Abstract
Objectives: The aim of this study was to evaluate the expression of RBM38 protein in gastric cancer patients and to explore its association with clinical pathological characteristics and prognosis. Materials and methods: A total of 120 pairs of gastric cancer tissues and non-cancerous gastric mucosa from 120 patients who underwent gastrectomy for gastric cancer were included in the current study. RBM38 protein expression levels were detected in all tissue specimens by immunohistochemistry staining. The positive rate of RBM38 was compared between cancer tissue and normal tissue, and its association with the clinical pathological characteristics and prognosis was elucidated. Results: RBM38 protein was predominantly expressed in the cytoplasm of epithelial cells. The percentage of tissues with high RBM38 protein expression level was significantly lower (χ2=28.972, P<0.001) in gastric cancer tissues compared with adjacent non-cancerous gastric mucosal tissues. The expression level of RBM38 protein was associated with tumor size (P=0.028), depth of invasion (P<0.001), lymph node metastasis (P<0.001), TNM stage (P<0.001) and Lauren classification of the tumor (P=0.001), whereas it was not associated with gender (P=0.066) and age (P=0.6) of patients. Moreover, we noticed that the low expression level of RBM38 protein was also associated with poor prognosis in gastric cancer patients (log rank =5.325; P=0.021). Conclusion: Overall, our findings indicated that RBM38 may play a vital role as a tumor suppressor, which may be a potential marker in the diagnosis and prognosis of gastric cancer.
Keywords: RBM38, gastric cancer, tumor suppressor, prognosis
Introduction
Gastric cancer (GC) is one of the most common cancers and a prime cause of cancer-related mortalities worldwide [1]. In 2015, about 679,100 new cases were diagnosed and 498,000 deaths were reported due to GC in China [2]. Due to considerable advances made towards the early diagnosis and treatment of GC, a significant decrease in its incidence and mortality was observed in China [2], but the prognosis is still far from optimistic. A comprehensive understanding of molecular mechanisms underlying GC may improve prognosis, and also help to identify more useful prognostic biomarkers and provide novel chemotherapeutic targets.
RNA-binding proteins (RBPs) play crucial roles in post-transcriptional regulation in gene expression, and regulate all aspects of RNA biology, such as polyadenylation, RNA splicing, transport, stability and translation [3]. One such mechanism is through microRNAs, belonging to a class of small noncoding RNAs that negatively regulate protein expression by binding protein-coding mRNAs and repressing translation [4]. Dysfunctional or mutated RBPs can cause numerous human diseases ranging from metabolic disorders to cancer [5-8].
RBM38 belongs to the family of RBPs, which play pivotal role in regulating wide biological processes, ranging from cell proliferation, cell cycle arrest to cell myogenic differentiation [9,10]. The role of RBM38 as a potential oncogene in tumorigenesis was previously identified in prostate cancer [11], colorectal cancer [12] and esophageal cancer [13]. Moreover, its critical function as a tumor suppressor gene is elucidated in many malignancies, such as breast cancer [14], hepatocellular carcinoma [15] and renal cell carcinoma [16]. Although the expression of RBM38 has been studied in several types of cancer, the expression and biologic functions of RBM38 in gastric cancer is still ambiguous.
In this study, we investigated the critical expression of RBM38 effector protein in gastric cancer and adjacent normal gastric specimens, and analyzed its expression level with the clinicopathological characters. A lower level of RBM38 was expressed in gastric cancer compared to adjacent gastric tissue. The expression of RBM38 was not correlated with gender and age in GC but significantly correlated with tumor size, depth of invasion, lymph node metastasis, TNM stage, and Lauren classification. The low expression level of RBM38 protein was also associated with poor prognosis in gastric cancer patients. It showed that RBM38 may function as a tumor suppressor in gastric cancer, which may be a potentially useful independent biomarker for prognosis in GC patients.
