Abstract
Sphingosine-1-phosphate receptor 1 (S1PR1) is abnormally expressed in a variety of tumors. However, the clinical implications and biological roles of S1PR1 in esophageal squamous cell carcinoma (ESCC) remain unknown. In this study, we have focused on ESCC, and analyzed the expression of S1PR1 in human specimens at various histological grades of ESCC and the role of S1PR1 in Eca109 cells. Using human ESCC tissue microarray and immunohistochemistry, we found S1PR1 protein mainly located in the cytoplasm of cancer cells and normal esophageal mucosal epithelial cells, and small amounts in the plasma membrane. The levels of cytoplasmic S1PR1 in ESCC tissues were significantly higher than those in adjacent non-cancerous tissues. Cytoplasmic S1PR1 exhibited higher expression in ESCC tissues with poor differentiation than those with well differentiation. Conversely, the positive expression of plasma membrane S1PR1 was correlated with well differentiation. Kaplan-Meier survival analysis showed that patients with positive membrane S1PR1 expression tended to have longer survival time. Univariate and multivariate Cox regression analysis revealed that membrane S1PR1 expression was an independent prognostic factor for ESCC patients. Furthermore, overexpression of cytoplasmic S1PR1 promoted Eca109 cells from G1 phase to S phase and plasma membrane S1PR1 as the opposite, which may be associated with p21. Cytoplasmic S1PR1 signaling also promoted Eca109 cells migration. Our findings demonstrate that cytoplasmic S1PR1 plays an important role in the malignant behavior of human ESCC and may serve as a new target for ESCC therapy.
Keywords: S1PR1, ESCC, cell cycle, migration, p21
Introduction
Esophageal cancer is the eighth most common cancer and the sixth leading cause of cancer-related mortality worldwide [1,2]. In China, esophageal cancer is the third most common cancer and has been ranked as the fourth leading cause of cancer death [3]. The two primary histologic types of esophageal cancer are esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). In the highest-risk area, which stretches from Northern Iran through the Central Asian republics to North-Central China, 90% of cases are ESCC [4]. Despite the development of multimodal therapies, the 5 year overall survival is approximately 15-25% [2]. It is essential to understand better the molecular characteristics of ESCC for developing novel biomarkers and targeted therapies.
Sphingosine kinases (SKs, two isoforms called SK1 and SK2) phosphorylate sphingosine to generate sphingosine 1-phosphate (S1P), a pleiotropic phospholipid mediator that is involved in a diverse array of cellular processes including proliferation, differentiation, adhesion, apoptosis, calcium homeostasis, angiogenesis, vascular and neuronal maturation, cell migration and immune responses [5,6]. Generally, S1P binds to its 5-related G-protein coupled receptors (GPCRs), named as S1PR1-5 (also known as S1P1-5, and originally termed EDG-1, 3, 5, 6, and 8, respectively), to induce cellular responses [5]. S1P can also act as a second messenger to elicit various intracellular targets [6]. Recent, growing evidence suggests SKs/S1P/S1PRs signaling axis plays important roles in growth, infiltration, metastasis, angiogenesis, autophagy of various types of tumors [7-10]. SK1 is overexpressed in a variety of cancers and possesses many characteristics of an oncogene [7]. In contrast, the connection between SK2 and cancer is still not well defined [11]. S1P stimulates S1PRs and have a significant role in cancer progression [12].
S1PR1 is the first identified S1P receptor, and is ubiquitously expressed. Increasing evidence has demonstrated S1PR1 is abnormally expressed in a variety of tumors. S1PR1 is associated with proliferation, migration, invasion of tumor cells, and also correlated with the survival, drug resistance in tumor patients [9,10,12-15]. We have previously reported both ESCC cell line Eca109 and normal human esophageal mucosal epithelium express S1PR1 [5]. The role of S1PR1 in the development of ESCC remains obscure. In the present study, we examine the expression of S1PR1 in ESCC tissues and adjacent non-cancerous tissues, and analyze the correlations between S1PR1 expressions and various clinicopathological characteristics. Furthermore, we overexpress S1PR1 in Eca109 cells to explore the roles of S1PR1 in ESCC malignancy.
Materials and methods
Reagents and cell culture
Fibronectin and fatty acid-free bovine serum albumin (FAF-BSA) were purchased from Sigma (St Louis, MO, USA). Endo-Free plasmid midi kit was from Omega (Beijing, China). Lipofectamine 2000 was obtained from Invitrogen (Carlsbad, CA, USA). Propidium iodide was from Solarbio (Shanghai, China). SV total RNA isolation system was purchased from Promega (Madison, WI, USA). PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) and SYBR®Premix Ex Taq™ II (Tli RNaseH Plus) kit were from TaKaRa Biotech (Dalian, China). Anti-S1PR1 (ab23695) antibody was obtained from Abcam (Cambridge, MA, USA). Anti-p21 antibody (#2947) and horseradish peroxidase-conjugated goat anti-rabbit IgG (#7074) were obtained from Cell Signaling Technology (Danvers, CO, USA). Goat anti-mouse IgG conjugated to horseradish peroxidase (A0216), anti-β-actin antibody (AA128), RIPA buffer, BCA protein assay kit, BeyoECL Plus, and 4’-6-diamidino-2-phenylindole (DAPI) were supplied from Beyotime (Shanghai, China). Human ESCC cell line Eca109 was purchased from Cell Bank of the Chinese Academy of Science (Shanghai, China). Eca109 cells were maintained in RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum (FBS), 100 U/ml penicillin, and 100 U/ml streptomycin in a 5% CO2 humidified incubator containing at 37°C. All tissue culture reagents were purchased from Invitrogen (Carlsbad, CA, USA).
