Abstract
Tumorigenesis research has focused on the roles of deregulated microRNAs for many years. The aberrant expression of miR-124 in many tumors has been widely reported, yet its role in glioma formation still needs further research. In this study, the expression and mechanisms of miR-124 in glioma development were explored. We found that glioma cell lines and tumor tissues demonstrated downregulated miR-124 expression, and that cell proliferation, migration, and invasion were reduced when miR-124 was restored. Furthermore, a bioinformatic analysis indicated that Smad2 was a putative target of miR-124, and we confirmed that miR-124 directly targets Smad2 in a luciferase reporter assay system. These results indicate that glioma cell growth is suppressed by miR-124 through its negative regulation of Smad2 expression. Our findings disclose a critical role of miR-124 in glioma pathogenesis, and suggest its potential application for glioma therapy.
Keywords: MiR-124, glioma, Smad2, migration, invasion
Introduction
Gliomas are characteristically aggressive, and are also the most common primary pernicious brain tumors diagnosed in adults throughout the world. The glioma prognosis remains bleak, with a survival period of only 15-17 months following diagnosis. Great progress has been reported, however, with the use of chemical drug therapy and molecular targets [1-3]. The mechanism of glioma development and progression warrants further exploration, as it shows great promise as a target for new therapy attempts.
MicroRNAs (miRNAs) are small, noncoding RNAs that contain 22 nucleotides and regulate gene expression post-transcriptionally through binding to various target mRNAs [4-6]. MiRNAs, as considerable epigenetic regulators, are strongly associated with human diseases processes. Several studies have disclosed that disordered miRNAs are significantly correlated to carcinogenesis and tumor progression [7,8]. Among gliomas, tumor prognosis and progression have been closely associated with expression levels of miR-21, miR-105, miR-143, and miR-197 [4,9-11]. MiRNAs disrupt carcinogenesis and can serve as oncogene suppressors or tumor inhibitors by downregulating expression of their target genes [5]. Therefore, exploration of disordered miRNA expression levels in glioma might result in the discovery of novel miRNA biomarkers and glioma therapy targets [12].
The transforming growth factor-beta (TGF-β) superfamily performs a crucial role in embryonic development and postnatal tissue and organ homeostasis by regulating cell proliferation, differentiation, and migration [13,14]. A canonical TGF-β signaling is mediated through an ordinary linear cascade, which begins with TGF-β type I and II ligand-bound receptor activation [15]. The receptor-activated Smad2 and Smad3 are then phosphorylated, and form a heterodimer with the common mediator Smads (co-Smads) [16]. The resulting heterodimer complex then enters into the nucleus and regulates its target genes, with the help of various other transcriptional factors [15]. The Smads bind to the 12-O-tetradecanoyl-13-acetate responsive element of the promoters of many crucial genes that play a major role in tumor development [17]. The Smads arguably play a critical role in the TGF-β signaling pathway, and an exploration of the Smads regulators is critical to a study of the TGF-β signaling pathway.
In this current study, we explored the downregulation of miR-124 in gliomas, and the overexpression of miR-124 inhibited glioma cell proliferation, migration, and invasion in vitro. Moreover, we examined how miR-124 targets Smad2 and suppresses its expression at mRNA and protein levels. Taken together, we set out to confirm that miR-124 functions as a glioma suppressor through direct of targeting Smad2 and through the regulation of the TGF-β signaling pathway, in the hopes that miR-124 can be utilized as a potential tumor suppressor in glioma therapy.
Materials and methods
Human tissue specimens
Glioma tissues and normal brain tissues were obtained from patients undergoing surgery at the Third Affiliated Hospital of Harbin Medical University, China. Collected tissues were immediately snap-frozen and stored at -80°C. Informed consent was obtained from each patient to approve the use of their tissues for research purposes.
