Abstract
In this study, we investigated the levels of homeobox A13 (HOXA13) and the mechanisms underlying the co-expressed genes of HOXA13 in lung squamous cancer (LUSC), the signaling pathways in which the co-expressed genes of HOXA13 are involved and their functional roles in LUSC. The clinical significance of 23 paired LUSC tissues and adjacent non-tumor tissues were gathered. HOXA13 levels in LUSC were detected by quantitative real-time polymerase chain reaction (qRT-PCR). HOXA13 levels in LUSC from The Cancer Genome Atlas (TCGA) and Oncomine were analyzed. We performed receiver operator characteristic (ROC) curves of various clinicopathological features of LUSC. Co-expressed of HOXA13 were collected from MEM, cBioPortal and GEPIA. The functions and pathways of the most reliable overlapped genes were achieved from the Gene Otology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases, respectively. The protein-protein interaction (PPI) networks were mapped using STRING. HOXA13 in LUSC were markedly upregulated compared with those in the non-cancerous controls as demonstrated by qRT-PCR (LUSC: 0.330±0.360; CONTROLS: 0.155±0.142; P=0.021). TCGA (LUSC: 6.388±2.097, CONTROLS: 1.157±0.719; P<0.001) and Hou’s study from Oncomine (LUSC: 1.154±0.260; CONTROLS: 0.957±0.065; P=0.001) showed the same tendency. Meanwhile, the area under the curve (AUC) of TNM was calculated as 0.877 with P=0.002. Based on the HOXA13 expression data from TCGA, the ROC of the tissue types was calculated as AUC=0.971 (P<0.001). In addition, 506 genes were filtered as co-expression genes of HOXA13. The 3 most significant KEGG pathways were metabolic pathways (P=5.41E-15), the calcium signaling pathway (P=3.01E-11), and the cAMP signaling pathway (P=5.63E-11). MAPK1, GNG7, GNG12, PRKCA were selected as the hub genes. In conclusion, HOXA13 was upregulated and related to the TNM stage in LUSC. The expression of hub genes in LUSC might be deregulated by HOXA13. Moreover, the 4 co-expressed hub genes of HOXA13 might be crucial biomarkers for the diagnosis and prognosis of LUSC, as well as the development of novel therapeutic targets against LUSC.
Keywords: HOXA13, lung squamous cancer, potential mechanisms, bioinformatics
Introduction
Lung cancer is one of the most frequent cancers occurring in men and women in 2017, as well as a worldwide leading cause of cancer deaths [1]. As a life-threatening disease, it is categorized into two different major groups: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). At least 80%-85% of all lung tumors belong to NSCLC. LUSC and lung adenocarcinomas (LUAD) are the most common histological subtypes of NSCLC [2-5]. In addition, due to the high prevalence of smoking, about 30% of lung cancer patients were diagnosed as the squamous histopathological subtype [6]. The survival rate of advanced LUSC patients remains poor, only less than 5% of patients surviving 5 years after chemotherapeutic regimens [7]. Although tobacco smoke causes lung cancer was well established, not all lung cancer patients respond to it. Dysregulated genetic factors are speculated to play an important role in lung cancer susceptibility.
The homeobox genes were discovered in Drosophila firstly, where their mutations could cause the malformations of body parts in inappropriate contexts [8,9]. Both class I (HOX) and class II (non-HOX) are generally belong to homeobox genes. A total of 39 HOX genes have been identified in humans and are clustered into 4 groups named A, B, C and D, respectively. In addition, each member of HOX genes in a cluster is named from 1 to 13 [10,11]. Different clusters of homeobox genes play essential roles in embryonic morphogenesis, including organ systems, skeletal, limb and craniofacial region. In adults, homeobox genes also regulate tissue regeneration and play fundamental roles for the control of the self-renewal and differentiation of hematopoietic progenitors [8,12,13]. A lot of studies have reported that abnormal levels of HOX genes in certain organs could either suppress tumors or promote tumors, including the prostate [14], ovary [15], breast [16], kidney [17], and lung [18,19]. Altered expression of HOXA genes were reported in breast and ovarian cancers. In colon cancers, both HOXB and HOXD genes were dysregulated. Moreover, aberrantly expressed HOXC genes were indicated in prostate and lung cancers [20]. Based on the gene amplification, loss of heterozygosity, histone deacetylation and CpG island promoter hypermethylation, dysregulated HOX genes would facilitate the development and progression of cancers consequently [21]. As a member of the homeobox genes, HOXA13 has an oncogene-like character in multiple cancers. HOXA13 can enhance gastric cancer cell invasion and the epithelial-to-mesenchymal transition (EMT) via the TGF-β signaling pathway. Upregulation of HOXA13 is related to a poorer prognosis of gastric cancer patients [22,23]. In prostate cancer, both cell growth and cell cycle were closely associated with downregulated HOXA13. Lower HOXA13 exerts a role of tumor-suppressor and provides a potential therapeutic approach against this malignancy [24]. In part, HOXA13 promotes progression of glioma and would be helpful to diagnose glioblastoma. HOXA13 was also revealed to be an independent prognostic factor especially in high-grade glioma [25].
However, the probable mechanisms and pathways of HOXA13 in lung cancer have rarely been reported. Sang Y et al. found that HOTTIP was a transcriptional modulator of HOXA13 and partly regulated HOXA13 to promote cell proliferation, migration, and inhibit apoptosis of lung cancer cell [26]. Kang JU et al. demonstrated that rearranged 7p arm of HOXA13 (7p15.2) may be a valuable potential target and a therapeutic target for LUAD [27]. In this study, we aimed to investigate the potential roles of HOXA13 in LUSC. The clinical significance of HOXA13 in LUSC was gathered from qRT-PCR results and the TCGA and Oncomine databases. Meanwhile, the potential mechanisms and pathways of the co-expressed genes of HOXA13 in LUSC were predicted. Moreover, the correlated functions between HOXA13 and its several pivotal co-expressed genes in LUSC were studied based on the aforementioned information.
Methods and materials
Clinicopathological significance
Sample collection
23 paired LUSC and adjacent non-cancerous lung tissues were obtained from patients who performed surgery at The First Affiliated Hospital of Guangxi Medical University between January 2012 and February 2014 and were histopathologically diagnosed with LUSC. The clinicopathological information of the patients is shown in Table 1. The study was undertaken with the understanding and written consent of each patient.
Table 1.
The clinical significance of 23 LUSC patients
| Clinical significance | N | |
|---|---|---|
| Clinicopathological types | LUSC | 23 |
| Adjacent non-cancerous tissues | 23 | |
| Tumor size | ≤3 cm | 7 |
| >3 cm | 16 | |
| TNM stage | I-II | 10 |
| III-IV | 13 | |
| Gender | Male | 18 |
| Female | 5 | |
| Age | <60 years | 15 |
| ≥60 years | 8 | |
| Smoking | Yes | 11 |
| No | 12 | |
| Vascular invasion | Yes | 3 |
| No | 20 | |
| Grading | I | 0 |
| II | 16 | |
| III | 7 | |
Note: TNM, tumor-node-metastasis.