Tissue microarray (TMA) and immunohistochemistry (IHC)
Human ESCC tissue microarray (HESo-Squ180Sur-03, Shanghai Outdo Biotech Co. Ltd., Shanghai, China) was used in this study, which contained a total of 87 pairs of ESCC tissues and matched adjacent non-cancerous tissues, and 6 non-matched ESCC tissues. The expression of S1PR1 in the tissues was evaluated by immunohistochemical staining. Briefly, formalin-fixed, paraffin-embedded sections were deparaffinized and rehydrated. Antigen retrieval was conducted at high temperature under high pressure in sodium citrate buffer (pH 6) for 10 minutes. After quenching of endogenous peroxidase activity and blocking of nonspecific binding, sections were incubated with antibody against S1PR1 (1:500) at 4°C overnight. Subsequently, the sections were serially rinsed. EnVision FLEX Visualization System (K8000; Dako, Glostrup, Denmark) was utilized for visualization. 3,3’-diaminobenzidine was used as a substrate. Finally, sections were counterstained with hematoxylin.
The staining was semi-quantitatively scored according to the staining intensity and positive rate by two pathologists in a blinded manner. The percentages of stained cells were scored from 0, 1 to 9, corresponding to the ranges from 0-5%, 5-15% to the final 85-100%. The intensity of staining was graded on the following scale: 0, no visual staining; 1, weak staining; 2, moderate staining; or 3, strong staining; the mixed state between them was recorded as 0.5. In this context, the relative levels of protein expression at histological levels were scored by multiplying the percentage score by the intensity value, i.e. by the so-called Q-score (Q = percentage × intensity; minimum = 0 and maximum = 27) proposed by Liu for TMA [16]. For data analysis, Q-score ≤13.5 were considered as low expression, and Q-score >13.5 were considered as high expression.
Immunocytochemistry (ICC)
Eca109 cells were gently placed on poly-L-lysine coated coverslips in the bottom of each well of 6-well culture plates. The cells were then incubated at 37°C in 5% CO2. When cells on the coverslips reached 50-80% confluence, the media in the wells were removed, and cells on the coverslips were washed with phosphate-buffered saline (PBS) 3 times, fixed with 4% (w/v) paraformaldehyde/PBS for 15 min and washed with PBS 3 times again. To perform ICC, the cells on the coverslips were incubated with permeabilizing solution (0.5% Triton X-100 in PBS) for 20 min and washed with PBS 3 times. The immunostaining protocol was the same as described for immunohistochemistry. Under a light microscope, the extent and subcellular localization of S1PR1 were observed.
Plasmids and transfection
Plasmid S1PR1-EGFP encoding a human S1PR1 and EGFP fusion protein was a kind gift from Dr. Meixiong Wu (Harvard Medical School, MA, USA) and has been described previously [17]. The control plasmid Control-EGFP was constructed as previously described [5]. Transfection was performed using Lipofectamine 2000 according to the manufacturer’s protocols.
Confocal microscopy
3.5×105 Eca109 cells were plated onto glass coverslips in six-well dishes. 1 day later, cells were grown to 80-90% confluence and transfected with either S1PR1-EGFP plasmid or Control-EGFP plasmid. Eca109 cells were incubated in RPMI 1640 medium containing 0.1% FAF-BSA for 12 h, 24 h and 48 h, and fixed with 4% (w/v) paraformaldehyde/PBS for 15 min at room temperature. Cell nuclei were stained with DAPI. Coverslips were mounted using an antifade medium. The fluorescence was revealed on a Leica SP5 confocal microscope (Leica Microsystems, Wetzlar, Germany).
Cell cycle analysis by flow cytometry
3.5×105 Eca109 cells were in six-well dishes. 1 day later, cells were grown to 80-90% confluence and transfected with either S1PR1-EGFP plasmid or Control-EGFP plasmid. Eca109 cells were incubated in RPMI 1640 medium containing 0.1% FAF-BSA for 12 h or 48 h. Transfected Eca109 cells were harvested by trypsinisation and washed twice in ice-cold PBS. After resuspension in ice-cold PBS, cells were fixed in a final concentration of 0.5% (w/v) ice-cold paraformaldehyde/PBS for 30 min at 4°C. Following brief centrifugation, cells were washed in ice-cold PBS then resuspended in ice-cold 70% (v/v) ethanol in PBS at -20°C overnight. Cells were centrifuged and stained with 50 μg/ml of propidium iodide in PBS containing 0.1% (v/v) Triton X-100 and 50 μg/ml RNase A for 30 min at 4°C in the dark. Finally cells were analyzed by flow cytometry (Guava easyCyte, Merck Millipore, MA, USA), and cell cycle profiles were evaluated using ModFit LT software (Verity Software House, Topsham, ME, USA). The proliferation index (PI) was calculated according to the following equation: (S+G2)/G1.