Cell lines
The human glioma cell lines U87, U251, U373, and T98G were obtained from the American Type Culture Collection (ATCC, Rockville, MD). These cell lines were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen, Grand Island, NY). Normal human astrocytes (NHA) cell line was obtained from the Lonza group (Lonza, Basel, Switzerland) and cultured according to the manufacturer’s instructions.
MiRNAs and siRNA transfection
Negative control (NC) miRNAs and miR-124 mimics (mimics) were purchased from GenePharma (Shanghai, China). The cells were transfected with NC and mimics using Lipofectamine 2000 (Invitrogen, USA), following the manufacturer’s protocol. Transfection efficiency was monitored by qRT-PCR.
Plasmids construction
Human Smad2 3’-untranslated region (UTR), containing putative miR-124 binding sites, was amplified by polymerase chain reaction (PCR) from human genomic DNA. Fragments were double digested with restricted enzyme NotI and XbaI, and then cloned into pRL-TK (Promega). MiR-124 binding sequences in the Smad2 luciferase reporter were mutated using KOD-Plus-Mutagenesis Kit (SMK-101, Toyobo). Smad2 cDNA was amplified by RT-PCR using the whole RNA extracted from U87 and cloned into 3× Flag-pCMV 12.0, the expression vector. Mature miR-124 sequences were cloned into the retroviral vector pSuper-puro. Retroviral miR-124 or control vector (ctrl) plasmids were transfected into the phoenix cells and the supernatant were harvested 48 h post-transfection. For infection, U87 cells were incubated for 16 h at 37°C with filtered viral supernatants supplemented with polybrene (8 µg/ml). Stable cell lines were established using puromycin selection. Cells with restored expression of miR-124 were designated as U87miR-124; the respective control cells infected with siRNA control vectors were designated as U87siRNA control.
Luciferase reporter gene assays
The 3’-UTR of Smad2 containing the putative binding site of miR-124 was amplified and subcloned into pGL3 luciferase promoter vector (Promega, Madison, WI, USA). The vector was co-transfected with miR-124 mimics into HEK293T cells for 48 h. The cells were harvested and relative luciferase activity was detected using a dual-luciferase reporter assay kit (Promega), according to the manufacturer’s instructions. All experiments were performed at least three times.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from tissues and cell lines using the miRNeasy Mini Kit (Qiagen). The miRNA Q-PCR Detection Kit (GeneCopoeia) was used for quantification of miRNA levels according to the manufacturer’s protocol. For quantification of PRMT1 mRNA levels, the RT reactions were conducted with the RevertAid TM H Minus First Strand cDNA Synthesis Kit (Fermentas). QRT-PCR was performed using an ABI 7900 System (Bio-Rad). RNU6B and β-actin were used as normalizing controls for miRNA and mRNA quantification, respectively. The 2-ΔΔCt method was employed to calculate the relative expression levels. The primers were as follows: MiR-124, forward primer: 5’-ACACTCCAGCTGGGTAAGGCACGCGGTGA-3’, and reverse primer: 5’-TGGTGTCGTGGAGTCG-3’; Smad2, forward primer: 5’-CCGACACACCGAGATCCTAAC-3’, and reverse primer: 5’-GAGGTGGCGTTTCTGGAATATAA-3’.
Western blotting analysis
Whole cell extracts were prepared with a cell lysis reagent (Sigma-Aldrich, St. Louis, MO, USA) according to the manual, and then, the protein was quantified by a BCA assay (Pierce, Rockford, IL, USA). The protein samples were then separated by SDS-PAGE (10%) and detected by western blot, using polyclonal (rabbit) anti-Smad2 (Santa Cruz Bio-technology, Santa Cruz, CA, USA). Goat anti-rabbit IgG (Pierce, Rockford, IL, USA) secondary antibody conjugated to horseradish peroxidase and ECL detection systems (SuperSignal West Femto, Pierce) were used for detection.