Patients and methods
Patient selection
The present study was approved by the Institutional Review Board of Jiangsu Shengze Hospital (Suzhou, China). Written informed consent was obtained from all patients prior to enrollment in the present study. All specimens were anonymously handled in accordance with the Declaration of Helsinki and legal standards.
A total of 120 patients who underwent gastrectomy for gastric cancer at Jiangsu Shengze Hospital (Suzhou, China) between January 2004 and January 2012 were included in the current study. All these enrolled cases were previously diagnosed by histopathological analysis by two senior pathologists using the surgical specimens. The patient cohort consisted of 88 males and 32 females, with a median age of 61.8±9.85 years (range: 30-91 years). Among the 120 gastric cancer cases, 28 were from the cardia, 51 from the body and 41 from the gastric antrum.
According to the World Health Organization histological classification [17] of gastric carcinoma, 49 cases were identified as tubular, 33 papillary, 18 signet-ring cell and 20 mucinous adenocarcinomas. Among them 34 were well differentiated, 39 moderately differentiated, 43 poorly differentiated and 4 undifferentiated adenocarcinomas. On the basis of the Lauren classification of gastric cancer, 60 cases were diffuse-type, 53 intestinal-type and 7 were mixed type. 109 cases in this group showed lymph node metastasis but no case with distant metastasis. In terms of the 7th edition of the Union for International Cancer Control Tumor-Node-Metastasis (UICC-TNM) classification system for gastric cancer [18]; 18 cases were categorized as stage I, 22 as stage II, 80 as stage III and no case as stage IV. A total of 120 pairs of gastric cancer tissues and adjacent non-cancerous gastric mucosa were collected following gastrectomy and formalin-fixed and paraffin-embedded (FFPE) for further study. Following surgery, routine chemotherapy was administered to patients with advanced disease, and no radiation treatment was administered to any of the patients.
Follow-up
All patients had a follow-up record for ≥5 years. The retrospective design of the study data were acquired through medical record review and direct phone-interview with patients, relatives, or general practitioners. The last follow-up was in January 2017. The survival time was determined from the date of surgery to the follow-up deadline or date of mortality.
Tissue microarray (TMA) construction and immunohistochemistry
TMA blocks containing 120 pairs of gastric cancer tissues and non-cancerous gastric mucosa were prepared using the following method: tissue cylinders 2 mm in diameter were punched from the targeted area of each donor block and precisely arrayed into a recipient block using a TMA instrument (no. HM315R; GMI, Inc., Ramsey, MN, USA). Each TMA block contained six non-cancerous gastric mucosal tissues as the controls. Consecutive 4 µm thick sections were cut from each of the resulting TMA blocks, and one section from each block was H&E stained for histological verification for the adequacy of the arrayed tumor tissues. Eligible sections were those in which the tumor tissue occupied >10% of the core area. Sections were then placed on microscope slides for further analysis.
Immunohistochemical staining was performed on the TMA slides using the following stepwise method: (1) Baking the TMA slides at a temperature of 60°C for 2 h, dewaxing with xylene and rehydrating in graded ethanol sequentially (100, 95 and 80%, v/v); (2) Incubating the slides with 10% normal goat serum (Beijing Solarbio Science & Technology Co., Ltd.,) at room temperature for 10 min to reduce nonspecific reactions; (3) Incubating the slides with the primary antibody (Rabbit polyclonal to RBM38-C-terminal (ab200403); dilution, 1/25-1/100; Abcam) in a moist chamber at 4°C over-night; (4) Washing three times with 0.01 M phosphate buffer (Beijing Solarbio Science & Technology Co., Ltd.; pH, 7.2), then incubating the TMA slides with secondary antibody (horseradish peroxidase-conjugated mouse monoclonal anti-rabbit immunoglobulin; cat. no. M0737; dilution, 1:1; Dako; Agilent Technologies Inc., Santa Clara, CA, USA) for 20 min at room temperature and stained with diaminobenzidine (DAB)-H2O2; (5) Counterstaining the TMA slides with hematoxylin (0.5%, w/v), which was dehydrated and mounted on a coverslip with neutral balsam (Shanghai Specimen and Model Factory, Shanghai, China) and subsequently viewed under an optical microscope.