RNA extraction and quantitative reverse transcript-PCR (qRT-PCR) analysis
Total RNA was extracted using SV total RNA isolation system according to the manufacturer’s instructions. The RNA was reverse transcribed using PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time). The qRT-PCR analysis of the samples was performed using a LightCycler 96 System (Roche Diagnostics, Mannheim, Germany) with SYBR®Premix Ex Taq™ II (Tli RNaseH Plus) kit. The following primer sequences were used: for S1PR1, 5’-ATCGTCCTGAGCGTCTTCAT-3’ (Forward), 5’-CCAGGAAGTACTCCGCTCTG-3’ (Reverse); for GAPDH, 5’-ACCCACTCCTCCACCTTTG-3’ (Forward), 5’-CTCTTGTGCTCTTGCTGGG-3’ (Reverse). Amplifications were performed starting with 10 min of denaturation at 95°C, then 40 cycles of 10 s denaturation at 95°C, and 40 s of primer specific annealing and specific extension at 60°C. All reactions were run in triplicate. The results were normalized using the 2-ΔΔCT method.
Western blot analysis
Cells were lysed with ice-cold RIPA buffer, containing protease inhibitor mixture. Protein concentration was determined using BCA protein assay kit. Equal amounts of protein were separated in 12% SDS-PAGE gels, and then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, USA). After being blocked in Tris-based saline-Tween 20 (TBST) containing 5% (w/v) non-fat milk, the membranes were incubated with specific primary antibodies at 4°C overnight and then with horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibody for 70 min at room temperature. The signals were visualized using BeyoECL Plus and imaged using Fusion Fx7 imaging system (Vilber Lourmat, Marne-la-Vallée, France).
Migration assay
Migration assay was performed on a 6.5 mm-diameter transwell chamber with an 8 μm pore size (Corning, NY, USA). The underside of transwell membranes was precoated with fibronectin (10 μg/ml) and allowed to dry. Cells were trypsinized at 48 h post-transfection, washed and resuspended in RPMI 1640 medium with 0.1% FAF-BSA at 1×106 cells/ml. Cells (1×105 cells/well) were added into the upper chamber of transwell chambers, and the lower chamber was filled with RPMI 1640 medium with 0.1% FAF-BSA. Following 24 h incubation, nonmigrating cells on the upper surface of membranes were removed with cotton swabs, fixed and stained with 0.1% crystal violet in PBS at the ambient temperature, and then destained with PBS. Migrating cells were counted in 5 randomly selected high-power fields per membrane under an Olympus IX71 inverted microscope.
Statistical analysis
All group data were presented as mean ± standard deviation (SD) from at least three independent experiments. Analysis was performed with SPSS 16.0 software for windows (IBM Corp., Armonk, NY, USA). Graphical representations were carried out with GraphPad Prism 6 software (San Diego, CA, USA). The difference in the expression level of S1PR1 between ESCC tissues and adjacent non-cancerous tissues were analyzed by Wilcoxon test. The relationship between S1PR1 and clinicopathological characteristics was tested by Chi-square test or Fisher’s exact test. The survival calculations were illustrated with Kaplan-Meier curves and differences between survival curves were tested by the log-rank test. Cox regression was used for univariate and multivariate analysis. The student’s t-test was used for comparison between two groups depending on distribution. P values <0.05 were considered statistically significant.
Results
Cytoplasmic S1PR1 is highly expressed in human ESCC tissues and correlated with poor differentiation
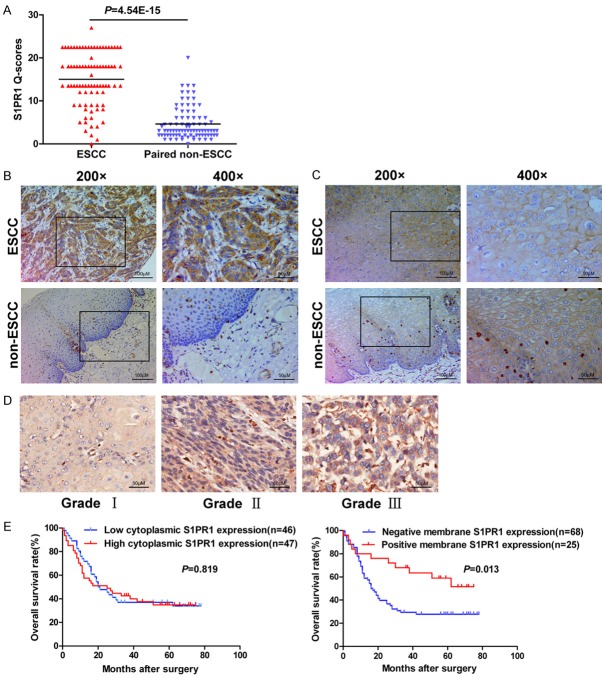
S1PR1 protein expression mainly located in the cytoplasm of cancer cells and normal esophageal mucosal epithelial cells, and small amounts in the plasma membrane. The levels of cytoplasmic S1PR1 in ESCC tissues were significantly higher than those in paired, adjacent non-cancerous tissues (P<0.001, Figure 1A and 1B). Positive plasma membrane expression of S1PR1 was rare in ESCC tissues and adjacent non-cancerous tissues. In ESCC tissues, membrane S1PR1 mainly expressed in well differentiated cancer cells, including keratin pearl. In normal human esophageal mucosal epithelium, S1PR1 expressed in the cytoplasm of the basal layer epithelial cells, which are mitotically active, in contrast, in the membrane of polyhedral higher layer cells and flattened surface cells (Figure 1C).