RNA extraction and qRT-PCR assay
Total RNA was extracted from FFPE tissues using the RNeasy reagent (QIAGEN, Shanghai, China) according to the manufacturer’s instructions. We used the ND-2000 NanoDrop system (Thermo Scientific, USA) to detect the concentration and purity of RNA, and the A260/280 ratio was 1.8-2.0. According to the manufacturer’s instructions, total RNA was reverse transcribed in a final volume of 10 µl using a reverse transcription kit (ABI, Life Technologies, USA). Fluorochrome SYBR Green I Master was used to establish a 20-μL real-time fluorescence PCR system. The PCR procedure was described as follows: initial denaturation at 95°C for 10 min, denaturation at 95°C for 10 s; refolding for 5 s at annealing temperature 60°C; extension at 72°C for 5 s (a total of 40 cycles). The specific primers used were as follows: HOXA13 forward primer: 5’-GAACGGCCAAATGTACTGCC-3’, reverse primer: 5’-CGCCTCCGTTTGTCCTTAGT-3’. GAPDH (internal control) forward primer: 5’-TGCACCACCAACTGCTTA-3’, reverse primer: 5’-GGATGCAGGGATGATGTTC-3’. The expression difference was calculated using the 2-ΔXθ method [28,29].
TCGA data downloading
From TCGA (http://cancergenome.nih.gov), the raw count of mRNAs (level 3) and clinical parameters of LUSC patients were downloaded. The HOXA13 levels in LUSC and AUC values of ROC were calculated based on the TCGA data. AUC values of ROC were calculated using GraphPad Prism 5. Meanwhile, the overall survival (OS) and disease free survival (DFS) of LUSC patients were evaluated by SPSS 22.0.
Oncomine data mining
To further verify the level of HOXA13 in LUSC, we amplified multiple Oncomine expression analyses for HOXA13 in LUSC datasets with expression levels (https://www.oncomine.org). HOXA13 levels in different cases were shown in scatter diagrams using GraphPad Prism 5.0.
Bioinformatics analysis
Genes extraction and KEGG pathway prediction
Genes expressed similarly to HOXA13 were extracted from 3 different datasets, including MEM (http://biit.cs.ut.ee/mem), cBioPortal (http://www.cbioportal.org), and GEPIA (http://gepia.cancer-pku.cn). The overlapped genes in any 2 or 3 datasets were chosen. For a higher quality of results, the intersected genes were used to analyze KEGG pathways by DAVID (https://david.ncifcrf.gov) [30,31]. The genes involved in various significant pathways were selected as co-expression genes of HOXA13 for further analysis.
Enrichment analysis and PPI networks
Based on the co-expression genes of HOXA13, GO term enrichment analysis was performed using Cytoscape 3.5.1. The PPI networks were mapped by STRING (http://www.string-db.org). In addition, the genes acting on others over 50 times would be selected as our hub genes in the current study. The correlations between hub genes and HOXA13 were evaluated by GraphPad Prism 5.0.
Statistical analysis
All statistical analyses were performed using SPSS 22.0 statistics software (SPSS, Chicago, USA). The clinical follow-up data of HOXA13 levels in LUSC provided by PCR and TCGA were listed as the means ± standard deviation (SD). For the parametric data of clinical features, the means of 2 continuous variables were compared using independent samples Student’s t-test. The means of 2 paired variables were calculated by paired Student’s t-test. Comparison of more than two different groups was performed by one-way ANOVA. The OS and DFS rates were plotted utilizing the Kaplan-Meier method. The correlations between HOXA13 and hub genes were calculated by Pearson Correlation. A P-value <0.05 indicated statistical significance.
Results
Clinical significance
PCR data
The results of qRT-PCR are shown in Table 2. HOXA13 was expressed at a significantly higher level in LUSC tissues (0.330±0.360) than in non-cancerous tissues (0.155±0.142) (P=0.021). In TNM I-II (0.112±0.072), the levels of HOXA13 were significantly lower than those in TNM III-IV (0.499±0.404) (P=0.005). Meanwhile, the AUC of TNM was calculated as 0.877 with P=0.002 (Figure 1). HOXA13 expression conditions for other clinical features showed no significant outcomes.
Table 2.
Correlations between clinical significance and expression of HOXA13 in LUSC
| Clinical significance | N | LUSC expression condition (2-ΔXθ) | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Mean | SD | t (or F) | P value | |||
| Clinicopathological types | LUSC | 23 | 0.330 | 0.360 | 2.480 | 0.021 |
| Adjacent non-cancerous tissues | 23 | 0.155 | 0.142 | |||
| Tumor size | ≤3 cm | 7 | 0.414 | 0.484 | 0.732 | 0.472 |
| >3 cm | 16 | 0.294 | 0.303 | |||
| TNM stage | I-II | 10 | 0.112 | 0.072 | -3.385 | 0.005 |
| III-IV | 13 | 0.499 | 0.404 | |||
| Gender | Male | 18 | 0.349 | 0.394 | 0.461 | 0.650 |
| Female | 5 | 0.264 | 0.212 | |||
| Age | <60 years | 15 | 0.340 | 0.368 | 0.170 | 0.867 |
| ≥60 years | 8 | 0.313 | 0.369 | |||
| Smoking | Yes | 11 | 0.472 | 0.461 | -1.846 | 0.089 |
| No | 12 | 0.201 | 0.166 | |||
| Vascular invasion | Yes | 3 | 0.466 | 0.176 | -0.693 | 0.496 |
| No | 20 | 0.310 | 0.379 | |||
| Grading | I | 0 | - | - | 0.788 | 0.385 |
| II | 16 | 0.375 | 0.412 | |||
| III | 7 | 0.229 | 0.183 | |||
Note: t, Student’s t-test; F, one-way ANOVA; SD, standard deviation; TNM, tumor-node-metastasis.
Figure 1.
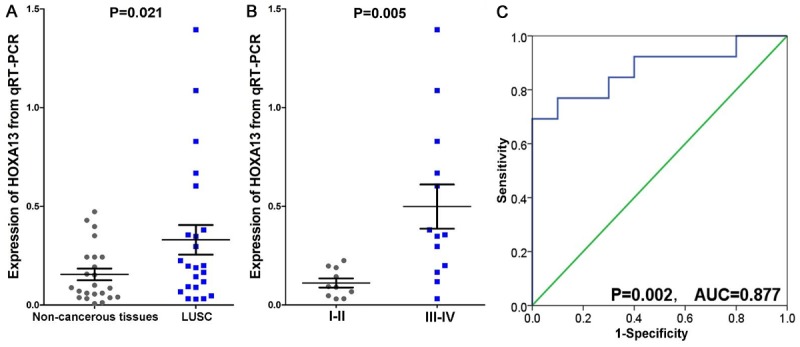
qRT-PCR results of HOXA13 levels in LUSC. A. Expression of HOXA13 in LUSC and non-cancerous tissues from qRT-PCR; B. HOXA13 levels in different TNM stages from qRT-PCR; C. ROC of HOXA13 in different TNM stages.
TCGA data
In the TCGA datasets, the HOXA13 levels in LUSC tissues and non-cancerous controls were 6.387±2.097 and 1.157±0.719, respectively, with a P value <0.001. Among the three races, HOXA13 was expressed the highest in Black individuals (6.304±1.585), followed by White individuals (5.465±2.051) and Asian individuals (4.264±2.662) (P=0.021) (Table 3). The ROC of the tissue types was calculated as AUC=0.971 (P<0.001). Unfortunately, no significance was found in both survival rate of OS and DFS (Figure 2).
Table 3.
