Abstract
Epidermal growth factor receptor (EGFR), androgen receptor (AR) and 14-3-3 sigma have been reported to be implicated in breast tumorigenesis. Their correlations, however, remain elusive in this condition. In order to examine the correlation of EGFR, AR and 14-3-3 sigma in breast cancer, and analyze their relationships with molecular subtypes of breast cancer and their impacts on overall survival, we immunohistochemistrically detected EGFR, AR and 14-3-3 sigma expression in 139 cases of breast cancer. We found that EGFR expression was negatively correlated with AR (r=-0.223, P=0.008) and positively with 14-3-3 sigma expression (r=0.181, P=0.033). There were significant differences in EGFR and AR expression between different molecular subtypes (P=0.000 and P=0.000 respectively). Kaplan-Meier cumulative survival analysis showed that none of the three biomarkers had significant impacts on overall survival of breast cancer patients (P=0.315, P=0.709, P=0.789 respectively). Univariate survival analysis revealed that tumor size (P=0.044), lymph node status (P=0.006) and clinical stage (P=0.008) were significantly associated with overall survival. Multivariate analysis demonstrated that lymph node status was the only statistically significant independent prognostic factor for overall survival [P=0.006, exp (B) =1.511, CI (1.124-2.032)]. In conclusion, EGFR expression is negatively correlated with AR and positively with 14-3-3 sigma expression in breast cancer. Furthermore, there are significant differences in EGFR and AR expression between various molecular subtypes of breast cancer. Lastly, EGFR, AR and 14-3-3 sigma have no significant impacts on overall survival of breast cancer patients.
Keywords: Correlation, EGFR, AR, 14-3-3 sigma, breast cancer, immunohistochemistry
Introduction
Breast cancer is the most commonly diagnosed cancer among women worldwide and it is also the leading cause of cancer-related deaths among women [1].
Epidermal growth factor receptor (EGFR), a prototypic cell-surface receptor belonging to the ErbB/HER oncogene family, has been known to play an important role in the regulation of cell proliferation, differentiation, and migration [2]. Its involvement in breast cancer has been extensively investigated. There is however no consensus as to the relation of EGER to the aggressive behaviors of breast cancer [3,4]. Moreover, it is questionable whether EGFR is truly a predictive biomarker for breast cancer [5].
As a receptor of precursor ligands of the estrogen receptor (ER), several groups have illuminated the role of the androgen receptor (AR) in mammary cancer since the mid-1970s [6]. Nevertheless, the potential role of AR in the development of breast cancer is inconclusive [7-9], and reports regarding the influence of AR on tumor progression were conflicting. Furthermore, the correlation between AR expression and prognosis of breast cancer patients remains unclear [10,11].
14-3-3 sigma belongs to the 14-3-3 protein family and regulates numerous cellular processes that are important to cancer development. It is the predominant isoform expressed in epithelial cells [12] and can bind to several steroid hormone receptors, including the AR [13]. Depending on the specific context and protein pathway interactions, 14-3-3 sigma may appear to have some tumor-suppressing or oncogenic properties [14]. It was reported to be directly implicated in the tumorigenesis of breast cancer [15]. However, 14-3-3 sigma has been recently documented to be a very significant prognostic indicator for breast cancer, which is the opposite of its previously known function as a tumor suppressor. All of this suggests a different role of 14-3-3 sigma in breast cancer [16].
In the present study, we attempted to examine the correlation of EGFR, AR and 14-3-3 sigma in breast cancer, and analyze their relationships with molecular subtypes of breast cancer and their impacts on overall survival.
Materials and methods
Patients and tissue microarray
Breast cancer tissue microarray (TMA) containing 150 cancer cases (HBre-Duc150Sur-02) was purchased from Shanghai Outdo Biotech Company (Shanghai, China). The recorded clinic pathological data were also obtained from the TMA, including patients’ age, sex, tumor size, pathological grade, lymph node status, distant metastasis status, clinical stage, immunohistochemical markers (ER, PR, HER-2, Ki67) and follow-up data (from August 2004 to August 2014). The overall survival time ranged from 2 to 119 months, with a median of 82 months. Detailed information is shown in Table 1. Eleven cases were excluded due to lack of complete clinical data or loss of tissue section from the glass slide. In the 139 cases of breast tumors that were eventually included, the major histological type was invasive ductal carcinoma (120 cases). The rest cases were such histological types as lobular carcinoma and mucinous carcinoma. The tissue samples on the TMA that we used in this study were collected from Tai Zhou Hospital of Zhejiang Province, China. Informed consents were obtained from all the patients, and the collection of tissue samples for research was approved by the Ethics Committee of Tai Zhou Hospital.
Table 1.