Figure 1.
S1PR1 protein expression is assessed in ESCC tissues and adjacent non-cancerous tissues. A, B. Significant differential expression of cytoplasmic S1PR1 was confirmed between paired ESCC tissues and their adjacent non-cancerous tissues by Q-scores, with representative images. C. Plasma membrane S1PR1 expressed in ESCC tissues and adjacent non-cancerous tissues. D. Representative IHC images for S1PR1 with different histological grade were shown. E. Kaplan-Meier curves for patients with ESCC indicated no significant association between the expression level of cytoplasmic S1PR1 and overall survival (P>0.05), while positive expression of plasma membrane S1PR1 significantly associated with better survival (P = 0.013).
The correlation between S1PR1 protein expression and the clinicopathological characteristics of the patients with ESCC was investigated (Table 1). The high expression of cytoplasmic S1PR1 in cancer cells was significantly correlated with gender (P = 0.031) and histological grade (P = 0.030), but not with other factors, including age, tumor size, pathologic type, lymph node metastasis, infiltration degree, and TNM stage. The expression of plasma membrane S1PR1 was significantly correlated with pathologic type (P = 0.044) and histological grade (P<0.001), but not with other factors. Collectively, cytoplasmic S1PR1 exhibited higher expression in ESCC tissues with poor differentiation than those with well differentiation. Conversely, the positive expression of plasma membrane S1PR1 was correlated with well differentiation. Representative IHC images for S1PR1 with different histological grade were shown in Figure 1D.
Table 1.
Association between the expression of S1PR1 and clinicopathological characteristics in ESCC (n = 93)
| Variables | Cytoplasmic S1PR1 expression | Membrane S1PR1 expression | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Low | High | P value | Negative | Positive | P value | |
| Age/year | 0.871 | 0.894 | ||||
| ≤60 | 13 | 14 | 20 | 7 | ||
| >60 | 33 | 33 | 48 | 18 | ||
| Gender | 0.031 | 0.122 | ||||
| Male | 42 | 35 | 59 | 18 | ||
| Female | 4 | 12 | 9 | 7 | ||
| Size (cm) | 0.248 | 0.793 | ||||
| ≤4 | 18 | 24 | 32 | 11 | ||
| >4 | 28 | 23 | 36 | 14 | ||
| Pathologic type | 0.595 | 0.044 | ||||
| Medullary | 10 | 14 | 20 | 4 | ||
| Ulcerative | 26 | 22 | 37 | 11 | ||
| Others | 10 | 11 | 11 | 10 | ||
| Lymph node metastasis | 0.470 | 0.068 | ||||
| Negative | 24 | 21 | 29 | 16 | ||
| Positive | 22 | 26 | 39 | 9 | ||
| Infiltration degree | 0.704 | 0.115 | ||||
| T0-2 | 11 | 11 | 13 | 9 | ||
| T3-4 | 29 | 35 | 49 | 15 | ||
| Miss | 6 | 1 | 6 | 1 | ||
| Histological grade | 0.030 | <0.001 | ||||
| I, I-II | 27 | 17 | 22 | 22 | ||
| II, II-III, III | 19 | 30 | 46 | 3 | ||
| TNM stage (AJCC) | 1.000 | 0.064 | ||||
| I | 1 | 2 | 1 | 2 | ||
| II | 21 | 24 | 30 | 15 | ||
| III | 19 | 21 | 33 | 7 | ||
| IV | 0 | 0 | 0 | 0 | ||
| Miss | 5 | 0 | 4 | 1 | ||
Positive expression of plasma membrane S1PR1 is associated with better overall survival
Survival analysis was performed to identify whether S1PR1 has prognostic role in ESCC patients. Significant association was observed between the positive expression of plasma membrane S1PR1 and better overall survival (P = 0.013). However, there was no statistically significant relationship in overall survival between patients with high level and low level of cytoplasmic S1PR1 (P>0.05) (Figure 1E). Furthermore, multivariate Cox regression analysis revealed that membrane S1PR1 was an independent prognostic factor for ESCC patients (hazard ratio = 0.415, 95% confident interval: 0.208-0.830, P = 0.013) (Table 2).
Table 2.
Univariate and multivariate analysis of the overall survival in ESCC
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Cytoplasmic S1PR1 expression | 1.060 (0.639-1.760) | 0.821 | ||
| Membrane S1PR1 expression | 0.446 (0.231-0.862) | 0.016 | 0.415 (0.208-0.830) | 0.013 |
| Age/year | 1.013 (0.985-1.043) | 0.362 | ||
| Gender | 0.556 (0.252-1.225) | 0.145 | ||
| Size (cm) | 1.070 (0.642-1.783) | 0.797 | ||
| Pathologic type | 1.009 (0.692-1.470) | 0.963 | ||
| Lymph node metastasis | 1.737 (1.037-2.912) | 0.036 | ||
| Infiltration degree | 2.038 (1.025-4.056) | 0.042 | ||
| Histological grade | 1.444 (0.865-2.411) | 0.160 | ||
| TNM stage (AJCC) | 2.148 (1.273-3.627) | 0.004 | 2.024 (1.187-3.451) | 0.010 |
HR, hazard ratio; CI, confident interval.