Clinicopathological features of HOXA13 in LUSC based on TCGA
| Clinicopathological features | N | HOXA13 expression | |||
|---|---|---|---|---|---|
|
| |||||
| Mean ± SD | t (or F) | P value | |||
| Tissues | Cancer | 470 | 6.387±2.097 | -19.232 | <0.001 |
| Non-cancerous | 8 | 1.157±0.719 | |||
| Race | White | 331 | 5.465±2.051 | F=3.892 | 0.021 |
| Asian | 9 | 4.264±2.662 | |||
| Black | 28 | 6.304±1.585 | |||
| AGE | ≥60 years | 198 | 6.341±2.105 | -0.345 | 0.730 |
| <60 years | 40 | 6.470±2.377 | |||
| Gender | Male | 349 | 6.471±2.081 | 1.475 | 0.141 |
| Female | 121 | 6.145±2.133 | |||
| Status | Dead | 201 | 6.272±2.154 | -1.030 | 0.304 |
| Alive | 269 | 6.473±2.054 | |||
| Neoplasm Cancer Status | With tumor | 105 | 6.247±2.268 | -0.678 | 0.500 |
| Tumor free | 299 | 6.410±2.0725 | |||
| Stage | I-II | 382 | 6.492±2.040 | 1.798 | 0.073 |
| III-IV | 84 | 6.043±2.211 | |||
| M | M0 | 390 | 5.549±2.069 | F=0.224 | 0.800 |
| M1 | 4 | 6.041±2.528 | |||
| MX | 73 | 5.431±1.974 | |||
| T | T1-T2 | 385 | 6.432±2.015 | 0.881 | 0.380 |
| T3-T4 | 85 | 6.182±2.439 | |||
| N | N0-N1 | 424 | 5.525±2.050 | F=2.566 | 0.078 |
| N2-N3 | 45 | 5.780±1.974 | |||
| NX | 4 | 3.369±2.596 | |||
| Recurrence | Distant metastasis | 36 | 5.524±2.346 | F=0.565 | 0.571 |
| New primary tumor | 12 | 5.457±2.031 | |||
| Locoregional recurrence | 30 | 4.959±2.135 | |||
Note: t, Student’s t-test; F, one-way ANOVA; SD, standard deviation; T, tumor; N, lymph node; M, metastasis.
Figure 2.
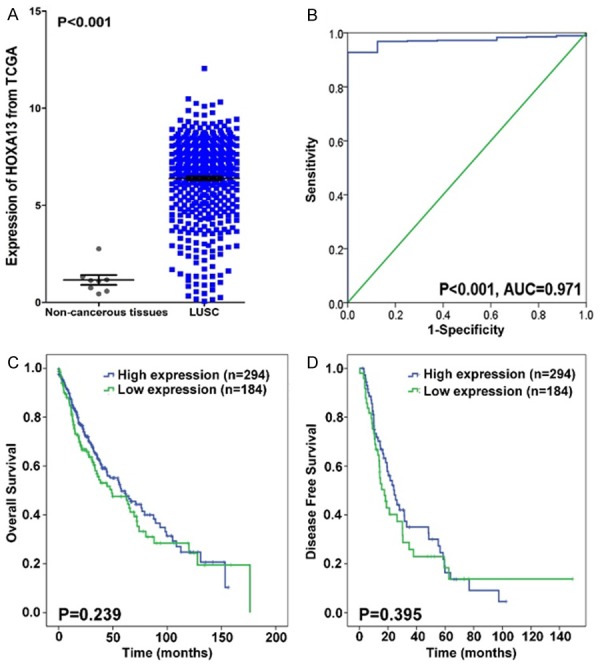
Data of HOXA13 expression levels and KM curves from TCGA. A. Expression of HOXA13 from TCGA database; B. HOXA13’s ROC curve in TCGA; C. Kaplan-Meier plots of overall survival (months); D. Kaplan-Meier plots of disease free survival (months).
Oncomine data
Two datasets related to HOXA13 and LUSC were extracted. In Hou’s study [32], 65 controls and 27 LUSC cases were found. Meanwhile, 6 controls and 14 LUSC samples were obtained from Garber’s study [33]. HOXA13 levels increased in LUSC tissues in Hou’s study (P<0.001). By contrast, Garber reported that HOXA13 levels were decreased in LUSC tissues compared with those in the controls (P=0.012) (Figure 3).
Figure 3.
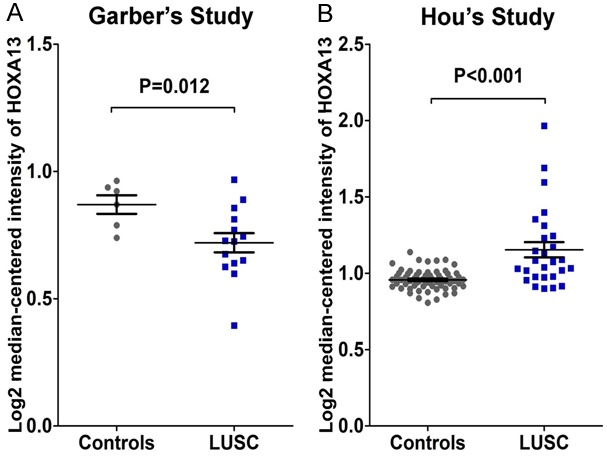
Two related studies of HOXA13 levels in LUSC. A. HOXA13 in LUSC in Garber’s study; B. Levels of HOXA13 in LUSC from Hou’s study.
Bioinformatics analysis
Extraction of similarly expressed genes of HOXA13
In MEM, 4 different probesets of HOXA13 in the human genome were filtered. The output limitation of the genes was set as 1,500 per probesets. Finally, 2,456 different genes were obtained. Meanwhile, 20,434 genes, whose expressions were similar to that of HOXA13 in LUSC, were obtained from cBioPortal. From GEPIA, the 200 most similarly expressed genes associated with HOXA13 in LUSC were selected. A total of 2,565 intersected genes in 2 or 3 datasets were extracted as shown in Veen diagram (Figure 4). After removing the repetitive genes, 2,247 genes were saved for further analysis.
Figure 4.
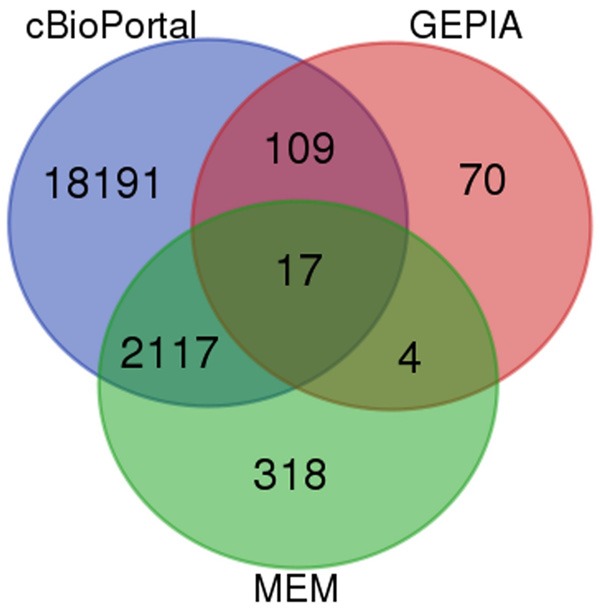
Intersected genes achieved from Venn. Each circle filled with different color represents a different platform. The numbers are the counts of genes.
KEGG pathway annotation and co-expression gene collection
Based on 2,247 selected genes, KEGG pathway annotation was performed. As shown in Table 4, the first three significant pathways were calcium signaling pathway (P=1.01E-05), cAMP signaling pathway (P=2.36E-05), and proteoglycans in cancer (P=6.73E-05). For a higher quality of results, the genes involved in all significant KEGG pathways were pooled together. Ultimately, 506 more reliable co-expression genes of HOXA13 in LUSC were selected, and KEGG pathway annotation was reperformed (Table 5). The first three pathways were metabolic pathways (P=5.41E-15), the calcium signaling pathway (P=3.01E-11), and the cAMP signaling pathway (P=5.63E-11).
Table 4.