Clinical and pathological features
| Variables | No. of cases | Percentage (%) |
|---|---|---|
| Age | ||
| ≤49 years | 37 | 26.6% |
| >49 years | 102 | 73.4% |
| Tumor size (cm) | ||
| ≤2 | 38 | 27.3% |
| 2< size ≤5 | 89 | 64% |
| >5 | 12 | 8.6% |
| Lymph node status | ||
| N0 | 73 | 52.5% |
| N1 | 36 | 25.9% |
| N2 | 19 | 13.7% |
| N3 | 11 | 7.9% |
| Pathological grade | ||
| I | 10 | 7.2% |
| II | 94 | 67.6% |
| III | 35 | 25.2% |
| Clinical stage | ||
| 0 | 1 | 0.7% |
| I | 23 | 16.5% |
| II | 79 | 56.8% |
| III | 36 | 25.9% |
| ER | ||
| Positive | 77 | 55.4% |
| Negative | 62 | 44.6% |
| PR | ||
| Positive | 34 | 24.5% |
| Negative | 105 | 75.5% |
| HER2 | ||
| Positive | 43 | 30.9% |
| Negative | 96 | 69.1% |
| Ki-67 | ||
| >14% | 41 | 29.5% |
| ≤14% | 98 | 70.5% |
| AR | ||
| Positive | 94 | 67.6% |
| Negative | 45 | 32.4% |
| EGFR | ||
| Positive | 21 | 15.1% |
| Negative | 118 | 84.9% |
| 14-3-3 sigma | ||
| Positive | 63 | 45.3% |
| Negative | 76 | 54.7% |
| Subtype | ||
| Luminal A | 51 | 36.7% |
| Luminal B | ||
| Her-2 (-) | 13 | 9.4% |
| Her-2 (+) | 15 | 10.8% |
| Erb-B2 overexpression | 27 | 19.4% |
| TNBC (Triple negative) | 33 | 23.7% |
Immunohistochemistry
Briefly, the TMA sections were deparaffinized in xylene and rehydrated in a graded series of ethanol. After that, they were subjected to 5-minute high pressure for antigen retrieval. Endogenous peroxidase activity was blocked using 100 μl of peroxidase solution for 10 min. The sections were subsequently incubated overnight at 4°C with primary antibodies as follows: EGFR (Maixin, prediluted antibody ready-to-use, RMA-0554, Fuzhou Maixin Biotechnology, Fuzhou, China), AR (Maixin, prediluted antibody ready-to-use, RMA-0073, Fuzhou Maixin Biotechnology, Fuzhou, China), 14-3-3 sigma (dilution 1:2000, sc-100638, Santa Cruz Biotechnology, Dallas, Texas, U.S.A.). After washes in 1× phosphate buffered saline (PBS), the sections were incubated with biotinylated secondary antibodies (UltraSensitive TM S-P Allergic kits, Fuzhou Maixin Biotechnology, Fuzhou, China) for 30 min at room temperature, followed by incubation with streptavidin biotin peroxidase complex (UltraSensitive TM S-P Allergic kits, Fuzhou Maixin Biotechnology, Fuzhou, China). Finally, sections were incubated with DAB (DAB Detection kits, Fuzhou Maixin Biotechnology, Fuzhou, China) for 2 min. Immunostained slides were analyzed by the light microscopy.
Scoring of immunohistochemistry
All of the samples were analyzed by two pathologists who were blinded to the clinic pathologic information of the patients. EGFR positivity was defined as moderate to strong membrane staining in more than 10% of tumor cells [17]. AR positivity was defined as the presence of 10% or more positively stained nuclei in ten high-power fields [18]. More than 10% tumor cells positive for cytoplasmic staining of 14-3-3 sigma were considered 14-3-3 sigma positive [19].
Statistical analysis
Correlation between any two of the three biomarkers (EGFR, AR and 14-3-3 sigma expression) was calculated using Spearman’s correlation. That between clinical pathological variables and expression levels of EGFR, AR or 14-3-3 sigma was also determined using Spearman’s correlation. Comparisons between molecular subtypes and clinical pathological variables were carried out using Kruskal-Wallis tests. Those between clinical pathological variables and expression levels of EGFR, AR or 14-3-3 sigma were performed using Kruskal-Wallis tests which were also used to compare different clinical pathological variables. Overall survival (OS) was calculated from the diagnostic biopsy date until the date of death of any cause or last documented follow-up. Death was scored as an event, whereas patients who were alive at the time of last follow-up were censored. OS curves were visualized using the Kaplan-Meier method and the difference between survival distributions tested using the log-rank test. The Cox proportional hazards model was used to test the statistical significance of predictors in a univariate and multivariate setting. All statistical analyses were performed with the SPSS system (version 18.0 for windows; SPSS INC., Chicago, IL) and P<0.05 was regarded as statistically significant.
Results
Expression characteristics of EGFR, AR and 14-3-3 sigma
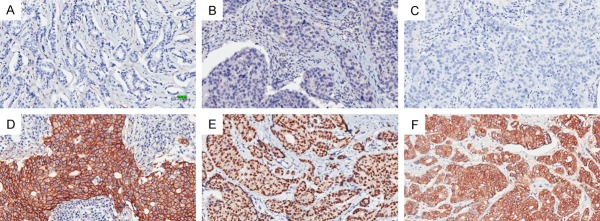
The malignant epithelial cells that were positive for EGFR showed clear membrane staining (Figure 1). AR was predominantly localized in the nuclei of cancer cells. 14-3-3 sigma was mainly expressed in the cytoplasm of glandular epithelia and surrounding myoepithelial cells. Representative images of immunohistochemical staining are shown in Figure 1.
Figure 1.
Immunostaining of EGFR, AR and 14-3-3 sigma. Negative expression of EGFR, AR and 14-3-3 sigma is shown in (A-C) respectively, and strong positive expression of EGFR, AR and 14-3-3 sigma in (D-F) respectively. (Bar=50 μm).
Clinical and pathological features
The ages of the 139 patients ranged from 33 to 88 years (median, 57 years). Other clinical and pathological characteristics are shown in Table 1. Briefly, the expression rate of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER-2) was 55.4% (77/139), 24.5% (34/139) and 30.9% (43/139) respectively. Positive immunostaining for EGFR, AR and 14-3-3 sigma was present in 15.1% (21/139), 67.6% (94/139) and 45.3% (63/139), respectively. All the cases fell into luminal A, luminal B, Erb-B2-enriched (Erb-B2 overexpression), or triple negative breast cancer (TNBC) based on the presence or absence of ER, PR, HER2, and Ki-67 [20]. The luminal A breast cancer accounted for the highest proportion (36.7%, 51/139) of the 139 cases of breast cancer. The second most common subtype was TNBC, accounting for 23.7% (33/139), which was followed by the luminal B subtype (20.2%, 28/139), among which the HER2-negative type was 9.4% (13/139) and the HER2-positive type 10.8% (15/139). The Erb-B2-enriched breast cancer made up 19.4% (27/139) (Table 1).