The expression and subcellular localization of S1PR1 is different in transiently transfected Eca109 cells at different time points
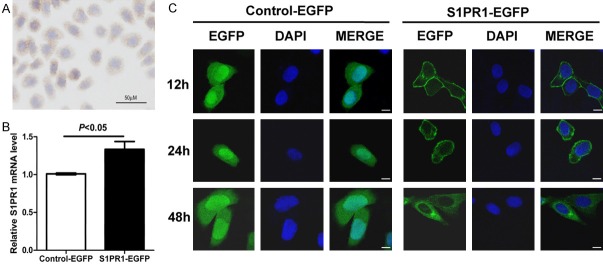
Because the mRNA expression level of S1PR1 is low in human ESCC Eca109 cells [5], the protein expression level and subcellular localization was detected by ICC. Shown in Figure 2A is representative ICC for S1PR1 protein expression in Eca109 cells. S1PR1 was weakly expressed in the cytoplasm of Eca109 cells. Thus, we overexpressed S1PR1 in Eca109 cell line to examine the biological function. S1PR1-EGFP plasmid or Control-EGFP plasmid was transiently transfected into Eca109 cells. A significant increase of S1PR1 mRNA in the S1PR1-EGFP-transfected Eca109 cells was confirmed by qRT-PCR (P<0.05, Figure 2B). Using confocal microscopy, the expression and subcellular localization of S1PR1-EGFP and Control-EGFP fusion protein were analyzed at 12 h, 24 h and 48 h post-transfection. The green fluorescence of S1PR1-EGFP fusion protein was predominantly localized in the plasma membrane, plasma membrane/cytoplasm, and cytoplasm at 12 h, 24 h and 48 h post-transfection, respectively. At all three time points, Control-EGFP fusion protein was localized in the cytoplasm and nucleus (Figure 2C). At 48 h post-transfection, the subcellular localization of S1PR1 was consistent with the results of IHC and ICC. Therefore, we chose 48 h post-transfection as the main time point.
Figure 2.
The expression and subcellular localization of S1PR1 is different in transiently transfected Eca109 cells at different time points. A. S1PR1 protein was weakly expressed in the cytoplasm of Eca109 cells. B. A significant increase of S1PR1 mRNA in the S1PR1-EGFP-transfected Eca109 cells was confirmed by qRT-PCR. C. Cellular localization pattern of S1PR1-EGFP and Control-EGFP fusion protein was analyzed by confocal microscopy. Scale bar, 10 µm.
Differential subcellular localization of S1PR1 affects cell cycle distribution and the expression of p21 in Eca109 cells
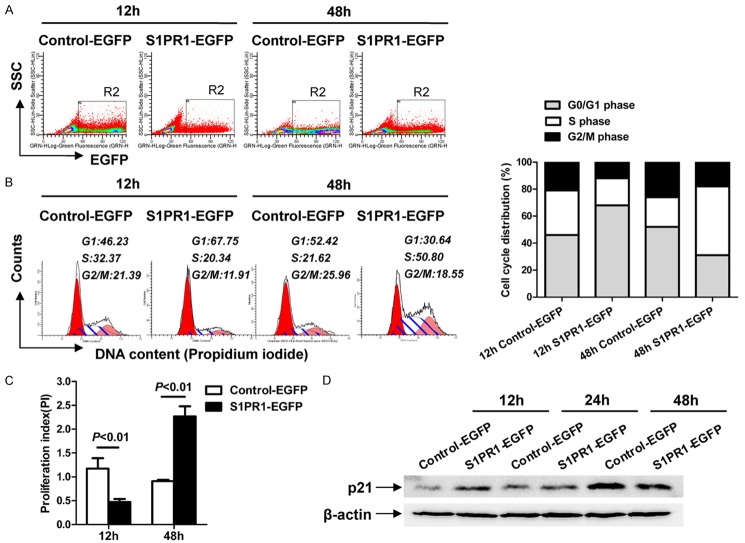
To clarify the effects of S1PR1 overexpression on the cell cycle progression in Eca109 cells, we performed flow cytometry analysis to detect cell cycle. At 12 h post-transfection, a significantly higher percentage of G1-phase cells (67.75% vs. 46.23%) were detected in S1PR1-EGFP-transfected cells compared to Control-EGFP-transfected cells (Figure 3A and 3B). The proliferation index (PI) of S1PR1-EGFP-transfected cells was apparently lower than that of Control-EGFP-transfected cells (P<0.01) (Figure 3C). By contrast, at 48 h post-transfection, a significantly lower percentage of G1-phase cells (30.64% vs. 52.42%) were detected in S1PR1-EGFP-transfected cells compared to Control-EGFP-transfected cells (Figure 3A and 3B). The proliferation index of S1PR1-EGFP-transfected cells was apparently higher than that of Control-EGFP-transfected cells (P<0.01) (Figure 3C). These data indicate that cytoplasmic S1PR1 promotes G1/S-phase transition and increases the proliferation index, whereas plasma membrane S1PR1 transduces opposite effects.