Top 10 significant KEGG pathways of 2,247 co-expressed genes of HOXA13
| Term | Count | P Value | Genes |
|---|---|---|---|
| Calcium signaling pathway | 41 | 1.01E-05 | ADCY1, GNA11, CAMK2G, DRD5, OXTR, BDKRB2, ITPKA, ATP2B1, PRKACG, ATP2B2 etc. |
| cAMP signaling pathway | 43 | 2.36E-05 | PPARA, ATP1B1, ADCY1, ATP1B3, GNAI1, CAMK2G, DRD5, ADCY6, OXTR, GRIN3B etc. |
| Proteoglycans in cancer | 42 | 6.73E-05 | WNT5B, LUM, PPP1R12B, CAMK2G, PPP1R12C, TLR4, PDCD4, ITGB1, HOXD10, PRKACG etc. |
| Peroxisome | 23 | 7.31E-05 | ACOX2, NUDT19, EHHADH, AMACR, CRAT, PEX11G, PEX11A, PEX11B, FAR2, MLYCD etc. |
| Glutamatergic synapse | 28 | 9.83E-05 | SLC38A3, ADCY1, GNAI1, ADCY6, GNG13, GRIK5, GRIN3B, GNG12, KCNJ3, PRKACG etc. |
| Axon guidance | 30 | 1.11E-04 | NRP1, GNAI1, EFNA2, EFNA3, L1CAM, EPHB3, EPHB4, ITGB1, SEMA5A, SEMA7A etc. |
| Cholinergic synapse | 27 | 1.60E-04 | ADCY1, GNAI1, CAMK2G, GNA11, ADCY6, GNG13, GNG12, KCNJ3, PRKACG, KCNQ4 etc. |
| Endocrine and other factor-regulated calcium reabsorption | 15 | 2.46E-04 | PRKCA, ATP1B1, CLTB, ATP1B3, KLK2, ADCY6, PRKCG, BDKRB2, KLK1, PRKACG, VDR etc. |
| Insulin secretion | 22 | 3.09E-04 | PRKCA, TRPM4, ATP1B1, ADCY1, ATP1B3, CAMK2G, GNA11, ADCY6, FFAR1, PRKCG etc. |
| Pancreatic secretion | 23 | 4.33E-04 | PRKCA, ATP1B1, PNLIPRP1, ADCY1, ATP1B3, SLC12A2, ADCY6, PRKCG, ITPR3, ATP2B1 etc. |
Table 5.
Top 10 significant KEGG pathways annotations of 506 higher qualified co-expressing genes of HOXA13
| Term | Count | P Value | Genes |
|---|---|---|---|
| Metabolic pathways | 156 | 5.41E-15 | ALAD, SGMS2, NT5C3A, EHHADH, PTGS1, PPCS, PI4K2B, ACSS2, NMRK1, ITPKA etc. |
| Calcium signaling pathway | 41 | 3.01E-11 | ADCY1, GNA11, CAMK2G, DRD5, OXTR, BDKRB2, ITPKA, ATP2B1, PRKACG, ATP2B2 etc. |
| cAMP signaling pathway | 43 | 5.63E-11 | PPARA, ATP1B1, ADCY1, ATP1B3, GNAI1, CAMK2G, DRD5, ADCY6, OXTR, GRIN3B etc. |
| Pathways in cancer | 64 | 2.92E-10 | FGF6, ADCY1, PPARD, GNA11, ADCY6, PPARG, LPAR1, PTEN, PRKACG, PLCB3 etc. |
| Proteoglycans in cancer | 42 | 2.95E-10 | WNT5B, LUM, PPP1R12B, CAMK2G, PPP1R12C, TLR4, PDCD4, ITGB1, HOXD10, PRKACG etc. |
| Endocytosis | 46 | 9.05E-09 | CLTB, CHMP3, TSG101, CHMP4B, CAPZA2, STAM2, KIAA0196, ASAP2, PIP5K1B, ASAP1 etc. |
| Axon guidance | 30 | 1.02E-08 | NRP1, GNAI1, EFNA2, EFNA3, L1CAM, EPHB3, ITGB1, EPHB4, SEMA5A, SEMA7A etc. |
| Glutamatergic synapse | 28 | 1.45E-08 | SLC38A3, ADCY1, GNAI1, ADCY6, GNG13, GRIK5, GRIN3B, GNG12, KCNJ3, PRKACG etc. |
| Focal adhesion | 39 | 2.93E-08 | TLN2, PPP1R12B, PPP1R12C, ARHGAP35, MYL10, PTEN, ITGB1, LAMB3, ARHGAP5, ITGB8 etc. |
| Neuroactive ligand-receptor interaction | 47 | 2.98E-08 | TSPO, THRA, THRB, DRD5, GRIK5, OXTR, GRIN3B, BDKRB2, LPAR1, VIPR1 etc. |
Enrichment analysis
Five hundred six co-expression genes of HOXA13 were annotated in GO. There were three categories of GO, including biological process (BP), molecular function (MF), and cellular component (CC). The top 5 most significantly enriched pathways of each category are shown in Table 6. In the BP category, the most significant pathway was axon guidance (6.63E-09). Additionally, in the MF and CC categories, the most significantly enriched pathways were ATP binding (8.62E-08) and cytosol (1.08E-17), respectively.
Table 6.
Top 5 most significant enriched pathways of BP, MF and CC categories
| Category | Term | Count | P Value | Genes |
|---|---|---|---|---|
| GOTERM_BP_DIRECT | Axon guidance | 22 | 6.63E-09 | NRP1, KIF5B, EFNA2, EFNB2, EFNA3, NTN4, ARHGAP35, L1CAM, EPHB3, SLIT2 etc. |
| GOTERM_BP_DIRECT | Positive regulation of cell migration | 23 | 1.87E-08 | PRKCA, EGFR, BMP4, WNT5B, SMAD3, HGF, SEMA5A, MAPK1, SEMA6B, SEMA6C etc. |
| GOTERM_BP_DIRECT | Semaphorin-plexin signaling pathway | 11 | 1.91E-08 | SEMA5A, SEMA6B, NRP1, SEMA6C, SEMA4G, SEMA3F, SEMA7A, RAC1, MET, SEMA3C, SEMA3B |
| GOTERM_BP_DIRECT | Neural crest cell migration | 12 | 4.45E-08 | SEMA5A, SEMA6B, SEMA6C, SEMA4G, SEMA3F, SEMA7A, KITLG, SEMA3C, SEMA3B, ISL1, HTR2B, ACVR1 |
| GOTERM_BP_DIRECT | Response to drug | 29 | 7.15E-08 | ALAD, TSPO, ADCY1, ASS1, PPARG, OXTR, PDX1, PTEN, MTHFR, GATA4 etc. |
| GOTERM_MF_DIRECT | Atp binding | 80 | 8.62E-08 | ATP1B1, ADCY1, ADCY6, PIP5K1B, HLCS, PI4K2B, ACSS2, ITPKA, NMRK1, INSRR etc. |
| GOTERM_MF_DIRECT | Semaphorin receptor binding | 8 | 2.61E-06 | SEMA5A, SEMA6B, SEMA6C, SEMA4G, SEMA3F, SEMA7A, SEMA3C, SEMA3B |
| GOTERM_MF_DIRECT | Chemorepellent activity | 8 | 8.57E-06 | SEMA5A, SEMA6B, SEMA6C, SEMA4G, SEMA3F, SEMA7A, SEMA3C, SEMA3B |
| GOTERM_MF_DIRECT | Calmodulin binding | 19 | 9.50E-06 | TRPM4, EGFR, SLC8A1, ADCY1, CAMK2G, GRIN1, IQGAP3, ITPKA, ATP2B1, ATP2B2 etc. |
| GOTERM_MF_DIRECT | Drug binding | 12 | 1.08E-05 | P2RX4, PPARA, ATP1B1, PPARD, PPARG, CHRNB2, PDE4D, PPP3CA, RARB, HTR2B, AOC1, CHRNA2 |
| GOTERM_CC_DIRECT | Cytosol | 167 | 1.08E-17 | ALAD, CHMP3, THRA, NT5C3A, CHMP4B, EHHADH, CAPZA2, PPP2R5C, IQGAP3, PPCS etc. |
| GOTERM_CC_DIRECT | Plasma membrane | 187 | 3.88E-15 | ATP1B1, ADCY1, CHMP3, ATP1B3, SGMS2, EFNA2, GNA11, EFNA3, GDF5, ADCY6 etc. |
| GOTERM_CC_DIRECT | Extracellular exosome | 133 | 1.79E-11 | ATP1B1, TSPO, ALAD, ADCY1, CHMP3, ATP1B3, THRB, CHMP4B, CAPZA2, GNA11 etc. |
| GOTERM_CC_DIRECT | Peroxisome | 20 | 3.48E-11 | ACOX2, EHHADH, AMACR, CRAT, PEX11G, PEX11A, PEX11B, FAR2, MLYCD, GSTK1 etc. |
| GOTERM_CC_DIRECT | Peroxisomal matrix | 13 | 2.02E-09 | ACOX2, FAR2, NUDT19, MLYCD, EHHADH, AMACR, IDH1, ABCD3, CAT, CRAT, SCP2, CROT, ACAA1 |
Note: BP, biological process; MF, molecular function; CC, cellular component.