The RBM38 protein was immunohistochemically stained and independently examined under a light microscope by two pathologists who were blinded to the clinical data. The immunoreactivity was evaluated by applying a scoring system combining the intensity of immunostaining with the proportion of immunoreactive cells as followed: the intensity of immunostaining was scored as 0 (no staining), 1 (weak staining, light yellow), 2 (moderate staining, yellowish brown) and 3 (intense staining, brown), and the proportion of immunoreactive cells was scored as 0 (≤5% positive cells), 1 (6-25% positive cells), 2 (26-50% positive cells) and 3 (≥51% positive cells). In the case of a discrepancy, a consensus score was selected. The product of the scores for intensity and proportion was used to signify the level of protein expression. The expression level of RBM38 was considered low if the product was ≤2, and high if the product was ≥3.
Statistical analysis
The data were analyzed using the SPSS 13.0 software (SPSS, Chicago, IL, USA). All p values were two sided, and P<0.05 was considered to indicate a statistically significant difference. Quantitative data are presented as the mean ± standard deviation. Data were analyzed using the Student’s t test, whereas categorical data were assessed using the χ2 test or Fisher’s exact test. The Kaplan Meier method was used to plot the survival curve and extract the cumulative survival rate and mean survival time. Multivariate survival analysis was carried out using the Cox proportional hazards model, and variables that were significant in the univariate analysis were included in the model with the Enter method.
Results
Expression levels of RBM38 protein in gastric cancer and non-cancerous gastric mucosa
RBM38 protein was predominantly expressed in the cytoplasm of epithelial cells (Figure 2). RBM38 protein was weakly expressed (26/120) and not expressed (43/120) in the patients with gastric cancer, whereas RBM38 protein expression level was high (82/120) in the adjacent non-cancerous gastric mucosal tissues. The percentage of tissues with high RBM38 protein expression level was significantly lower (χ2=28.972, P<0.001) in gastric cancer tissues compared with adjacent non-cancerous gastric mucosal tissues.
Figure 2.
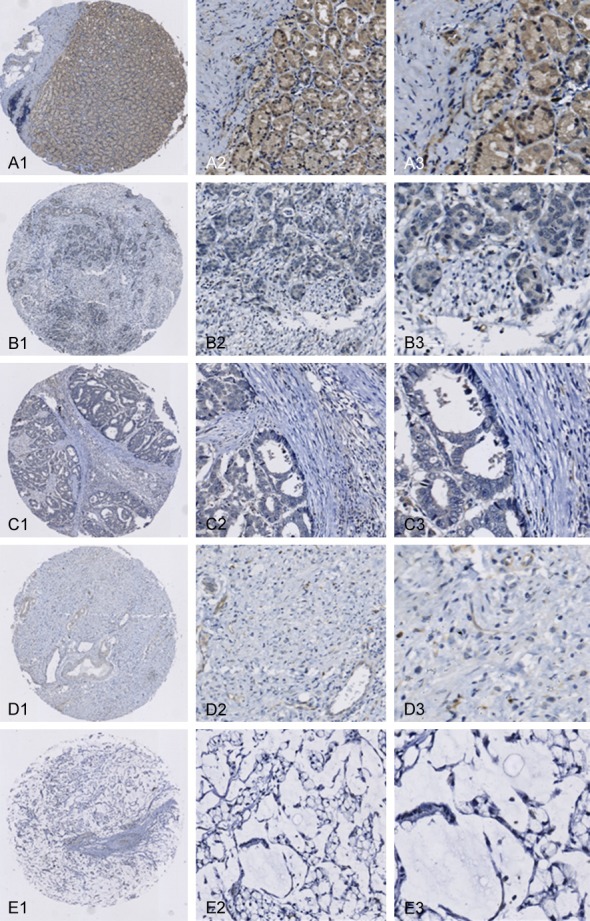
Representative immunohistochemical staining for RBM38 protein in cancerous and non-cancerous gastric tissues (EnVisionTM method). (A1-A3) Intense staining in adjacent non-cancerous gastric mucosa; (B1-B3) No staining in highly differentiated adenocarcinoma; (C1-C3) No staining in moderately differentiated adenocarcinoma; (D1-D3) No staining in poorly differentiated adenocarcinoma; (E1-E3) No staining in mucinous adenocarcinoma. Magnification: (A1, B1, C1, D1 and E1) ×40; (A2, B2, C2, D2 and E2) ×100; (A3, B3, C3, D3 and E3) ×400.