Association between EGFR, AR and 14-3-3 sigma and molecular subtype
Of all clinic pathological variables, EGFR, AR expression and pathological grade were demonstrated to have significant differences between various molecular subtypes. More specifically, significant difference in EGFR expression was observed in patients with different molecular subtypes (χ2=29.284, P=0.000). EGFR expression was highest in TNBC and lowest in luminal B breast cancer. The EGFR expression rate was 1.96% (1/51) in luminal A breast cancer, 0% (0/28) in luminal B breast cancer, 25.93% (7/27) in Erb-B2-enriched cancer, and 39.39% (13/33) in TNBC. EGFR expression from high to low in the order in various subtypes was TNBC, Erb-B2-enriched, luminal A, luminal B respectively. Similarly, there was significant difference in AR expression between different molecular subtypes (χ2=27.458, P=0.000). The AR expression rate was 86.27% (44/51) in luminal A, 76.92% (10/13) in HER2-negative luminal B, 80% (12/15) in HER2-positive luminal B, 62.96% (17/27) in Erb-B2-enriched, 33.33% (11/33) in TNBC. AR expression was highest in luminal A and lowest in TNBC. AR expression from high to low in the order in various subtypes was luminal A, luminal B, Erb-B2-enriched and TNBC, respectively. However, no significant difference was noted in 14-3-3 sigma expression between various molecular subtypes (χ2=4.437, P=0.350). 14-3-3 sigma expression from high to low in the order in various subtypes was TNBC, luminal B, Erb-B2 overexpression, luminal A, respectively. Additionally, the pathologic grade was highest in TNBC, while lowest in Luminal A (Table 2).
Table 2.
Association between molecular subtypes and clinical pathological variables
| Variables | Luminal A | Luminal B | Erb-B2 overexpression | TNBC | χ2 | P-value | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Her-2 (-) | Her-2 (+) | ||||||
| Total cases | 51 | 13 | 15 | 27 | 33 | ||
| Age | 4.159 | 0.385 | |||||
| ≤49 years | 12 | 2 | 3 | 11 | 9 | ||
| >49 years | 39 | 11 | 12 | 16 | 24 | ||
| Tumor size (cm) | 3.838 | 0.428 | |||||
| ≤2 | 18 | 5 | 4 | 5 | 6 | ||
| 2< size ≤5 | 28 | 7 | 11 | 19 | 24 | ||
| >5 | 5 | 1 | 0 | 3 | 3 | ||
| Lymph node status | 8.371 | 0.079 | |||||
| N0 | 30 | 7 | 7 | 9 | 20 | ||
| N1 | 13 | 6 | 3 | 7 | 7 | ||
| N2 | 6 | 0 | 4 | 7 | 2 | ||
| N3 | 2 | 0 | 1 | 4 | 4 | ||
| Pathological grade | 15.477 | 0.004 | |||||
| I | 8 | 0 | 2 | 0 | 0 | ||
| II | 38 | 9 | 7 | 19 | 21 | ||
| III | 5 | 4 | 6 | 8 | 12 | ||
| Clinical stage | 7.331 | 0.119 | |||||
| 0 | 1 | 0 | 0 | 0 | 0 | ||
| I | 9 | 3 | 2 | 3 | 6 | ||
| II | 31 | 9 | 8 | 12 | 19 | ||
| III | 10 | 1 | 5 | 12 | 8 | ||
| AR | 27.458 | 0.000 | |||||
| Negative | 7 | 3 | 3 | 10 | 22 | ||
| Positive | 44 | 10 | 12 | 17 | 11 | ||
| EGFR | 29.284 | 0.000 | |||||
| Negative | 50 | 13 | 15 | 20 | 20 | ||
| Positive | 1 | 0 | 0 | 7 | 13 | ||
| 14-3-3 sigma | 4.437 | 0.350 | |||||
| Negative | 33 | 6 | 8 | 15 | 14 | ||
| Positive | 18 | 7 | 7 | 12 | 19 | ||
Correlation of EGFR, AR and 14-3-3 sigma expression in breast cancer
EGFR expression was negatively correlated with AR expression (r=-0.223, P=0.008) and positively with 14-3-3 sigma expression (r=0.181, P=0.033) (Table 3).
Table 3.
Correlation of EGFR, AR and 14-3-3 sigma expression in breast cancer
| Variables | EGFR | |
|---|---|---|
|
| ||
| r | P-value | |
| AR | -0.223 | 0.008 |
| 14-3-3 sigma | 0.181 | 0.033 |
Correlation between EGFR and other clinical pathological variables
Table 4 shows the associations between the EGFR expression and other clinical pathological variables. EGFR expression was significantly associated with ER (r=-0.430, P=0.000) and PR (r=-0.240, P=0.004) status in a negative manner and with Ki67 level (r=0.212, P=0.012) positively, while no association was found between EGFR expression and age, tumor size, pathologic stage or lymph node status (Table 4). It was shown that the expression of ER, PR, Ki67, AR and 14-3-3 sigma was significantly different in patients with different expression levels of EGFR (Table 7). The expression of ER, PR and AR was profoundly higher in EGFR-negative patients than in EGFR-positive patients (χ2=25.483, P=0.000; χ2=7.953, P=0.005; χ2=6.882, P=0.009 respectively). The expression of ki67 and 14-3-3 sigma was much lower in EGFR-negative patients than in EGFR-positive patients (χ2=6.185, P=0.013; χ2=4.514, P=0.034, respectively) (Table 7).
Table 4.
Correlation between EGFR and other clinical pathological variables
| Variables | EGFR | |
|---|---|---|
|
| ||
| r | P-value | |
| Age | 0.027 | 0.754 |
| Tumor size (cm) | 0.109 | 0.203 |
| Lymph node status | -0.126 | 0.140 |
| Pathological grade | 0.155 | 0.069 |
| Clinical stage | -0.063 | 0.458 |
| ER | -0.430 | 0.000 |
| PR | -0.240 | 0.004 |
| HER2 | 0.022 | 0.798 |
| Ki67 | 0.212 | 0.012 |
Table 7.