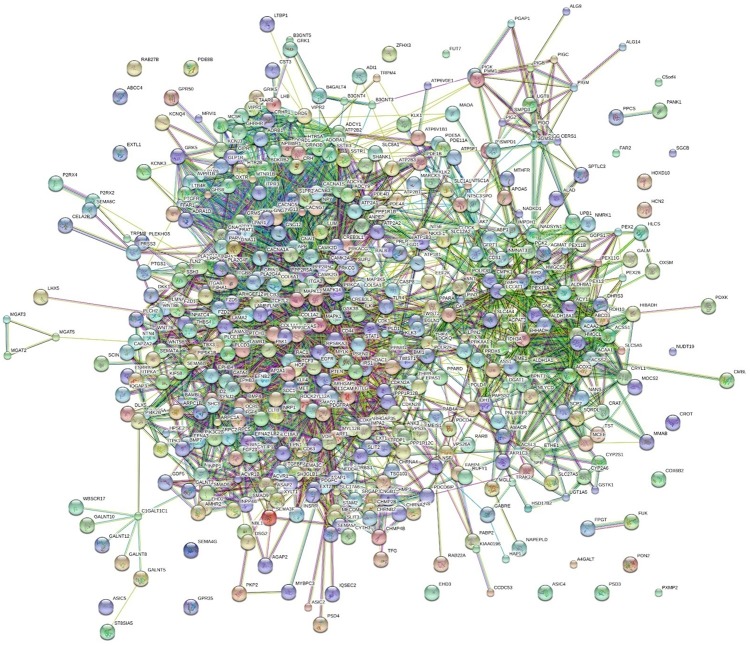
PPI network
The PPI network contained 485 items (Figure 5). The top 10 interacting gene pairs with the highest combined score are listed in Table 7. Moreover, MAPK1, GNG7, GNG12, PRKCA were selected as hub genes in this study. In addition to MAPK1, the remaining three hub genes were all significantly decreased in LUSC tissues in Figure 6 (P<0.001). In addition, there was no significant correlations between the levels of hub genes and HOXA13 in LUSC (Figure 7).
Figure 5.
PPI networks of co-expressed genes of HOXA13 in LUSC. Network nodes represent different genes. Edges represent protein-protein associations.
Table 7.
Top 10 interacted genes pairs with the highest combined score
| Node 1 | Node 2 | Neighborhood_on_chromosome | Gene_fusion | Phylogenetic_cooccurrence | Homology | Coexpression | Experimentally_determined_interaction | Database_annotated | Automated_textmining | Combined_score |
|---|---|---|---|---|---|---|---|---|---|---|
| VPS35 | VPS26A | 0 | 0 | 0 | 0 | 0.198 | 0.996 | 0.9 | 0.961 | 0.999 |
| EPHB4 | EFNB2 | 0 | 0 | 0 | 0 | 0.405 | 0.651 | 0.9 | 0.961 | 0.999 |
| ACAA1 | EHHADH | 0.505 | 0.117 | 0 | 0 | 0.678 | 0.94 | 0.951 | 0.832 | 0.999 |
| CHMP3 | CHMP4B | 0 | 0 | 0 | 0 | 0.225 | 0.977 | 0.9 | 0.587 | 0.999 |
| PARVA | ILK | 0 | 0 | 0 | 0 | 0.12 | 0.993 | 0.9 | 0.941 | 0.999 |
| CAMK2A | CAMK2D | 0 | 0 | 0.527 | 0.987 | 0 | 0.993 | 0.9 | 0.389 | 0.999 |
| ARPC5 | ARPC2 | 0 | 0 | 0 | 0 | 0.222 | 0.984 | 0.9 | 0.905 | 0.999 |
| PSEN1 | APP | 0 | 0 | 0 | 0 | 0.048 | 0.406 | 0.9 | 0.987 | 0.999 |
| GMPS | IMPDH1 | 0.505 | 0 | 0.352 | 0 | 0.952 | 0.928 | 0.951 | 0.689 | 0.999 |
| ARPC2 | ARPC1B | 0 | 0 | 0 | 0 | 0.149 | 0.968 | 0.9 | 0.878 | 0.999 |
Figure 6.
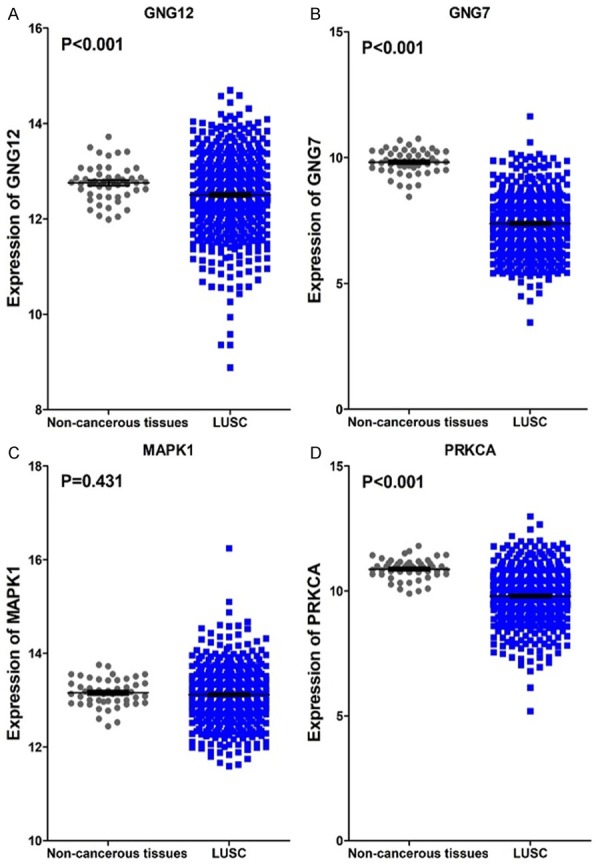
The 4 hub genes expression levels in LUSC. A. GNG12 levels in tissues of LUSC and non-tumor tissues; B. The gene GNG7 expressing conditions in LUSC and non-cancerous tissues; C. Comparison of MAPK1 levels in LUSC and controls; D. Expression of PRKCA in LUSC and normal tissues.
Figure 7.