Correlation of RBM38 protein expression with clinicopathological parameters
A correlation between RBM38 protein expression and clinicopathological characteristics of patients with gastric cancer was performed to assess the outcome and significance of expression on clinical parameters. The expression level of RBM38 protein in gastric cancer was associated with tumor size (P=0.028), depth of invasion (P<0.001), lymph node metastasis (P<0.001), TNM stage (P<0.001) and Lauren classification of the tumor (P=0.001), whereas it was not associated with gender (P=0.066) and age (P=0.6; Table 1).
Table 1.
Association of RBM38 protein expression level with clinicopathological parameters of patients with gastric cancer (n=120)
| Clinicopathological parameters | Total no. patients | RBM38 protein expression level | χ2 | P value | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Low (n, %) | High (n, %) | ||||||
| Gender | 3.376 | 0.066 | |||||
| Male | 88 | 55 | 62.5% | 33 | 37.5% | ||
| Female | 32 | 14 | 43.8% | 18 | 56.2% | ||
| Age range | 0.274 | 0.6 | |||||
| ≤55 years | 36 | 22 | 61.1% | 14 | 38.9% | ||
| >55 years | 84 | 47 | 56.0% | 37 | 44.0% | ||
| Tumor size | 4.831 | 0.028 | |||||
| ≤5 cm | 81 | 41 | 50.6% | 40 | 49.4% | ||
| >5 cm | 39 | 28 | 71.8% | 11 | 28.2% | ||
| Depth of invasion | 24.884 | <0.001 | |||||
| T1 | 17 | 3 | 17.6% | 14 | 82.4% | ||
| T2 | 19 | 6 | 31.6% | 13 | 68.4% | ||
| T3 | 46 | 36 | 78.3% | 10 | 21.7% | ||
| T4 | 38 | 24 | 63.2% | 14 | 36.8% | ||
| Lymph node metastasis | 21.64 | <0.001 | |||||
| N0 | 11 | 2 | 18.2% | 9 | 81.8% | ||
| N1 | 43 | 17 | 39.5% | 26 | 60.5% | ||
| N2 | 33 | 25 | 75.8% | 8 | 24.2% | ||
| N3 | 33 | 25 | 75.8% | 8 | 24.2% | ||
| TNM stage | 26.864 | <0.001 | |||||
| I | 18 | 3 | 16.7% | 15 | 83.3% | ||
| II | 22 | 7 | 31.8% | 15 | 68.2% | ||
| III | 80 | 59 | 73.8% | 21 | 26.2% | ||
| Lauren classification | 13.048 | 0.001 | |||||
| Diffuse | 60 | 42 | 70.0% | 18 | 30.0% | ||
| Mix | 7 | 6 | 85.7% | 1 | 14.3% | ||
| Intestinal | 53 | 21 | 39.6% | 32 | 60.4% | ||
Correlation between RBM38 protein expression level and prognosis of patients with gastric cancer
Univariate survival analysis indicated that low expression levels of RBM38 protein were associated with poor prognosis of patients with gastric cancer (log rank =5.325; P=0.021) (Figure 1). The 1, 3 and 5 years cumulative survival rates were 88.3, 59.9 and 44.0% for patients with low RBM38 expression levels, and 94.1, 85.8 and 52.6% for patients with high RBM38 expression levels, respectively. The mean survival time for patients with high expression levels of RBM38 was 68±9.66 months, which was significantly higher (P<0.05) compared with 49±12.87 months for patients with low RBM38 expression levels. It was also revealed that tumor size and lymph node metastasis were significantly associated with the survival of patients with gastric cancer, whereas gender, age, depth of invasion, TNM stage and Lauren classification of the tumor were not significantly associated with their survival (Table 2).