Figure 3.
Differential subcellular localization of S1PR1 affects cell cycle distribution and the expression of p21 in Eca109 cells. A. Side scatter versus EGFP fluorescence dot plot of Control-EGFP-transfected and S1PR1-EGFP-transfected cells incubated for the indicated time showing the position of gate R2. B. Histogram and the percentages of cells in the G1, S, and G2/M phases of Control-EGFP-transfected and S1PR1-EGFP-transfected cells incubated for the indicated time within gate R2. C. The proliferation index (PI) was calculated using the following equation: PI = (S+G2)/G1, where S, G2, and G1 are the percentages of cells in the S phase, G2/M phase, and G1 phase, respectively. D. The expression of p21 was measured in Control-EGFP or S1PR1-EGFP transfected Eca109 cells.
Cell cycle progression is governed by cyclins, cyclin-dependent kinases (CDKs) and CDK inhibitors (CKIs). Cyclins facilitate S phase entry, whereas CKIs (e.g., p21) keep cells arrested in G1 phase [18]. To study whether S1PR1 promotes cell cycle through the expression regulation of p21, Eca109 cells were transiently transfected with either S1PR1-EGFP or Control-EGFP plasmid for 12 h, 24 h and 48 h, followed by Western blot analysis for the levels of p21. The expression level of p21 in S1PR1-EGFP-transfected Eca109 cells was significantly higher than Control-EGFP-transfected Eca109 cells at 12 h post-transfection, Conversely, it was significantly lower at 48 h post-transfection (Figure 3D). The results show that cytoplasmic S1PR1 can significantly promote G1/S-phase transition, and plasma membrane S1PR1 inhibits G1/S-phase transition, which may be associated with the down-regulation or up-regulation of p21.
Overexpression of cytoplasmic S1PR1 enhances cell motility in Eca109 cells
The migration capability of Eca109 cells at 48 h post-transfection was detected by transwell assay. The average cell counts crossing fibronectin-coated membrane in one high power field was 67.00±11.00 for S1PR1-EGFP-transfected Eca109 cells and 39.33±6.43 for Control-EGFP-transfected Eca109 cells. The migrated number of S1PR1-EGFP-transfected Eca109 cells was more than that of Control-EGFP-transfected Eca109 cells at 48 h post-transfection (Figure 4, P<0.05). These results indicate that cytoplasmic S1PR1 promotes Eca109 cells migration.
Figure 4.
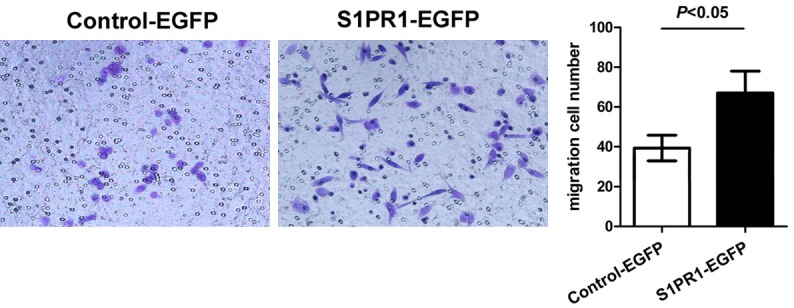
Overexpression of cytoplasmic S1PR1 promotes migration of Eca109 cells. Left panel, the migration capability of Eca109 cells was detected using transwell assay. Right panel, quantitative results were illustrated for left panels.
Discussion
Previous studies have shown S1PR1 is localized in various cellular compartments such as plasma membrane, cytoplasm and nucleus [19,20]. In our study, S1PR1 protein mainly locates in the cytoplasm of cancer cells and normal esophageal mucosal epithelial cells, and small amounts in the plasma membrane. The levels of cytoplasmic S1PR1 in ESCC tissues are significantly higher than those in adjacent non-cancerous tissues. Cytoplasmic S1PR1 exhibits higher expression in ESCC tissues with poor differentiation than those with well differentiation. The results suggest that cytoplasmic S1PR1 could be important in promoting malignant transformation in ESCC and might be a potential marker for the diagnosis of ESCC. Unexpectedly, although the expression of plasma membrane S1PR1 is rare in ESCC cancer cells, the analysis shows the positive expression of plasma membrane S1PR1 is correlated with well differentiation. The plasma membrane S1PR1 is closely linked to better prognosis and is an independent prognostic factor for ESCC patients. These results suggest plasma membrane S1PR1 could serve as a biomarker for ESCC diagnosis and prognosis. S1PR1 translocation from plasma membrane to cytoplasm might be one reason for increased S1PR1 in cytoplasm where it obtains protumor function.