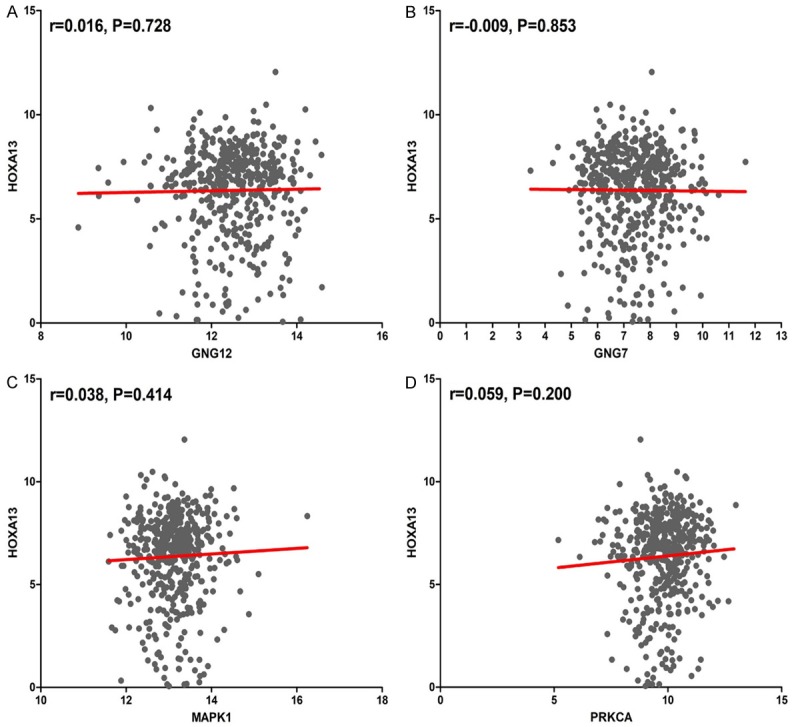
Correlations of 4 hub genes and HOXA13 in LUSC. A. Correlations between HOXA13 and GNG12; B. Correlations between HOXA13 and GNG7; C. Correlations between HOXA13 and MAPK1; D. Correlations between HOXA13 and PRCKA.
Discussion
HOX family members were known as a class of remarkable development-associated genes, aberrant expression of HOX genes have been discovered in many studies related to various solid tumors. In the present study, an increased level of HOXA13 in LUSC tissues was verified by qRT-PCR. The same expression trends were found in TCGA and Hou’s study [32] from Oncomine. However, Garber’s study [33] from Oncomine showed an inverse trend in the HOXA13 level such that it was expressed higher in controls than in LUSC tissues. Abate-Shen et al. proposed three mechanisms that drive the deregulation of aberrant HOX genes expression, including temporospatial deregulation, gene dominance and epigenetic deregulation. The first two mechanisms were demonstrated to be generally occurred in tumorigenesis which caused by HOX genes. Inversely, epigenetic deregulation could be found evidently in tissues when HOX genes function as tumor suppressor [34]. High HOXA13 in LUSC might be closely associated with temporospatial deregulation and gene dominance. Moreover, based on the results from qRT-PCR, HOXA13 was significantly higher in TNM III-IV than in TNM I-II. Our findings suggest that HOXA13 is likely involved in the tumorigenesis and progression of LUSC.
Among all the pathways predicted by the co-expression genes of HOXA13, “metabolic pathways” was the most significant. Metabolism is involved directly or indirectly in essentially all activities of a cell. In human lung cancer cell, metabolic reprograming of tumor cell directly connects to tumor evasion caused by the immune response [35]. It is concluded that most NSCLC cases are characterized by an orchestrated activation of glucose absorption and metabolism toward anaerobic pathways [36]. Moreover, phosphoglycerate dehydrogenase describes a unique metabolic program which is activated in a lung adenocarcinoma subset. This metabolic program could confer growth of malignancy and survival and may have therapeutic implications [37]. As a specific metabolism feature, upregulated GLUT1 and glycolytic metabolism could accelerate targeted treatment of LUSC. It also has been shown to be related to define histological subtypes of metabolic dependencies within NSCLC [5]. Broadly, vast factors were reported to be associated with metabolism in NSCLC. However, no clear mechanisms of metabolism have been described thus far in LUSC. Therefore, more comprehensive evidence of metabolic factors in carcinogenesis and development of LUSC should be validated. In addition, previous studies have reported relevant functions of the calcium signaling pathway. In the trisphosphate/calcium signaling pathway, generated Ca2+ could directly control metabolism, proliferation, smooth muscle contraction and so on [38]. Radioresistance of NSCLC cells could also be promoted by intracellular Ca2+ through the phosphorylation of the Akt signaling pathway [39]. Furthermore, tumor-related calcium signal transducer 2 (trop2) was identified to be correlated with tumor proliferation and invasion of NSCLC. Trop2 promoted the neovascularization of NSCLC via the activation of the ERK1/2 signaling pathway [40]. In the future, protein A2 combined with S100 calciummay serve as a dependable biomarker for the diagnosis and prognosis of NSCLC [41]. We deduced that Ca2+ might regulate multiple clinicopathological features of LUSC, such as tumorsize and vascular invasion conditions. Additionally, proteins and pathways related to Ca2+ might provide a novel strategy for LUSC treatment. As a second messenger that mobilize signaling pathways, 3’,5’-Cyclic adenosine monophosphate (cAMP) regulates various cellular functions, such as metabolism, cell proliferation, gene transcription, and cell migration [42]. The cAMP signaling system could induce the apoptosis of lung cancer cells [43]. cAMP signaling also modulates human lung cancer cell death induced by anticancer drugs [44]. Thus, deeper insights into the mechanisms of cAMP signaling pathways in NSCLC would be beneficial to LUSC treatment.
MAPK1, GNG7, GNG12, and PRKCA are the primary genes closely related to HOXA13 in LUSC. These 4 hub genes were all involved in the significant annotation of ‘pathways in cancers’. Both guanine nucleotide binding protein 7 (GNG7) and 12 (GNG12) are members of the large G protein gamma family. Previous studies have discovered that GNG7 might block proliferation of cells in multicellular organisms [45]. Downregulated GNG7 was found in many cancers, such as pancreatic cancer [46], esophageal cancer [47], and gastrointestinal tract cancer [45]. These results were consistent with a lower level of GNG7 in LUSC in this study. Therefore, we speculated that GNG7 might play a novel therapeutic role in LUSC by inhibiting the growth of cancer cells. Moreover, in living cells, protein kinase C phosphorylates GNG12 when receptors and G proteins are activated [48]. The regulator of G protein signaling 20 (RGS20) and G protein-coupled receptor 56 (GPR56) could both enhance cell invasion [49,50]. Downregulated GNG12 might be an important oncogene in LUSC due to the deregulation and functions of various G proteins. In addition, MAPK1 might be responsible for the development of smoking-induced lung cancer [51]. In LUSC cell lines which derived from the tongue, larynx and lung, MAPK1 was indicated as a crucial downstream signal transducer [52]. Additionally, it would help to predict the outcomes of patients with LUSC and improve the treatment strategies [53]. Furthermore, the amoeboid morphology of tumor cells could be maintained by PRKCA. Serum-activated PRKCA is a reliable biomarker applicable to lung cancer diagnosis [54,55]. Meanwhile, PRKCA is identified as an effective therapeutic target for anti-metastasis and lung cancer treatment due to its important role in both mesenchymal and amoeboid invasiveness [56,57]. In LUSC, GNG7, GNG12 and PRKCA showed an inverse trend in LUSC compared with HOXA13. They were significantly decreased in LUSC tissues. Therefore, we hypothesized that HOXA13 might hinder their transcription via binding to their promoters or key signaling pathways. However, no significant direct evidence between HOXA13 and the hub genes in LUSC have been reported. The relationship between HOXA13 and the hub genes is still needed to be validated by more experiments in the future.