Comparison between EGFR and other clinical pathological variables
| Variables | EGFR | χ2 | P-value | ||
|---|---|---|---|---|---|
|
| |||||
| Negative | Positive | ||||
| Age | ≤49 years | 32 | 5 | 0.099 | 0.753 |
| >49 years | 86 | 16 | |||
| Tumor size (cm) | ≤2 | 35 | 3 | 1.626 | 0.202 |
| 2< size ≤5 | 73 | 16 | |||
| >5 | 10 | 2 | |||
| Lymph node status | N0 | 60 | 13 | 2.182 | 0.140 |
| N1 | 29 | 7 | |||
| N2 | 18 | 1 | |||
| N3 | 11 | 0 | |||
| Pathology grade | I | 10 | 0 | 3.296 | 0.069 |
| II | 81 | 13 | |||
| III | 27 | 8 | |||
| Clinical stage | 0 | 1 | 0 | 5.556 | 0.456 |
| I | 21 | 2 | |||
| II | 62 | 17 | |||
| III | 34 | 2 | |||
| ER | Negative | 42 | 20 | 25.483 | 0.000 |
| Positive | 76 | 1 | |||
| PR | Negative | 84 | 21 | 7.953 | 0.005 |
| Positive | 34 | 0 | |||
| HER2 | Negative | 82 | 14 | 0.066 | 0.797 |
| Positive | 36 | 7 | |||
| Ki67 | ≤14% | 88 | 10 | 6.185 | 0.013 |
| >14% | 30 | 11 | |||
| AR | Negative | 33 | 12 | 6.882 | 0.009 |
| Positive | 85 | 9 | |||
| 14-3-3 sigma | Negative | 69 | 7 | 4.514 | 0.034 |
| Positive | 49 | 14 | |||
Correlation between AR and other clinical pathological variables
As shown in Table 5, AR expression was positively associated with ER (r=0.400, P=0.000), PR (r=0.358, P=0.000) status and age (r=0.175, P=0.040), negatively with tumor size (r=-0.241, P=0.004) and pathologic grade (r=-0.195, P=0.022). No association was found between AR expression and lymph node status, HER-2 or Ki67 level (Table 5). Table 8 shows that tumor size, pathological grade, age and expression of ER and PR were significantly different in patients with different expression levels of AR. The expression of ER and PR was substantially lower in AR-negative patients than in AR-positive patients (χ2=22.067, P=0.000; χ2=17.682, P=0.000 respectively). The AR expression was markedly higher in patients older than 49 years than in those equal to or younger than 49 years old (χ2=4.212, P=0.040). It was significantly increased in patients with smaller tumor size, as compared with those having larger tumor sizes (χ2=7.990, P=0.005). It was obviously higher in patients with lower pathologic grade than in those with higher pathologic grade (χ2=5.237, P=0.022).
Table 5.
Correlation between AR and other clinical pathological variables
| Variables | AR | |
|---|---|---|
|
| ||
| r | P-value | |
| Age | 0.175 | 0.040 |
| Tumor size (cm) | -0.241 | 0.004 |
| Lymph node status | 0.002 | 0.978 |
| Pathological grade | -0.195 | 0.022 |
| Clinical stage | -0.074 | 0.388 |
| ER | 0.400 | 0.000 |
| PR | 0.358 | 0.000 |
| HER2 | -0.003 | 0.975 |
| Ki67 | -0.126 | 0.141 |
Table 8.
Comparison between AR and other clinical pathological variables
| Variables | AR | χ2 | P-value | ||
|---|---|---|---|---|---|
|
| |||||
| Negative | Positive | ||||
| Age | ≤49 years | 17 | 20 | 4.212 | 0.040 |
| >49 years | 28 | 74 | |||
| Tumor sizes (cm) | ≤2 | 8 | 30 | 7.990 | 0.005 |
| 2< sizes ≤5 | 28 | 61 | |||
| >5 | 9 | 3 | |||
| Lymph node status | N0 | 24 | 49 | 0.001 | 0.978 |
| N1 | 11 | 25 | |||
| N2 | 6 | 13 | |||
| N3 | 4 | 7 | |||
| Pathology grade | I | 1 | 9 | 5.237 | 0.022 |
| II | 28 | 66 | |||
| III | 16 | 19 | |||
| Clinical stage | 0 | 0 | 1 | 0.750 | 0.386 |
| I | 6 | 17 | |||
| II | 26 | 53 | |||
| III | 13 | 23 | |||
| ER | Negative | 33 | 29 | 22.067 | 0.000 |
| Positive | 12 | 65 | |||
| PR | Negative | 44 | 61 | 17.682 | 0.000 |
| Positive | 1 | 33 | |||
| HER2 | Negative | 31 | 65 | 0.001 | 0.975 |
| Positive | 14 | 29 | |||
| Ki67 | ≤14% | 28 | 70 | 2.179 | 0.140 |
| >14% | 17 | 24 | |||
| EGFR | Negative | 33 | 85 | 6.882 | 0.009 |
| Positive | 12 | 9 | |||
| 14-3-3 sigma | Negative | 27 | 49 | 0.756 | 0.385 |
| Positive | 18 | 45 | |||
Correlation between 14-3-3 sigma and other clinical pathological variables
The 14-3-3 sigma expression was found not to be significantly associated with any of the known clinical-pathological parameters except for EGFR in invasive lesions (Tables 3, 6). EGFR expression significantly differed in patients with various levels of 14-3-3 sigma. It was much higher in patients with increased expression levels of 14-3-3 sigma than in those with less14-3-3 sigma expression (χ2=4.514, P=0.034). There were no significant differences in other clinical pathological variables except for EGFR expression between patients with different levels of 14-3-3 sigma (Table 9).
Table 6.
Correlation between 14-3-3 sigma and other clinical pathological variables
| Variables | 14-3-3 sigma | |
|---|---|---|
|
| ||
| r | P-value | |
| Age | 0.091 | 0.289 |
| Tumor size (cm) | -0.003 | 0.968 |
| Lymph node status | -0.107 | 0.208 |
| Pathological grade | 0.016 | 0.852 |
| Clinical stage | -0.103 | 0.228 |
| ER | -0.142 | 0.094 |
| PR | -0.014 | 0.872 |
| HER2 | 0.016 | 0.852 |
| Ki67 | 0.140 | 0.100 |
Table 9.