We have clearly shown that S1PR1 is highly expressed in cytoplasm of ESCC cancer cells from patient samples. This prompted us to examine the biological function of S1PR1 in greater detail through in vitro analysis of ESCC cell lines. In this study, cytoplasmic S1PR1 promotes Eca109 cells from G1 phase to S phase, and plasma membrane S1PR1 inhibits G1/S-phase transition, which may be associated with the down-regulation or up-regulation of p21. p21 regulates G1/S-phase transition, not only by inactivating G1-phase cyclins/CDKs complexes, but also through other processes including interaction with proliferating cell nuclear antigen (PCNA) or downregulation of inverted CCAAT box binding protein (ICBP90) to inhibit DNA replication [21]. S1PR1 can regulate cell cycle progression through p21, which possibly affect cell proliferation and tumorigenicity. It is important to note that there have been conflicting reports about the effects of S1PR1 on the proliferation. Yoshida et al. [22] reported overexpression of S1PR1 suppresses glioma cell proliferation and downregulation of S1PR1 promotes glioma cell proliferation. Molderings et al. [23] demonstrated that S1PR1 negatively regulates cell proliferation in pheochromocytoma cells. Conversely, Liu et al. [24] reported inhibiting S1PR1 expression down-regulates STAT3 activity and causes growth inhibition of the lymphoma tumor cells in vitro and in vivo. Xu et al. [25] reported S1PR1 inhibits the apoptosis of human myeloid leukemia cells and promotes their proliferation. The reason for this discrepancy is unclear. However, different tumor cells used and different subcellular localization of S1PR1 could be a probable cause. In the studies of S1PRs, in addition to analyzing the expression level of mRNA and protein, subcellular location also should be considered.
In this study, overexpression of cytoplasmic S1PR1 promoted the migration of Eca109 cells. For the most part, binding of S1PR1 has been shown to promote migration of several cancer cell lines, including follicular thyroid cancer [13], hepatocellular carcinoma [14], gastric cancer [15], glioblastoma [26], Wilms tumor [27], etc. These results suggest that the suppression of cytoplasmic S1PR1 expression has potential for antimetastatic therapy.
Because human ESCC tissues and cell lines express SK1 that forms S1P [28], it is possible that autocrine signaling by S1P affects ESCC cell biology. In summary, the present study firstly showed that cytoplasmic S1PR1 in ESCC tissues are significantly higher than those in adjacent non-cancerous tissues. Cytoplasmic S1PR1 exhibits higher expression in ESCC tissues with poor differentiation than those with well differentiation. We have also presented experimental evidence that overexpression of cytoplasmic S1PR1 promotes Eca109 cells from G1 phase to S phase with reduced expression of p21. Cytoplasmic S1PR1 signaling also promotes Eca109 cells migration. Based on these findings, we conclude that S1PR1 is functionally important in the development and progression of ESCC and may serve as a new target for ESCC therapy.
Acknowledgements
This study was supported by the Foundation of Science and Technology Department of Sichuan Province (No. 2011JYZ023), the Natural Science Foundation of the Education Department of Sichuan Province (No. 17ZB0180), and the foundation of North Sichuan Medical College (No. CBY12-A-ZD19).
Disclosure of conflict of interest
None.
References
- 1.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 2.Lin EW, Karakasheva TA, Hicks PD, Bass AJ, Rustgi AK. The tumor microenvironment in esophageal cancer. Oncogene. 2016;35:5337–5349. doi: 10.1038/onc.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 5.Hu WM, Li L, Jing BQ, Zhao YS, Wang CL, Feng L, Xie YE. Effect of S1P5 on proliferation and migration of human esophageal cancer cells. World J Gastroenterol. 2010;16:1859–1866. doi: 10.3748/wjg.v16.i15.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan H, Pitson SM. Post-translational regulation of sphingosine kinases. Biochim Biophys Acta. 2013;1831:147–156. doi: 10.1016/j.bbalip.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Yester JW, Tizazu E, Harikumar KB, Kordula T. Extracellular and intracellular sphingosine-1-phosphate in cancer. Cancer Metastasis Rev. 2011;30:577–597. doi: 10.1007/s10555-011-9305-0. [DOI] [PubMed] [Google Scholar]
- 8.Pyne NJ, Tonelli F, Lim KG, Long JS, Edwards J, Pyne S. Sphingosine 1-phosphate signalling in cancer. Biochem Soc Trans. 2012;40:94–100. doi: 10.1042/BST20110602. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez YI, Campos LE, Castro MG, Aladhami A, Oskeritzian CA, Alvarez SE. Sphingosine-1 phosphate: a new modulator of immune plasticity in the tumor microenvironment. Front Oncol. 2016;6:218. doi: 10.3389/fonc.2016.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patmanathan SN, Wang W, Yap LF, Herr DR, Paterson IC. Mechanisms of sphingosine 1-phosphate receptor signalling in cancer. Cell Signal. 2017;34:66–75. doi: 10.1016/j.cellsig.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Maceyka M, Harikumar KB, Milstien S, Spiegel S. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol. 2012;22:50–60. doi: 10.1016/j.tcb.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pyne NJ, Pyne S. Sphingosine 1-phosphate and cancer. Nat Rev Cancer. 2010;10:489–503. doi: 10.1038/nrc2875. [DOI] [PubMed] [Google Scholar]