Figure 1.
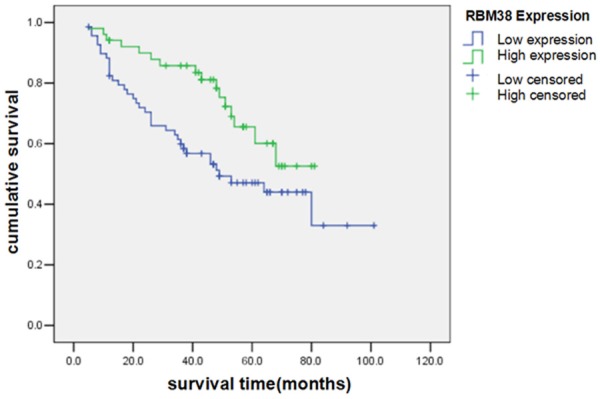
Kaplan Meier survival curves of patients with gastric cancer with high and low levels of RBM38 protein expression.
Table 2.
Univariate and multivariate analyses of factors associated with overall survival of gastric cancer patients (n=120)
| Clinical variables | Overall survival | ||
|---|---|---|---|
|
| |||
| HR (95% CI) | p value | ||
| Univariate analysis | |||
| Age (≤55 vs. >55) | 1.166 (0.650 to 2.090) | 0.607 | |
| Gender (male vs. female) | 1.084 (0.566 to 2.079) | 0.807 | |
| Tumor size (≤5 cm vs. >5 cm) | 0.429 (0.245 to 0.751) | 0.003 | |
| Lyn (N0-1 vs. N2-3) | 0.512 (0.285 to 0.920) | 0.025 | |
| Depth of invasion (T1/2 vs. T3/4) | 0.758 (0.402 to 1.431) | 0.393 | |
| TNM stage (I/II vs. III/IV) | 0.626 (0.332 to 1.183) | 0.149 | |
| Lauren (diffusion vs. intestine and mix) | 0.920 (0.530 to 1.596) | 0.767 | |
| RBM38 expression (low vs. high) | 2.106 (1.152 to 3.851) | 0.016 | |
| Multivariate analysis | |||
| Age (≤55 vs. >55) | 1.123 (0.623 to 2.024) | 0.700 | |
| Gender (male vs. female) | 1.082 (0.546 to 2.145) | 0.821 | |
| Tumor size (≤5 cm vs. >5 cm) | 0.464 (0.237 to 0.908) | 0.025 | |
| Lyn (N0-1 vs. N2-3) | 0.481 (0.163 to 1.422) | 0.186 | |
| Depth of invasion (T1/2 vs. T3/4) | 6370 (0.000 to 3E+061) | 0.897 | |
| TNM stage (I/II vs. III/IV) | 0.000 (0.000 to 2E+054) | 0.908 | |
| Lauren (diffusion vs. intestine and mix) | 0.741 (0.389 to 1.409) | 0.360 | |
| RBM38 expression (low vs. high) | 2.115 (1.044 to 4.286) | 0.038 | |
Discussion
In this study, we identified that the RBM38 protein expression was down regulated in gastric cancer tissues compared with non-cancerous gastric mucosa. Specifically, we showed that low expression levels of RBM38 protein were associated with poor prognosis of patients with gastric cancer. Moreover, we demonstrated that RBM38 might play a major role as tumor suppressor in gastric cancer.