MTT assay
The 3-(4,5-dimethylthiazal-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay was used to estimate cell viability. Briefly, cells were plated at a density of 1×104 cells per well in 96-well plates. After exposure to specific treatment, the cells were incubated with MTT at a final concentration of 0.5 mg/ml for 4 h at 37°C. After removal of the medium, 150 mM DMSO solution was added to dissolve the formazan crystals. The absorbance was read at 570 nm using a multi-well scanning spectrophotometer reader. Cells in the control group were considered 100% viable.
Migration assay
A wound healing assay was used to assess cell migration ability. U87 cells were cultured in a six-well plate for 24 h and transfected with miR-124 or siRNA control, and then maintained for 48 h until confluency was reached. A wound was created by manually scraping the cell monolayer with a 200-μl pipette tube, and the floating cells were removed with two phosphate-buffered saline washes. Cell migration was photographed at 48 h.
Invasion assay
Cell invasion capability was examined by transwell invasion assay. Cells were cultivated to 80% confluence on 12-well transwell invasion chamber plates then observed for 24 h. All experiments were performed in triplicate. The number of cells invading across the membrane were counted under a light microscope.
Statistical analysis
Data are expressed as mean ± SD and analyzed by Student’s t-test. Compared with respective controls, P values of <0.05 were considered statistically significant.
Results
MiR-124 expression in glioma tissues and cells
We first employed qRT-PCR to detect miR-124 levels in glioma tissues. As shown in Figure 1A, the expression level of miR-124 was markedly downregulated in glioma tissues compared with normal brain tissues. To determine the importance of the decreased miR-124 expression in glioma, we analyzed the relationship between miR-124 levels and the clinicopathological features of glioma tumor patients. For this purpose, the complete set of 84 different grade glioma samples were divided into three categories based on miR-124 levels: (1) “low”, values lower than or equal to the 25th percentile, (2) “medium”, values falling between the 25th and 75th percentile, and (3) “high”, values higher than or equal to the 75th percentile.
Figure 1.
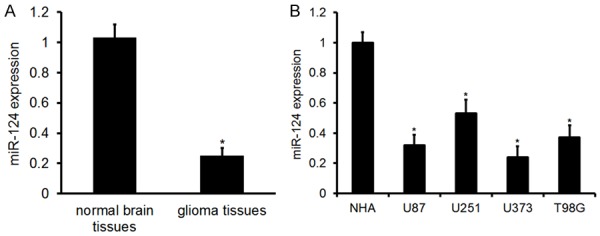
Expression of miR-124 in glioma tissues and cells. A. MiR-124 expression in glioma tissues and normal brain tissues. Error bars represent ± S.E. and *P<0.01 versus normal brain tissues. B. MiR-124 expression in glioma (U87, U251, U373, and T98G) and normal human astrocyte (NHA) cells. Error bars represent ± S.E. and *P<0.01 versus NHA cells.
According to the RT-PCR results, “low” mRNA levels of miR-124 were detected in 21 (25%) glioma patients, “medium” miR-124 levels in 40 (47.6%) patients, and “high” miR-124 levels in 23 (27.4%) glioma patients. As shown in Table 1, we found a significant positive relationship between miR-124 levels and low tumor malignancy (P<0.001). This suggests that miR-124 is negatively associated with glioma progression. MiR-124 levels were also associated with patient age (P<0.001), but not gender (P>0.05, Table 1).
Table 1.