Comparison between 14-3-3 sigma and other clinical pathological variables
| Variables | 14-3-3 sigma | χ2 | P-value | ||
|---|---|---|---|---|---|
|
| |||||
| Negative | Positive | ||||
| Age | ≤49 years | 23 | 14 | 1.132 | 0.287 |
| >49 years | 53 | 49 | |||
| Tumor sizes (cm) | ≤2 | 21 | 17 | 0.002 | 0.968 |
| 2< size ≤5 | 48 | 41 | |||
| >5 | 7 | 5 | |||
| Lymph node status | N0 | 37 | 36 | 1.592 | 0.207 |
| N1 | 19 | 17 | |||
| N2 | 13 | 6 | |||
| N3 | 7 | 4 | |||
| Pathology grade | I | 6 | 4 | 0.035 | 0.851 |
| II | 51 | 43 | |||
| III | 19 | 16 | |||
| Clinical stage | 0 | 0 | 1 | 1.459 | 0.227 |
| I | 12 | 11 | |||
| II | 41 | 38 | |||
| III | 23 | 13 | |||
| ER | Negative | 29 | 33 | 2.800 | 0.094 |
| Positive | 47 | 30 | |||
| PR | Negative | 57 | 48 | 0.026 | 0.871 |
| Positive | 19 | 15 | |||
| HER2 | Negative | 53 | 43 | 0.035 | 0.851 |
| Positive | 23 | 20 | |||
| Ki67 | ≤14% | 58 | 40 | 2.704 | 0.100 |
| >14% | 18 | 23 | |||
| EGFR | Negative | 69 | 49 | 4.514 | 0.034 |
| Positive | 7 | 14 | |||
| AR | Negative | 27 | 18 | 0.756 | 0.385 |
| Positive | 49 | 45 | |||
Impacts of EGFR, AR and 14-3-3 sigma on overall survival
In the present study, none of EGFR, AR and 14-3-3 sigma was found to have a significant impact on overall survival in patients with invasive breast cancer (Figure 2). Adjusting for EGFR, we found that both AR and 14-3-3 sigma had no significant impacts on overall survival (data not shown). Similarly, neither EGFR nor 14-3-3 sigma was revealed to have a significant impact on overall survival time after adjusting for AR (data not shown). EGFR and AR also failed to significantly affect the overall survival after adjusting for 14-3-3 sigma (data not shown).
Figure 2.
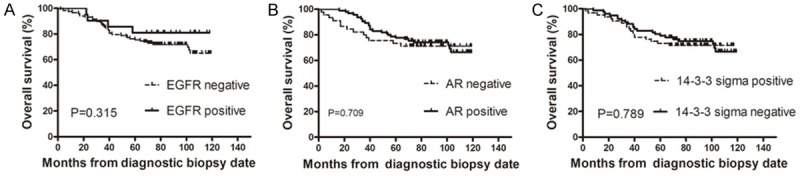
Kaplan-Meier cumulative survival analyses of EGFR, AR and 14-3-3 sigma status in breast cancer. The P value was calculated with use of the log rank test.
Cox regression analysis of clinical pathological variables
The univariate survival analysis revealed that tumor size (P=0.044), lymph node status (P=0.006) and clinical stage (P=0.008) were significantly associated with overall survival. Age (P=0.241), pathological grade (P=0.675), molecular subtype (P>0.05), EGFR (P=0.315), AR (P=0.709) and 14-3-3 sigma (P=0.789) status had no statistically significant associations with overall survival. In multivariate analysis, lymph node status was the only statistically significant independent prognostic factor for overall survival [P=0.006, exp (B)=1.511, CI (1.124-2.032)] (See Tables 10, 11).
Table 10.
Univariate cox analyses for overall survival in patients with breast cancer
| Variables | Overall survival | ||
|---|---|---|---|
|
| |||
| Exp (B) | (95% CI) | P-value | |
| Age | 0.673 | (0.347-1.305) | 0.241 |
| Tumor size | 1.779 | (1.016-3.115) | 0.044 |
| Pathologic grade | 0.884 | (0.496-1.573) | 0.675 |
| Lymph node status | 1.511 | (1.124-2.032) | 0.006 |
| Clinical stage | 1.977 | (1.196-3.269) | 0.008 |
| EGFR expression | 0.593 | (0.211-1.667) | 0.315 |
| AR expression | 0.882 | (0.455-1.710) | 0.709 |
| 14-3-3 sigma expression | 1.089 | (0.583-2.035) | 0.789 |
| Molecular subtype | >0.05 | ||
Table 11.
Multivariate cox analyses of overall survival in patients with breast cancer
| Variables | Overall survival | ||
|---|---|---|---|
|
| |||
| Exp (B) | (95% CI) | P-value | |
| Lymph node status | 1.511 | (1.124-2.032) | 0.006 |
Discussion
Our study demonstrates that EGFR expression is negatively correlated with AR expression and positively with 14-3-3 sigma expression. There are significant differences in EGFR and AR expression between various molecular subtypes. EGFR, AR and 14-3-3 sigma have no significant impacts on the overall survival of breast cancer patients.
A great number of studies have examined the involvement of the three biomarkers in carcinogenesis. AR expression in prostate cancer cells was found to be suppressed by the activation of EGFR and ErbB2, therefore leading to decreased AR activities [21]. Leotoing et al. believe that the crosstalk between AR and EGFR-signaling pathways is a molecular switch for epithelial cell differentiation [22]. The interaction between the two pathways may be crucial for the acquisition and the maintenance of androgen sensitivity. Bonaccorsi et al. had reported that AR was associated with EGFR in androgen-sensitive prostate cancer cells [23]. However, whether the interaction between AR and EGFR is due to direct binding of the two proteins or is mediated by another protein remains unclear. Some study documented that there was a crosstalk between the AR signaling pathway and peptide growth factor receptor pathways, suggesting that 14-3-3 sigma may be one of the mediators of common signaling events between AR and EGFR in human prostate cancer cells [24]. Whether the similar finding can be confirmed in human breast cancer cells needs to be further investigated. If so, the interaction of the two pathways may be crucial for the survival of breast cancer cells.