As described previously, RBM38 play an important role in tumorigenesis. RBM38 potentially serves as a diagnostic tool in breast and ovarian cancers and also as a predictive marker of poor prognosis [14,19]. RBM38 also reported to play a role in antiproliferation, which might exert its activity by regulating the function of a protein by directly involved in the cell cycle regulation [20]. Another previous study showed that it also could inhibit migration and invasion of breast cancer cells by regulating EMT [14]. Former study reported RBM38 as a target of the p53 family and exhibited a feedback regulatory loop with the p53 family proteins [8,20-23]. Missense mutation of p53 is a common event in gastric cancer [24] and is often associated with poor cancer outcome [24,25]. Moreover, this may explain the low expression of RBM38 in gastric cancer with poor prognosis. Missense mutation of p53 not only cause loss of tumor suppression function (LOF), but also causes gain of oncogenic function (GOF) [26]. Mutations of p53 tumor suppressor was often highly expressed and had a long half-life in various tumors [27].
In conclusion, we identified RBM38 as a potential diagnostic and prognostic target of GC in this study, which may function as a tumor suppressor. The complex regulatory mechanism of RBM38 in gastric tumorigenesis needs to be delineated in future to explore its exact role and relevance in gastric cancer and to implement it to serve as a tumor suppressive target in gastric cancer therapy.
Acknowledgements
This study was supported by Jiangsu cadre health care research project fund (BJ16008).
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends--an update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Kim MY, Hur J, Jeong S. Emerging roles of RNA and RNA-binding protein network in cancer cells. BMB Rep. 2009;42:125–130. doi: 10.5483/bmbrep.2009.42.3.125. [DOI] [PubMed] [Google Scholar]
- 4.Leveille N, Elkon R, Davalos V, Manoharan V, Hollingworth D, Oude Vrielink J, le Sage C, Melo CA, Horlings HM, Wesseling J, Ule J, Esteller M, Ramos A, Agami R. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat Commun. 2011;2:513. doi: 10.1038/ncomms1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fredericks AM, Cygan KJ, Brown BA, Fairbrother WG. RNA-binding proteins: splicing factors and disease. Biomolecules. 2015;5:893–909. doi: 10.3390/biom5020893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng F, Pan Y, Lu YM, Zhu L, Chen S. RNAbinding protein Dnd1 promotes breast cancer apoptosis by stabilizing the Bim mRNA in a miR-221 binding site. Biomed Res Int. 2017;2017:9596152. doi: 10.1155/2017/9596152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calaluce R, Gubin MM, Davis JW, Magee JD, Chen J, Kuwano Y, Gorospe M, Atasoy U. The RNA binding protein HuR differentially regulates unique subsets of mRNAs in estrogen receptor negative and estrogen receptor positive breast cancer. BMC Cancer. 2010;10:126. doi: 10.1186/1471-2407-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan W, Zhang J, Zhang Y, Jung YS, Chen X. p73 expression is regulated by RNPC1, a target of the p53 family, via mRNA stability. Mol Cell Biol. 2012;32:2336–2348. doi: 10.1128/MCB.00215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho SJ, Zhang J, Chen X. RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic Acids Res. 2010;38:2256–2267. doi: 10.1093/nar/gkp1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyamoto S, Hidaka K, Jin D, Morisaki T. RNAbinding proteins Rbm38 and Rbm24 regulate myogenic differentiation via p21-dependent and -independent regulatory pathways. Genes Cells. 2009;14:1241–1252. doi: 10.1111/j.1365-2443.2009.01347.x. [DOI] [PubMed] [Google Scholar]