In conclusion, our results showed that HOXA13 is upregulated and associated with the TNM stage in LUSC. The pathway annotations are helpful to elucidate the mechanisms of LUSC. HOXA13 might hinder the transcription of hub genes in LUSC. Moreover, the 4 crucial co-expression hub genes of HOXA13 might be potential biomarkers for the diagnosis and prognosis of LUSC, as well as the development of novel therapeutic targets against LUSC.
Acknowledgements
The present study was supported by the Fund of National Natural Science Foundation of China (NSFC81560386), the Natural Science Foundation of Guangxi, China (2016GXNSFBA380039) and the Promoting Project of Basic Capacity for University Young and Middle-aged Teachers in Guangxi (KY2016LX031). We also wish to thank the public databases, such as the Gene Expression Omnibus functional genomics data repository, The Cancer Genome Atlas dataset and so on.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Tiran V, Lindenmann J, Brcic L, Heitzer E, Stanzer S, Tabrizi-Wizsy NG, Stacher E, Stoeger H, Popper HH, Balic M, Dandachi N. Primary patient-derived lung adenocarcinoma cell culture challenges the association of cancer stem cells with epithelial-to-mesenchymal transition. Sci Rep. 2017;7:10040. doi: 10.1038/s41598-017-09929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang L, Zhu W, Streicher K, Morehouse C, Brohawn P, Ge X, Dong Z, Yin X, Zhu G, Gu Y, Ranade K, Higgs BW, Yao Y, Huang J. Increased IR-A/IR-B ratio in non-small cell lung cancers associates with lower epithelial-mesenchymal transition signature and longer survival in squamous cell lung carcinoma. BMC Cancer. 2014;14:131. doi: 10.1186/1471-2407-14-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Travis WD, Brambilla E, Geisinger KR. Histological grading in lung cancer: one system for all or separate systems for each histological type? Eur Respir J. 2016;47:720–723. doi: 10.1183/13993003.00035-2016. [DOI] [PubMed] [Google Scholar]
- 5.Goodwin J, Neugent ML, Lee SY, Choe JH, Choi H, Jenkins DMR, Ruthenborg RJ, Robinson MW, Jeong JY, Wake M, Abe H, Takeda N, Endo H, Inoue M, Xuan Z, Yoo H, Chen M, Ahn JM, Minna JD, Helke KL, Singh PK, Shackelford DB, Kim JW. The distinct metabolic phenotype of lung squamous cell carcinoma defines selective vulnerability to glycolytic inhibition. Nat Commun. 2017;8:15503. doi: 10.1038/ncomms15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Land SR, Liu Q, Wickerham DL, Costantino JP, Ganz PA. Cigarette smoking, physical activity, and alcohol consumption as predictors of cancer incidence among women at high risk of breast cancer in the NSABP P-1 trial. Cancer Epidemiol Biomarkers Prev. 2014;23:823–832. doi: 10.1158/1055-9965.EPI-13-1105-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drilon A, Rekhtman N, Ladanyi M, Paik P. Squamous-cell carcinomas of the lung: emerging biology, controversies, and the promise of targeted therapy. Lancet Oncol. 2012;13:e418–426. doi: 10.1016/S1470-2045(12)70291-7. [DOI] [PubMed] [Google Scholar]
- 8.Haria D, Naora H. Homeobox gene deregulation: impact on the hallmarks of cancer. Cancer Hallm. 2013;1:67–76. doi: 10.1166/ch.2013.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 10.Platais C, Hakami F, Darda L, Lambert DW, Morgan R, Hunter KD. The role of HOX genes in head and neck squamous cell carcinoma. J Oral Pathol Med. 2016;45:239–247. doi: 10.1111/jop.12388. [DOI] [PubMed] [Google Scholar]
- 11.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 12.Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- 13.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 14.Morgan R, Boxall A, Harrington KJ, Simpson GR, Michael A, Pandha HS. Targeting HOX transcription factors in prostate cancer. BMC Urol. 2014;14:17. doi: 10.1186/1471-2490-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan R, Plowright L, Harrington KJ, Michael A, Pandha HS. Targeting HOX and PBX transcription factors in ovarian cancer. BMC Cancer. 2010;10:89. doi: 10.1186/1471-2407-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashida T, Takahashi F, Chiba N, Brachtel E, Takahashi M, Godin-Heymann N, Gross KW, Vivanco M, Wijendran V, Shioda T, Sgroi D, Donahoe PK, Maheswaran S. HOXB9, a gene overexpressed in breast cancer, promotes tumorigenicity and lung metastasis. Proc Natl Acad Sci U S A. 2010;107:1100–1105. doi: 10.1073/pnas.0912710107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shears L, Plowright L, Harrington K, Pandha HS, Morgan R. Disrupting the interaction between HOX and PBX causes necrotic and apoptotic cell death in the renal cancer lines CaKi-2 and 769-P. J Urol. 2008;180:2196–2201. doi: 10.1016/j.juro.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 18.Morgan R, Simpson G, Gray S, Gillett C, Tabi Z, Spicer J, Harrington KJ, Pandha HS. HOX transcription factors are potential targets and markers in malignant mesothelioma. BMC Cancer. 2016;16:85. doi: 10.1186/s12885-016-2106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansour MA, Senga T. HOXD8 exerts a tumor-suppressing role in colorectal cancer as an apoptotic inducer. Int J Biochem Cell Biol. 2017;88:1–13. doi: 10.1016/j.biocel.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Bhatlekar S, Fields JZ, Boman BM. HOX genes and their role in the development of human cancers. J Mol Med (Berl) 2014;92:811–823. doi: 10.1007/s00109-014-1181-y. [DOI] [PubMed] [Google Scholar]
- 21.Joo MK, Park JJ, Chun HJ. Impact of homeobox genes in gastrointestinal cancer. World J Gastroenterol. 2016;22:8247–8256. doi: 10.3748/wjg.v22.i37.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He YX, Song XH, Zhao ZY, Zhao H. HOXA13 upregulation in gastric cancer is associated with enhanced cancer cell invasion and epithelial-to-mesenchymal transition. Eur Rev Med Pharmacol Sci. 2017;21:258–265. [PubMed] [Google Scholar]
- 23.Qu LP, Zhong YM, Zheng Z, Zhao RX. CDH17 is a downstream effector of HOXA13 in modulating the Wnt/beta-catenin signaling pathway in gastric cancer. Eur Rev Med Pharmacol Sci. 2017;21:1234–1241. [PubMed] [Google Scholar]
- 24.Zhang SR, Yang JK, Xie JK, Zhao LC. Long noncoding RNA HOTTIP contributes to the progression of prostate cancer by regulating HOXA13. Cell Mol Biol (Noisy-le-grand) 2016;62:84–88. [PubMed] [Google Scholar]
- 25.Duan R, Han L, Wang Q, Wei J, Chen L, Zhang J, Kang C, Wang L. HOXA13 is a potential GBM diagnostic marker and promotes glioma invasion by activating the Wnt and TGF-beta pathways. Oncotarget. 2015;6:27778–27793. doi: 10.18632/oncotarget.4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sang Y, Zhou F, Wang D, Bi X, Liu X, Hao Z, Li Q, Zhang W. Up-regulation of long non-coding HOTTIP functions as an oncogene by regulating HOXA13 in non-small cell lung cancer. Am J Transl Res. 2016;8:2022–2032. [PMC free article] [PubMed] [Google Scholar]