In addition, the current study revealed that there were significant differences in EGFR and AR expression rather than 14-3-3 sigma between different molecular subtypes of breast cancer.
EGFR expression was highest in TNBC and lowest in luminal B breast cancer. EGFR expression from high to low in the order in various subtypes was TNBC (39.39%), Erb-B2-enriched breast cancer (25.93%), luminal A (1.96%), luminal B breast cancer (0%) respectively. Meche et al. reported that EGFR was expressed in 41.66% basal-like carcinomas, in 50% luminal B carcinomas, and in 21.42% breast cancers with Erb-B2 overexpression [25]. The discrepancy, we assume, is due to the different sample selection criteria or immunohistochemical interpretation.
In the present study, AR expression was shown to be highest in luminal A breast cancer and lowest in TNBC. Expression of AR from high to low in the order in various subtypes was luminal A breast cancer (86.27%), HER-2-negative luminal B cancer (76.92%) HER-2-positive luminal B cancer (80%), ERb-B2-enriched breast cancer (62.96%) and TNBC (33.33%), respectively. Our findings are consistent with some studies which found that AR expression varied between the different subtypes, and AR expression was commonly seen in luminal A and luminal B types of invasive breast cancer [26,27].
Unlike EGFR and AR, no significant difference was noted in 14-3-3 sigma expression between various molecular subtypes in our study. The 14-3-3 sigma expression from high to low in the order in various subtypes was TNBC, luminal B cancer, cancer with Erb-B2 overexpression, luminal A cancer respectively. Nakamura et al. also revealed no significant associations between 14-3-3 sigma expression and intrinsic subtypes [28]. Ko et al. reported triple-negative tumors were more frequently positive for 14-3-3 sigma than receptor-positive tumors [16], which showed the agreement with our results.
EGFR is generally considered a negative prognostic factor for breast cancer [29,30]. In the present study, both univariate analysis and multivariate analysis revealed no prognostic significance of EGFR expression for breast cancer. Arnes et al. reported EGFR expression was prognostically significant among BRCA1 mutated cases only. But in the multivariate survival analysis of all cases, no independent effect was seen in their study [31]. Schroeder et al. found the prognostic significance of EGFR expression in the univariate analysis, which, however, could not be confirmed by multivariate analysis, either [32]. To date, there is no consistency for EGFR as a predictive or prognostic marker for breast cancer [33]. Standardized methods for the measurement of EGFR expression are required for further evaluation; thus far, EGFR expression alone in breast cancer bears no prognostic value in breast cancer [33]. But some researchers argue that the prognostic significance of EGFR in breast cancer should not be easily denied [33-35].
A majority of researches associated AR with favorable clinic pathological features [36-40]. Nevertheless, some studies have shown that AR levels fail to predict the overall survival [41,42]. In the current study, we also found that AR had no impact on overall survival both in univariate analysis and in multivariate analysis. The prognostic value of AR, therefore, needs to be further confirmed.
Additionally, Ko et al. had reported positive 14-3-3 σ expression was significantly correlated with poor prognosis [16]. By contrast, Yoon et al. reported 14-3-3 sigma expression at lower levels had a much worse prognosis than individuals who maintained higher or equal levels of 14-3-3 sigma in invasive ductal carcinoma [14]. The current study found no association of 14-3-3 σ expression with the prognosis of breast cancer patients, which is in accordance with Simpson et al. who also showed no statistical association between 14-3-3 sigma expression and overall survival (P=0.3421) by Kaplan-Meier analysis [43].
In conclusion, our study found an interesting correlation among EGFR, AR and 14-3-3 sigma in breast cancer-i.e. EGFR expression negatively correlated with AR and positively with 14-3-3 σ expression. In addition, there were significant differences in EGFR and AR expression between different molecular subtypes of breast cancer. Future studies are warranted on the mechanism underlying the complex relationships among them in breast cancers.
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Tebbutt N, Pedersen MW, Johns TG. Targeting the ERBB family in cancer: couples therapy. Nat Rev Cancer. 2013;13:663–673. doi: 10.1038/nrc3559. [DOI] [PubMed] [Google Scholar]
- 3.Tsutsui S, Ohno S, Murakami S, Hachitanda Y, Oda S. Prognostic value of epidermal growth factor receptor (EGFR) and its relationship to the estrogen receptor status in 1029 patients with breast cancer. Breast Cancer Res Treat. 2002;71:67–75. doi: 10.1023/a:1013397232011. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz TA, Tu X, Ang KK, Esteva FJ, Kuerer HM, Pusztai L, Cristofanilli M, Singletary SE, Hortobagyi GN, Sahin AA. Epidermal growth factor receptor expression correlates with poor survival in patients who have breast carcinoma treated with doxorubicin-based neoadjuvant chemotherapy. Cancer. 2005;104:676–681. doi: 10.1002/cncr.21217. [DOI] [PubMed] [Google Scholar]