- 11.Zheng SL, Xu J, Isaacs SD, Wiley K, Chang B, Bleecker ER, Walsh PC, Trent JM, Meyers DA, Isaacs WB. Evidence for a prostate cancer linkage to chromosome 20 in 159 hereditary prostate cancer families. Hum Genet. 2001;108:430–435. doi: 10.1007/s004390100513. [DOI] [PubMed] [Google Scholar]
- 12.Korn WM, Yasutake T, Kuo WL, Warren RS, Collins C, Tomita M, Gray J, Waldman FM. Chromosome arm 20q gains and other genomic alterations in colorectal cancer metastatic to liver, as analyzed by comparative genomic hybridization and fluorescence in situ hybridization. Genes Chromosomes Cancer. 1999;25:82–90. doi: 10.1002/(sici)1098-2264(199906)25:2<82::aid-gcc2>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 13.Hotte GJ, Linam-Lennon N, Reynolds JV, Maher SG. Radiation sensitivity of esophageal adenocarcinoma: the contribution of the RNA-binding protein RNPC1 and p21-mediated cell cycle arrest to radioresistance. Radiat Res. 2012;177:272–279. doi: 10.1667/rr2776.1. [DOI] [PubMed] [Google Scholar]
- 14.Xue JQ, Xia TS, Liang XQ, Zhou W, Cheng L, Shi L, Wang Y, Ding Q. RNA-binding protein RNPC1: acting as a tumor suppressor in breast cancer. BMC Cancer. 2014;14:322. doi: 10.1186/1471-2407-14-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding C, Cheng S, Yang Z, Lv Z, Xiao H, Du C, Peng C, Xie H, Zhou L, Wu J, Zheng S. Long noncoding RNA HOTAIR promotes cell migration and invasion via down-regulation of RNA binding motif protein 38 in hepatocellular carcinoma cells. Int J Mol Sci. 2014;15:4060–4076. doi: 10.3390/ijms15034060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang W, Wei XL, Ni W, Cao M, Meng L, Yang H. The expression of RNA-binding protein RBM38 decreased in renal cell carcinoma and represses renal cancer cell proliferation, migration, and invasion. Tumour Biol. 2017;39:1010428317701635. doi: 10.1177/1010428317701635. [DOI] [PubMed] [Google Scholar]
- 17.Bosman FT CF, Hruban RH, Theise ND. WHO Classification of Tumours of the Digestive System. 4th edition. 2010. p. 3. [Google Scholar]
- 18.Sobin L GM, Wittekind C, editors. TNM Classification of Malignant Tumours. 7th edition 2009. [Google Scholar]
- 19.Feldstein O, Ben-Hamo R, Bashari D, Efroni S, Ginsberg D. RBM38 is a direct transcriptional target of E2F1 that limits E2F1-induced proliferation. Mol Cancer Res. 2012;10:1169–1177. doi: 10.1158/1541-7786.MCR-12-0331. [DOI] [PubMed] [Google Scholar]
- 20.Shu L, Yan W, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev. 2006;20:2961–2972. doi: 10.1101/gad.1463306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu E, Zhang J, Chen X. MDM2 expression is repressed by the RNA-binding protein RNPC1 via mRNA stability. Oncogene. 2013;32:2169–2178. doi: 10.1038/onc.2012.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J, Zhou XJ, Sun X, Xia TS, Li XX, Shi L, Zhu L, Zhou WB, Wei JF, Ding Q. RBM38 is involved in TGF-beta-induced epithelial-to-mesenchymal transition by stabilising zonula occludens-1 mRNA in breast cancer. Br J Cancer. 2017;117:675–684. doi: 10.1038/bjc.2017.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Cho SJ, Shu L, Yan W, Guerrero T, Kent M, Skorupski K, Chen H, Chen X. Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev. 2011;25:1528–1543. doi: 10.1101/gad.2069311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Hu Y, Fang JY, Xu J. Gain-of-function miRNA signature by mutant p53 associates with poor cancer outcome. Oncotarget. 2016;7:11056–11066. doi: 10.18632/oncotarget.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blagosklonny MV. p53 from complexity to simplicity: mutant p53 stabilization, gain-of-function, and dominant-negative effect. Faseb J. 2000;14:1901–1907. doi: 10.1096/fj.99-1078rev. [DOI] [PubMed] [Google Scholar]
- 27.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]