Association between miR-124 expression and clinicopathological variables of the glioma patients
| Variables | Number of cases | miR-124 mRNA level | p-value | ||
|---|---|---|---|---|---|
|
|
|||||
| Low, n (%) | Medium, n (%) | High, n (%) | |||
| Overall | 84 | 21 (25) | 40 (47.6) | 23 (27.4) | |
| Age (Years) | <0.001 | ||||
| ≤50 | 43 | 23 (53.5) | 14 (32.6) | 6 (13.9) | |
| >50 | 41 | 18 (27.9) | 22 (53.6) | 1 (2.5) | |
| Gender | 0.784 | ||||
| Male | 45 | 14 (31.1) | 23 (51.1) | 8 (17.8) | |
| Female | 43 | 12 (27.9) | 18 (41.8) | 13 (30.3) | |
| Pathological grade (WHO) | <0.001 | ||||
| Grade I | 12 | 0 (0) | 8 (66.7) | 4 (33.3) | |
| Grade II | 13 | 6 (46.1) | 5 (38.5) | 2 (15.4) | |
| Grade III | 24 | 11 (45.8) | 10 (41.7) | 3 (12.5) | |
| Grade IV | 35 | 19 (54.3) | 14 (40.0) | 2 (5.7) | |
Real-time PCR analysis showed that the expression level of miR-124 was markedly downregulated in four of the glioma cell lines (U87, U251, U373, and T98G), in comparison with the expression levels in the NHA cell line (Figure 1B). Taken together, these results indicate that miR-124 may be a tumor inhibitor in the progression of glioma.
MiR-124 inhibited proliferation, invasion, and migration of glioma cells
To further verify the antitumor role of miR-124 in U87 cells, we then performed rescue experiments. The transient transfection of miR-124 mimics was used to restore miR-124 expression in glioma cells. As shown in Figure 2A, the expression level of miR-124 was greatly increased by miR-124 mimics. To further characterize the functional importance of miR-124 in glioma progression, we examined its effect on the proliferation and invasion of glioma cells using the MTT and transwell invasion assays. The results showed that miR-124 mimics decreased the proliferation of glioma cells (Figure 2B). Similar results were observed in invasion (Figure 2C) and migration (Figure 2D) assays of glioma cells. These findings demonstrate that miR-124 inhibits glioma cell proliferation and invasion in vitro.
Figure 2.
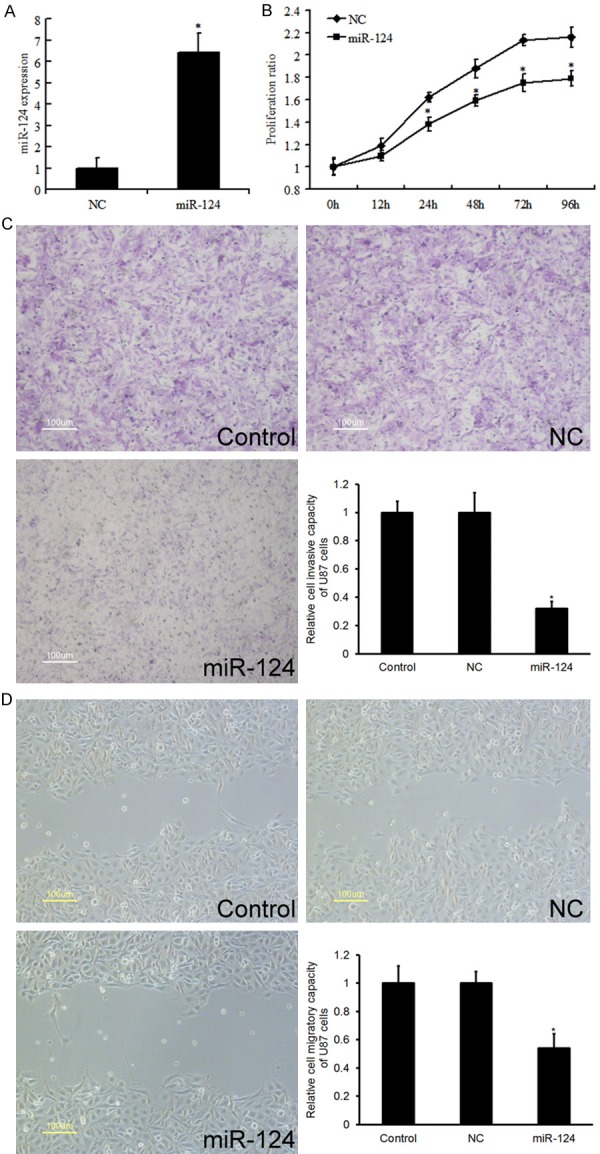
Effect of miR-124 in glioma cells on proliferation, invasion, and migration. (A) QRT-PCR analysis revealed the effects of miR-124 mimics on the expression level of miR-124. (B) MTT assays (B) and transwell assays (C) revealed the invasion ability of U87 cell transfected with miR-NC and miR-124. (D) MiR-124 inhibited cell migration as determined by wound healing assay. Data are the mean ± SD of duplicates from a representative of three independent experiments. *P<0.01 vs. NC group.