- 5.Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136:331–345. doi: 10.1007/s10549-012-2289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iacopetta D, Rechoum Y, Fuqua SA. The role of androgen receptor in breast cancer. Drug Discov Today Dis Mech. 2012;9:e19–e27. doi: 10.1016/j.ddmec.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaaks R, Berrino F, Key T, Rinaldi S, Dossus L, Biessy C, Secreto G, Amiano P, Bingham S, Boeing H, Bueno de Mesquita HB, Chang-Claude J, Clavel-Chapelon F, Fournier A, van Gils CH, Gonzalez CA, Gurrea AB, Critselis E, Khaw KT, Krogh V, Lahmann PH, Nagel G, Olsen A, Onland-Moret NC, Overvad K, Palli D, Panico S, Peeters P, Quiros JR, Roddam A, Thiebaut A, Tjonneland A, Chirlaque MD, Trichopoulou A, Trichopoulos D, Tumino R, Vineis P, Norat T, Ferrari P, Slimani N, Riboli E. Serum sex steroids in premenopausal women and breast cancer risk within the European prospective investigation into cancer and nutrition (EPIC) J Natl Cancer Inst. 2005;97:755–765. doi: 10.1093/jnci/dji132. [DOI] [PubMed] [Google Scholar]
- 8.Sieri S, Krogh V, Bolelli G, Abagnato CA, Grioni S, Pala V, Evangelista A, Allemani C, Micheli A, Tagliabue G, Schunemann HJ, Menard S, Berrino F, Muti P. Sex hormone levels, breast cancer risk, and cancer receptor status in postmenopausal women: the ORDET cohort. Cancer Epidemiol Biomarkers Prev. 2009;18:169–176. doi: 10.1158/1055-9965.EPI-08-0808. [DOI] [PubMed] [Google Scholar]
- 9.Baglietto L, Severi G, English DR, Krishnan K, Hopper JL, McLean C, Morris HA, Tilley WD, Giles GG. Circulating steroid hormone levels and risk of breast cancer for postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2010;19:492–502. doi: 10.1158/1055-9965.EPI-09-0532. [DOI] [PubMed] [Google Scholar]
- 10.McGhan LJ, McCullough AE, Protheroe CA, Dueck AC, Lee JJ, Nunez-Nateras R, Castle EP, Gray RJ, Wasif N, Goetz MP, Hawse JR, Henry TJ, Barrett MT, Cunliffe HE, Pockaj BA. Androgen receptor-positive triple negative breast cancer: a unique breast cancer subtype. Ann Surg Oncol. 2014;21:361–367. doi: 10.1245/s10434-013-3260-7. [DOI] [PubMed] [Google Scholar]
- 11.Fioretti FM, Sita-Lumsden A, Bevan CL, Brooke GN. Revising the role of the androgen receptor in breast cancer. J Mol Endocrinol. 2014;52:R257–265. doi: 10.1530/JME-14-0030. [DOI] [PubMed] [Google Scholar]
- 12.Yaffe MB. How do 14-3-3 proteins work?--Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–57. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- 13.Zilliacus J, Holter E, Wakui H, Tazawa H, Treuter E, Gustafsson JA. Regulation of glucocorticoid receptor activity by 14--3-3-dependent intracellular relocalization of the corepressor RIP140. Mol Endocrinol. 2001;15:501–511. doi: 10.1210/mend.15.4.0624. [DOI] [PubMed] [Google Scholar]
- 14.Yoon NK, Seligson DB, Chia D, Elshimali Y, Sulur G, Li A, Horvath S, Maresh E, Mah V, Bose S, Bonavida B, Goodglick L. Higher expression levels of 14-3-3 sigma in ductal carcinoma in situ of the breast predict poorer outcome. Cancer Biomark. 2009;5:215–224. doi: 10.3233/CBM-2009-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 16.Ko S, Kim JY, Jeong J, Lee JE, Yang WI, Jung WH. The role and regulatory mechanism of 14-3-3 sigma in human breast cancer. J Breast Cancer. 2014;17:207–218. doi: 10.4048/jbc.2014.17.3.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altaf FJ, Mokhtar GA, Emam E, Bokhary RY, Mahfouz NB, Al Amoudi S, Al-Gaithy ZK. Metaplastic carcinoma of the breast: an immunohistochemical study. Diagn Pathol. 2014;9:139. doi: 10.1186/1746-1596-9-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yenidunya S, Bayrak R, Haltas H. Predictive value of pathological and immunohistochemical parameters for axillary lymph node metastasis in breast carcinoma. Diagn Pathol. 2011;6:18. doi: 10.1186/1746-1596-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirza S, Sharma G, Parshad R, Srivastava A, Gupta SD, Ralhan R. Clinical significance of Stratifin, ERalpha and PR promoter methylation in tumor and serum DNA in Indian breast cancer patients. Clin Biochem. 2010;43:380–386. doi: 10.1016/j.clinbiochem.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 20.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen international expert consensus on the primary therapy of early breast cancer 2011. Ann Oncol. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai C, Portnoy DC, Wang H, Jiang X, Chen S, Balk SP. Androgen receptor expression in prostate cancer cells is suppressed by activation of epidermal growth factor receptor and ErbB2. Cancer Res. 2009;69:5202–5209. doi: 10.1158/0008-5472.CAN-09-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leotoing L, Manin M, Monte D, Baron S, Communal Y, Lours C, Veyssiere G, Morel L, Beaudoin C. Crosstalk between androgen receptor and epidermal growth factor receptorsignalling pathways: a molecular switch for epithelial cell differentiation. J Mol Endocrinol. 2007;39:151–162. doi: 10.1677/JME-07-0021. [DOI] [PubMed] [Google Scholar]
- 23.Bonaccorsi L, Muratori M, Carloni V, Marchiani S, Formigli L, Forti G, Baldi E. The androgen receptor associates with the epidermal growth factor receptor in androgen-sensitive prostate cancer cells. Steroids. 2004;69:549–552. doi: 10.1016/j.steroids.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 24.Huang D, Liu X, Plymate SR, Idowu M, Grimes M, Best AM, McKinney JL, Ware JL. Proteomic identification of 14-3-3 sigma as a common component of the androgen receptor and the epidermal growth factor receptor signaling pathways of the human prostate epithelial cell line M12. Oncogene. 2004;23:6881–6889. doi: 10.1038/sj.onc.1207788. [DOI] [PubMed] [Google Scholar]
