqbel R, Sykes BD, Rowe BH, Adamko DJ. Metabolomic profiling of asthma: diagnostic utility of urine nuclear magnetic resonance spectroscopy. J Allergy Clin Immunol. 2011;127:757–764. e751–756. doi: 10.1016/j.jaci.2010.12.1077. [DOI] [PubMed] [Google Scholar]
- 10.Qiu Y, Cai G, Su M, Chen T, Zheng X, Xu Y, Ni Y, Zhao A, Xu LX, Cai S, Jia W. Serum metabolite profiling of human colorectal cancer using GCTOFMS and UPLC-QTOFMS. J Proteome Res. 2009;8:4844–4850. doi: 10.1021/pr9004162. [DOI] [PubMed] [Google Scholar]
- 11.Mohamad N, Ismet RI, Rofiee M, Bannur Z, Hennessy T, Selvaraj M, Ahmad A, Nor F, Abdul Rahman T, Md Isa K, Ismail A, Teh LK, Salleh MZ. Metabolomics and partial least square discriminant analysis to predict history of myocardial infarction of self-claimed healthy subjects: validity and feasibility for clinical practice. J Clin Bioinforma. 2015;5:3. doi: 10.1186/s13336-015-0018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayr M, Chung YL, Mayr U, Yin X, Ly L, Troy H, Fredericks S, Hu Y, Griffiths JR, Xu Q. Proteomic and metabolomic analyses of atherosclerotic vessels from apolipoprotein E-deficient mice reveal alterations in inflammation, oxidative stress, and energy metabolism. Arterioscler Thromb Vasc Biol. 2005;25:2135–2142. doi: 10.1161/01.ATV.0000183928.25844.f6. [DOI] [PubMed] [Google Scholar]
- 13.Beger RD, Holland RD, Sun J, Schnackenberg LK, Moore PC, Dent CL, Devarajan P, Portilla D. Metabonomics of acute kidney injury in children after cardiac surgery. Pediatr Nephrol. 2008;23:977–984. doi: 10.1007/s00467-008-0756-7. [DOI] [PubMed] [Google Scholar]
- 14.Turi KN, Romick-Rosendale L, Ryckman KK, Hartert TV. A review of metabolomics approaches and their application in identifying causal pathways of childhood asthma. J Allergy Clin Immunol. 2017 doi: 10.1016/j.jaci.2017.04.021. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Checkley W, Deza MP, Klawitter J, Romero KM, Klawitter J, Pollard SL, Wise RA, Christians U, Hansel NN. Identifying biomarkers for asthma diagnosis using targeted metabolomics approaches. Respir Med. 2016;121:59–66. doi: 10.1016/j.rmed.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carraro S, Giordano G, Reniero F, Carpi D, Stocchero M, Sterk PJ, Baraldi E. Asthma severity in childhood and metabolomic profiling of breath condensate. Allergy. 2013;68:110–117. doi: 10.1111/all.12063. [DOI] [PubMed] [Google Scholar]
- 17.Mattarucchi E, Baraldi E, Guillou C. Metabolomics applied to urine samples in childhood asthma; differentiation between asthma phenotypes and identification of relevant metabolites. Biomed Chromatogr. 2012;26:89–94. doi: 10.1002/bmc.1631. [DOI] [PubMed] [Google Scholar]
- 18.Ried JS, Baurecht H, Stuckler F, Krumsiek J, Gieger C, Heinrich J, Kabesch M, Prehn C, Peters A, Rodriguez E, Schulz H, Strauch K, Suhre K, Wang-Sattler R, Wichmann HE, Theis FJ, Illig T, Adamski J, Weidinger S. Integrative genetic and metabolite profiling analysis suggests altered phosphatidylcholine metabolism in asthma. Allergy. 2013;68:629–636. doi: 10.1111/all.12110. [DOI] [PubMed] [Google Scholar]