MiR-124 directly targets Smad2 in glioma
Two miRNA target prediction websites, www.microRNA.org and TargetScan, demonstrated that the 3’-UTR of Smad2 mRNA contains a conserved miR-124-binding site (Figure 3A). To confirm this prediction and verify whether Smad2 is a direct target of miR-124, a dual-luciferase reporter system was employed by co-transfection of miR-124 and luciferase reporter plasmids containing the wild-type or mutated 3’UTR of Smad2 (the latter bearing deletions of the putative miR-124 target sites). Co-transfection of miR-124 mimics suppressed the luciferase activity of the reporter containing wild-type Smad2 3’UTR sequence by dual-luciferase reporter assay; however, miR-124 mimics did not have any effect on luciferase activity when target cells were transfected with mutated Smad2 (Figure 3B). The Smad2 protein level was markedly reduced in U87 cells transfected with miR-124 mimics (Figure 3C). These data suggest that Smad2 may be a direct functional target of miR-124 in glioma. Increased levels of Smad2 restored miR-124 and reduced U87 cell proliferation (Figure 3D), invasion (Figure 3E), and migration (Figure 3F).
Figure 3.
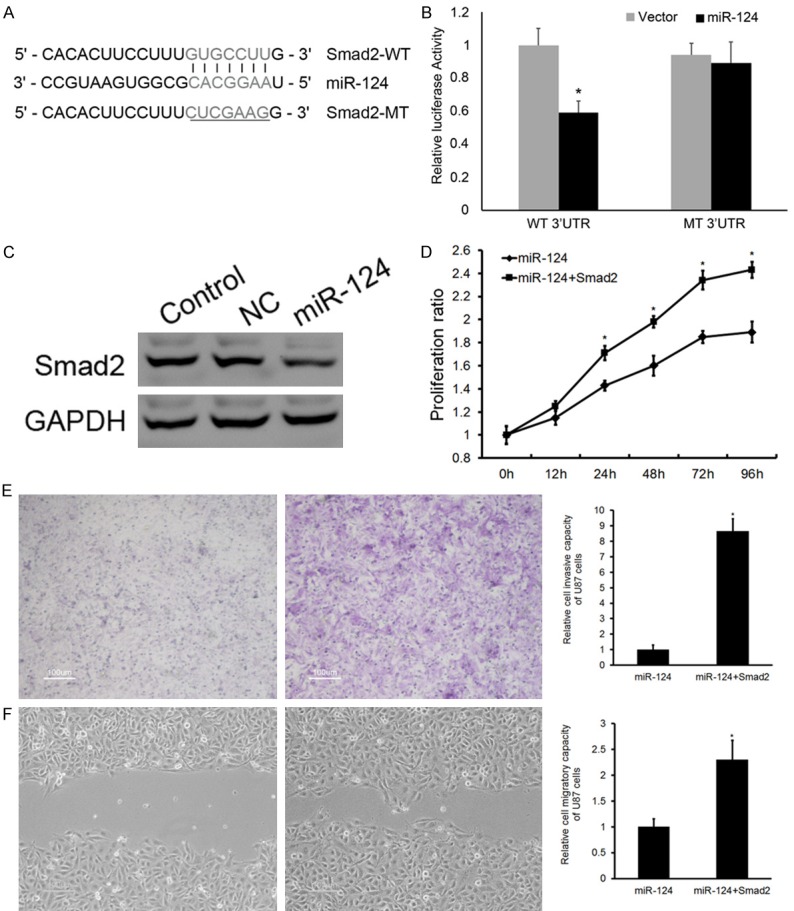
Smad2 is the direct target of miR-124 in glioma. A. Sequence alignment of miR-124 and 3’UTR of Smad2 using targetscan.org. B. Luciferase reporter assay with co-transfection of wild-type or mutant Smad2 and miR-124 mimics or miR-control in U87 cells. C. Western blot analysis revealed the effects of miR-124 mimics on the expression level of Smad2. D. Extra Smad2 restored miR-124 levels and reduced U87 cell proliferation. E. Invasion. F. Migration. Error bars represent ± S.E. and *P<0.01 versus negative control (NC).