- 13.Balthasar S, Samulin J, Ahlgren H, Bergelin N, Lundqvist M, Toescu EC, Eggo MC, Tornquist K. Sphingosine 1-phosphate receptor expression profile and regulation of migration in human thyroid cancer cells. Biochem J. 2006;398:547–556. doi: 10.1042/BJ20060299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bao M, Chen Z, Xu Y, Zhao Y, Zha R, Huang S, Liu L, Chen T, Li J, Tu H, He X. Sphingosine kinase 1 promotes tumour cell migration and invasion via the S1P/EDG1 axis in hepatocellular carcinoma. Liver Int. 2012;32:331–338. doi: 10.1111/j.1478-3231.2011.02666.x. [DOI] [PubMed] [Google Scholar]
- 15.Yamashita H, Kitayama J, Shida D, Yamaguchi H, Mori K, Osada M, Aoki S, Yatomi Y, Takuwa Y, Nagawa H. Sphingosine 1-phosphate receptor expression profile in human gastric cancer cells: differential regulation on the migration and proliferation. J Surg Res. 2006;130:80–87. doi: 10.1016/j.jss.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Li C, Xing Z, Yuan X, Wu Y, Xu M, Tu K, Li Q, Wu C, Zhao M, Zeng R. Proteomic mining in the dysplastic liver of WHV/c-myc mice--insights and indicators for early hepatocarcinogenesis. FEBS J. 2010;277:4039–4053. doi: 10.1111/j.1742-4658.2010.07795.x. [DOI] [PubMed] [Google Scholar]
- 17.Watterson KR, Johnston E, Chalmers C, Pronin A, Cook SJ, Benovic JL, Palmer TM. Dual regulation of EDG1/S1P(1) receptor phosphorylation and internalization by protein kinase C and G-protein-coupled receptor kinase 2. J Biol Chem. 2002;277:5767–5777. doi: 10.1074/jbc.M110647200. [DOI] [PubMed] [Google Scholar]
- 18.Starostina NG, Kipreos ET. Multiple degradation pathways regulate versatile CIP/KIP CDK inhibitors. Trends Cell Biol. 2012;22:33–41. doi: 10.1016/j.tcb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aarthi JJ, Darendeliler MA, Pushparaj PN. Dissecting the role of the S1P/S1PR axis in health and disease. J Dent Res. 2011;90:841–854. doi: 10.1177/0022034510389178. [DOI] [PubMed] [Google Scholar]
- 20.Wang C, Mao J, Redfield S, Mo Y, Lage JM, Zhou X. Systemic distribution, subcellular localization and differential expression of sphingosine-1-phosphate receptors in benign and malignant human tissues. Exp Mol Pathol. 2014;97:259–265. doi: 10.1016/j.yexmp.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cazzalini O, Scovassi AI, Savio M, Stivala LA, Prosperi E. Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat Res. 2010;704:12–20. doi: 10.1016/j.mrrev.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida Y, Nakada M, Harada T, Tanaka S, Furuta T, Hayashi Y, Kita D, Uchiyama N, Hayashi Y, Hamada J. The expression level of sphingosine-1-phosphate receptor type 1 is related to MIB-1 labeling index and predicts survival of glioblastoma patients. J Neurooncol. 2010;98:41–47. doi: 10.1007/s11060-009-0064-5. [DOI] [PubMed] [Google Scholar]
- 23.Molderings GJ, Bonisch H, Bruss M, Wolf C, von Kugelgen I, Gothert M. S1P-receptors in PC12 and transfected HEK293 cells: molecular targets of hypotensive imidazoline I(1) receptor ligands. Neurochem Int. 2007;51:476–485. doi: 10.1016/j.neuint.2007.04.022. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Deng J, Wang L, Lee H, Armstrong B, Scuto A, Kowolik C, Weiss LM, Forman S, Yu H. S1PR1 is an effective target to block STAT3 signaling in activated B cell-like diffuse large Bcell lymphoma. Blood. 2012;120:1458–1465. doi: 10.1182/blood-2011-12-399030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu XQ, Huang CM, Zhang YF, Chen L, Cheng H, Wang JM. S1PR1 mediates antiapoptotic/proproliferative processes in human acute myeloid leukemia cells. Mol Med Rep. 2016;14:3369–3375. doi: 10.3892/mmr.2016.5629. [DOI] [PubMed] [Google Scholar]
- 26.Bien-Moller S, Lange S, Holm T, Bohm A, Paland H, Kupper J, Herzog S, Weitmann K, Havemann C, Vogelgesang S, Marx S, Hoffmann W, Schroeder HW, Rauch BH. Expression of S1P metabolizing enzymes and receptors correlate with survival time and regulate cell migration in glioblastoma multiforme. Oncotarget. 2016;7:13031–13046. doi: 10.18632/oncotarget.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li MH, Sanchez T, Yamase H, Hla T, Oo ML, Pappalardo A, Lynch KR, Lin CY, Ferrer F. S1P/S1P1 signaling stimulates cell migration and invasion in Wilms tumor. Cancer Lett. 2009;276:171–179. doi: 10.1016/j.canlet.2008.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan J, Tao YF, Zhou Z, Cao BR, Wu SY, Zhang YL, Hu SY, Zhao WL, Wang J, Lou GL, Li Z, Feng X, Ni J. An novel role of sphingosine kinase-1 (SPHK1) in the invasion and metastasis of esophageal carcinoma. J Transl Med. 2011;9:157. doi: 10.1186/1479-5876-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]