- 19.Spahn JD. Asthma biomarkers in sputum. Immunol Allergy Clin North Am. 2007;27:607–622. doi: 10.1016/j.iac.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Ivanisevic J, Elias D, Deguchi H, Averell PM, Kurczy M, Johnson CH, Tautenhahn R, Zhu Z, Watrous J, Jain M, Griffin J, Patti GJ, Siuzdak G. Arteriovenous blood metabolomics: a readout of intra-tissue metabostasis. Sci Rep. 2015;5:12757. doi: 10.1038/srep12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Yang B, Sun H, Zhang A. Pattern recognition approaches and computational systems tools for ultra performance liquid chromatography-mass spectrometry-based comprehensive metabolomic profiling and pathways analysis of biological data sets. Anal Chem. 2012;84:428–439. doi: 10.1021/ac202828r. [DOI] [PubMed] [Google Scholar]
- 22.Akinbami LJ, Moorman JE, Bailey C, Zahran HS, King M, Johnson CA, Liu X. Trends in asthma prevalence, health care use, and mortality in the United States, 2001-2010. NCHS Data Brief. 2012:1–8. [PubMed] [Google Scholar]
- 23.Kelly RS, Virkud Y, Giorgio R, Celedon JC, Weiss ST, Lasky-Su J. Metabolomic profiling of lung function in Costa-Rican children with asthma. Biochim Biophys Acta. 2017;1863:1590–1595. doi: 10.1016/j.bbadis.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dzeja P, Terzic A. Adenylate kinase and AMP signaling networks: metabolic monitoring, signal communication and body energy sensing. Int J Mol Sci. 2009;10:1729–1772. doi: 10.3390/ijms10041729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carreno-Quintero N, Bouwmeester HJ, Keurentjes JJ. Genetic analysis of metabolome-phenotype interactions: from model to crop species. Trends Genet. 2013;29:41–50. doi: 10.1016/j.tig.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Carraro S, Rezzi S, Reniero F, Heberger K, Giordano G, Zanconato S, Guillou C, Baraldi E. Metabolomics applied to exhaled breath condensate in childhood asthma. Am J Respir Crit Care Med. 2007;175:986–990. doi: 10.1164/rccm.200606-769OC. [DOI] [PubMed] [Google Scholar]
- 27.Chen M, Zhao L, Jia W. Metabonomic study on the biochemical profiles of a hydrocortisoneinduced animal model. J Proteome Res. 2005;4:2391–2396. doi: 10.1021/pr050158o. [DOI] [PubMed] [Google Scholar]
- 28.Kim K, Aronov P, Zakharkin SO, Anderson D, Perroud B, Thompson IM, Weiss RH. Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol Cell Proteomics. 2009;8:558–570. doi: 10.1074/mcp.M800165-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wedes SH, Wu W, Comhair SA, McDowell KM, DiDonato JA, Erzurum SC, Hazen SL. Urinary bromotyrosine measures asthma control and predicts asthma exacerbations in children. J Pediatr. 2011;159:248–255. e241. doi: 10.1016/j.jpeds.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Z. The metabonomics study of Shaoyao-Gancao decoction on the acute exacerbation of asthma. Northwest Univ. 2015 [Google Scholar]
- 31.Quinn KD, Schedel M, Nkrumah-Elie Y, Joetham A, Armstrong M, Cruickshank-Quinn C, Reisdorph R, Gelfand EW, Reisdorph N. Dysregulation of metabolic pathways in a mouse model of allergic asthma. Allergy. 2017;72:1327–1337. doi: 10.1111/all.13144. [DOI] [PubMed] [Google Scholar]
- 32.Hughes AR, Horstman DA, Takemura H, Putney JW Jr. Inositol phosphate metabolism and signal transduction. Am Rev Respir Dis. 1990;141:S115–118. doi: 10.1164/ajrccm/141.3_Pt_2.S115. [DOI] [PubMed] [Google Scholar]
- 33.Duong QH, Clark KD, Lapsley KG, Pegg RB. Quantification of inositol phosphates in almond meal and almond brown skins by HPLC/ESI/MS. Food Chem. 2017;229:84–92. doi: 10.1016/j.foodchem.2017.02.031. [DOI] [PubMed] [Google Scholar]