- 25.Meche A, Cimpean AM, Raica M. Immunohistochemical expression and significance of epidermal growth factor receptor (EGFR) in breast cancer. Rom J Morphol Embryol. 2009;50:217–221. [PubMed] [Google Scholar]
- 26.Grogg A, Trippel M, Pfaltz K, Ladrach C, Droeser RA, Cihoric N, Salhia B, Zweifel M, Tapia C. Androgen receptor status is highly conserved during tumor progression of breast cancer. BMC Cancer. 2015;15:872. doi: 10.1186/s12885-015-1897-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins LC, Cole KS, Marotti JD, Hu R, Schnitt SJ, Tamimi RM. Androgen receptor expression in breast cancer in relation to molecular phenotype: results from the nurses’ health study. Mod Pathol. 2011;24:924–931. doi: 10.1038/modpathol.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura Y, Oshima K, Naoi Y, Nakayama T, Kim SJ, Shimazu K, Shimomura A, Maruyama N, Tamaki Y, Noguchi S. 14-3-3 sigma expression is associated with poor pathological complete response to neoadjuvant chemotherapy in human breast cancers. Breast Cancer Res Treat. 2012;134:229–236. doi: 10.1007/s10549-012-1976-x. [DOI] [PubMed] [Google Scholar]
- 29.Witton CJ, Reeves JR, Going JJ, Cooke TG, Bartlett JM. Expression of the HER1-4 family of receptor tyrosine kinases in breast cancer. J Pathol. 2003;200:290–297. doi: 10.1002/path.1370. [DOI] [PubMed] [Google Scholar]
- 30.Fuchs IB, Siemer I, Buhler H, Schmider A, Henrich W, Lichtenegger W, Schaller G, Kuemmel S. Epidermal growth factor receptor changes during breast cancer metastasis. Anticancer Res. 2006;26:4397–4401. [PubMed] [Google Scholar]
- 31.Arnes JB, Begin LR, Stefansson I, Brunet JS, Nielsen TO, Foulkes WD, Akslen LA. Expression of epidermal growth factor receptor in relation to BRCA1 status, basal-like markers and prognosis in breast cancer. J Clin Pathol. 2009;62:139–146. doi: 10.1136/jcp.2008.056291. [DOI] [PubMed] [Google Scholar]
- 32.Schroeder W, Biesterfeld S, Zillessen S, Rath W. Epidermal growth factor receptor-immunohistochemical detection and clinical significance for treatment of primary breast cancer. Anticancer Res. 1997;17:2799–2802. [PubMed] [Google Scholar]
- 33.Chan SK, Hill ME, Gullick WJ. The role of the epidermal growth factor receptor in breast cancer. J Mammary Gland Biol Neoplasia. 2006;11:3–11. doi: 10.1007/s10911-006-9008-2. [DOI] [PubMed] [Google Scholar]
- 34.Rampaul RS, Pinder SE, Nicholson RI, Gullick WJ, Robertson JF, Ellis IO. Clinical value of epidermal growth factor receptor expression in primary breast cancer. Adv Anat Pathol. 2005;12:271–273. doi: 10.1097/01.pap.0000184178.43048.80. [DOI] [PubMed] [Google Scholar]
- 35.Koutras AK, Evans TR. The epidermal growth factor receptor family in breast cancer. Onco Targets Ther. 2008;1:5–19. doi: 10.2147/ott.s3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsang JY, Ni YB, Chan SK, Shao MM, Law BK, Tan PH, Tse GM. Androgen receptor expression shows distinctive significance in ER positive and negative breast cancers. Ann Surg Oncol. 2014;21:2218–2228. doi: 10.1245/s10434-014-3629-2. [DOI] [PubMed] [Google Scholar]
- 37.Yu Q, Niu Y, Liu N, Zhang JZ, Liu TJ, Zhang RJ, Wang SL, Ding XM, Xiao XQ. Expression of androgen receptor in breast cancer and its significance as a prognostic factor. Ann Oncol. 2011;22:1288–1294. doi: 10.1093/annonc/mdq586. [DOI] [PubMed] [Google Scholar]
- 38.Castellano I, Allia E, Accortanzo V, Vandone AM, Chiusa L, Arisio R, Durando A, Donadio M, Bussolati G, Coates AS, Viale G, Sapino A. Androgen receptor expression is a significant prognostic factor in estrogen receptor positive breast cancers. Breast Cancer Res Treat. 2010;124:607–617. doi: 10.1007/s10549-010-0761-y. [DOI] [PubMed] [Google Scholar]
- 39.Hu R, Dawood S, Holmes MD, Collins LC, Schnitt SJ, Cole K, Marotti JD, Hankinson SE, Colditz GA, Tamimi RM. Androgen receptor expression and breast cancer survival in post-menopausal women. Clin Cancer Res. 2011;17:1867–1874. doi: 10.1158/1078-0432.CCR-10-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park S, Koo JS, Kim MS, Park HS, Lee JS, Kim SI, Park BW, Lee KS. Androgen receptor expression is significantly associated with better outcomes in estrogen receptor-positive breast cancers. Ann Oncol. 2011;22:1755–1762. doi: 10.1093/annonc/mdq678. [DOI] [PubMed] [Google Scholar]
- 41.Allegra JC, Lippman ME, Simon R, Thompson EB, Barlock A, Green L, Huff KK, Do HM, Aitken SC, Warren R. Association between steroid hormone receptor status and disease-free interval in breast cancer. Cancer Treat Rep. 1979;63:1271–1277. [PubMed] [Google Scholar]
- 42.Carreno G, Del Casar JM, Corte MD, Gonzalez LO, Bongera M, Merino AM, Juan G, Obregon R, Martinez E, Vizoso FJ. Local recurrence after mastectomy for breast cancer: analysis of clinicopathological, biological and prognostic characteristics. Breast Cancer Res Treat. 2007;102:61–73. doi: 10.1007/s10549-006-9310-0. [DOI] [PubMed] [Google Scholar]
- 43.Simpson PT, Gale T, Reis-Filho JS, Jones C, Parry S, Steele D, Cossu A, Budroni M, Palmieri G, Lakhani SR. Distribution and significance of 14-3-3 sigma, a novel myoepithelial marker, in normal, benign, and malignant breast tissue. J Pathol. 2004;202:274–285. doi: 10.1002/path.1530. [DOI] [PubMed] [Google Scholar]