- 27.Kang JU. Characterization of amplification patterns and target genes on the short arm of chromosome 7 in early-stage lung adenocarcinoma. Mol Med Rep. 2013;8:1373–1378. doi: 10.3892/mmr.2013.1686. [DOI] [PubMed] [Google Scholar]
- 28.Rong M, He R, Dang Y, Chen G. Expression and clinicopathological significance of miR-146a in hepatocellular carcinoma tissues. Ups J Med Sci. 2014;119:19–24. doi: 10.3109/03009734.2013.856970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen G, Kronenberger P, Umelo IA, Teugels E, De Greve J. Quantification of epidermal growth factor receptor T790M mutant transcripts in lung cancer cells by real-time reverse transcriptase-quantitative polymerase chain reaction. Anal Biochem. 2010;398:266–268. doi: 10.1016/j.ab.2009.11.034. [DOI] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, Grosveld F, Philipsen S. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, Altman RB, Brown PO, Botstein D, Petersen I. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 35.Feng J, Yang H, Zhang Y, Wei H, Zhu Z, Zhu B, Yang M, Cao W, Wang L, Wu Z. Tumor cellderived lactate induces TAZ-dependent upregulation of PD-L1 through GPR81 in human lung cancer cells. Oncogene. 2017;36:5829–5839. doi: 10.1038/onc.2017.188. [DOI] [PubMed] [Google Scholar]
- 36.Giatromanolaki A, Sivridis E, Arelaki S, Koukourakis MI. Expression of enzymes related to glucose metabolism in non-small cell lung cancer and prognosis. Exp Lung Res. 2017:1–8. doi: 10.1080/01902148.2017.1328714. [DOI] [PubMed] [Google Scholar]
- 37.Zhang B, Zheng A, Hydbring P, Ambroise G, Ouchida AT, Goiny M, Vakifahmetoglu-Norberg H, Norberg E. PHGDH defines a metabolic subtype in lung adenocarcinomas with poor prognosis. Cell Rep. 2017;19:2289–2303. doi: 10.1016/j.celrep.2017.05.067. [DOI] [PubMed] [Google Scholar]
- 38.Berridge MJ. The inositol trisphosphate/calcium signaling pathway in health and disease. Physiol Rev. 2016;96:1261–1296. doi: 10.1152/physrev.00006.2016. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, He J, Zhang S, Yang Q. Intracellular calcium promotes radioresistance of nonsmall cell lung cancer A549 cells through activating Akt signaling. Tumour Biol. 2017;39:1010428317695970. doi: 10.1177/1010428317695970. [DOI] [PubMed] [Google Scholar]
- 40.Guo X, Zhu X, Zhao L, Li X, Cheng D, Feng K. Tumor-associated calcium signal transducer 2 regulates neovascularization of non-smallcell lung cancer via activating ERK1/2 signaling pathway. Tumour Biol. 2017;39:1010428317694324. doi: 10.1177/1010428317694324. [DOI] [PubMed] [Google Scholar]
- 41.Wang T, Liang Y, Thakur A, Zhang S, Liu F, Khan H, Shi P, Wang N, Chen M, Ren H. Expression and clinicopathological significance of S100 calcium binding protein A2 in lung cancer patients of Chinese Han ethnicity. Clin Chim Acta. 2017;464:118–122. doi: 10.1016/j.cca.2016.11.027. [DOI] [PubMed] [Google Scholar]
- 42.Kim EJ, Juhnn YS. Cyclic AMP signaling reduces sirtuin 6 expression in non-small cell lung cancer cells by promoting ubiquitin-proteasomal degradation via inhibition of the Raf-MEK-ERK (Raf/mitogen-activated extracellular signal-regulated kinase/extracellular signalregulated kinase) pathway. J Biol Chem. 2015;290:9604–9613. doi: 10.1074/jbc.M114.633198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allam M, Bertrand R, Zhang-Sun G, Pappas J, Viallet J. Cholera toxin triggers apoptosis in human lung cancer cell lines. Cancer Res. 1997;57:2615–2618. [PubMed] [Google Scholar]
- 44.Choi YJ, Oh JM, Kim SY, Seo M, Juhnn YS. Stimulatory heterotrimeric GTP-binding protein augments cisplatin-induced apoptosis by upregulating Bak expression in human lung cancer cells. Cancer Sci. 2009;100:1069–1074. doi: 10.1111/j.1349-7006.2009.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibata K, Tanaka S, Shiraishi T, Kitano S, Mori M. G-protein gamma 7 is down-regulated in cancers and associated with p 27kip1-induced growth arrest. Cancer Res. 1999;59:1096–1101. [PubMed] [Google Scholar]
- 46.Shibata K, Mori M, Tanaka S, Kitano S, Akiyoshi T. Identification and cloning of human G-protein gamma 7, down-regulated in pancreatic cancer. Biochem Biophys Res Commun. 1998;246:205–209. doi: 10.1006/bbrc.1998.8581. [DOI] [PubMed] [Google Scholar]
- 47.Ohta M, Mimori K, Fukuyoshi Y, Kita Y, Motoyama K, Yamashita K, Ishii H, Inoue H, Mori M. Clinical significance of the reduced expression of G protein gamma 7 (GNG7) in oesophageal cancer. Br J Cancer. 2008;98:410–417. doi: 10.1038/sj.bjc.6604124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davis CA 3rd, Pearce WH, Haines GK, Shah M, Koch AE. Increased ICAM-1 expression in aortic disease. J Vasc Surg. 1993;18:875–880. [PubMed] [Google Scholar]
- 49.Yang L, Lee MM, Leung MM, Wong YH. Regulator of G protein signaling 20 enhances cancer cell aggregation, migration, invasion and adhesion. Cell Signal. 2016;28:1663–1672. doi: 10.1016/j.cellsig.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 50.Song Y, Li A, Zhang L, Duan L. Expression of G protein-coupled receptor 56 is associated with tumor progression in non-small-cell lung carcinoma patients. Onco Targets Ther. 2016;9:4105–4112. doi: 10.2147/OTT.S106907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Z, Zhuan B, Yan Y, Jiang S, Wang T. Identification of gene markers in the development of smoking-induced lung cancer. Gene. 2016;576:451–457. doi: 10.1016/j.gene.2015.10.060. [DOI] [PubMed] [Google Scholar]
- 52.Gusenbauer S, Zanucco E, Knyazev P, Ullrich A. Erk2 but not Erk1 regulates crosstalk between Met and EGFR in squamous cell carcinoma cell lines. Mol Cancer. 2015;14:54. doi: 10.1186/s12943-015-0319-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Wang J, Chen Y, Yang L, Chen S. A prognostic 4-gene expression signature for squamous cell lung carcinoma. J Cell Physiol. 2017;232:3702–3713. doi: 10.1002/jcp.25846. [DOI] [PubMed] [Google Scholar]
- 54.Kang JH, Mori T, Kitazaki H, Niidome T, Takayama K, Nakanishi Y, Katayama Y. Serum protein kinase Calpha as a diagnostic biomarker of cancers. Cancer Biomark. 2013;13:99–103. doi: 10.3233/CBM-130340. [DOI] [PubMed] [Google Scholar]
- 55.Kang JH, Mori T, Kitazaki H, Niidome T, Takayama K, Nakanishi Y, Katayama Y. Kinase activity of protein kinase calpha in serum as a diagnostic biomarker of human lung cancer. Anticancer Res. 2013;33:485–488. [PubMed] [Google Scholar]
- 56.Vaskovicova K, Szabadosova E, Cermak V, Gandalovicova A, Kasalova L, Rosel D, Brabek J. PKCalpha promotes the mesenchymal to amoeboid transition and increases cancer cell invasiveness. BMC Cancer. 2015;15:326. doi: 10.1186/s12885-015-1347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abera MB, Kazanietz MG. Protein kinase Calpha mediates erlotinib resistance in lung cancer cells. Mol Pharmacol. 2015;87:832–841. doi: 10.1124/mol.115.097725. [DOI] [PMC free article] [PubMed] [Google Scholar]