Discussion
Various studies have suggested that deregulated miRNAs play an important role in various human cancers, including glioma. Identifying the miRNAs that are essential to halting glioma progression and their targets may provide promising therapeutic opportunities [10-12]. In this study, we demonstrated that miR-124 is a tumor suppressor that inhibits the proliferation and invasion of glioma via targeting Smad2.
MiR-124 has been classified as a tumor suppressor in several types of human cancers [18,19]. Previous research has shown that miR-124 is abundantly expressed in normal brain tissue [20], is involved in embryonic neuronal differentiation [21], and is an important regulator of adult neurogenesis in the subventricular zone stem cell niche [22]. The present study determined that miR-124 expression is downregulated in human glioma tissues and negatively correlated with the pathological grade of glioma. Furthermore, the in vitro study revealed that miR-124 restoration inhibited glioma cell proliferation and invasion, illustrating its role as a tumor suppressor.
Smad2 is a pivotal transducer of the TGF-β pathway and plays complex and contradictory roles during tumorigenesis [23]. Accumulating evidence suggests that abnormal Smad2 expression is closely associated with a variety of human cancers, including bladder [24], ovarian [25], cervical [26] and colorectal [27] cancer. Smad2 must be strictly controlled to ensure the normal growth of cells. Our study identified Smad2 as a new target gene of miR-124, which halts glioma progression by regulating its proliferation, invasion, and migration.
In conclusion, our results demonstrated that miR-124 is downregulated in gliomas and this dysfunction plays a critical role in the growth and invasion of glioma cells. Smad2 is a direct target of miR-124 and the overexpression of miR-124 downregulates the expression of Smad2 proteins and mRNA simultaneously. Our results indicate that miR-124 functions as a tumor suppressor miRNA through the downregulation of Smad2, and shows great potential as a target in the gene therapy of glioma.
Acknowledgements
The present study was supported by the Youth Science Foundation of Heilongjiang Province (grant No. QC2013C093).
Disclosure of conflict of interest
None.
References
- 1.Bush NA, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev. 2017;40:1–14. doi: 10.1007/s10143-016-0709-8. [DOI] [PubMed] [Google Scholar]
- 2.Codrici E, Enciu AM, Popescu ID, Mihai S, Tanase C. Glioma stem cells and their microenvironments-providers of challenging therapeutic targets. Stem Cells Int. 2016;2016:5728438. doi: 10.1155/2016/5728438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, He D, Li Z, Zhang X, Pan D, Chen G. Overexpression of vascular endothelial growth factor indicates poor outcomes of glioma: a systematic review and meta-analysis. Int J Clin Exp Med. 2015;8:8709. [PMC free article] [PubMed] [Google Scholar]
- 4.Guan Y, Chen L, Bao Y, Li Z, Cui R, Li G, Wang Y. Identification of low miR-105 expression as a novel poor prognostic predictor for human glioma. Int J Clin Exp Med. 2015;8:10855. [PMC free article] [PubMed] [Google Scholar]
- 5.Wang BC, Ma J. Role of microRNAs in malignant glioma. Chin Med J (Engl) 2015;128:1238. doi: 10.4103/0366-6999.156141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuna M, Machado AS, Calin GA. Genetic and epigenetic alterations of microRNAs and implications for human cancers and other diseases. Genes Chromosomes Cancer. 2016;55:193–214. doi: 10.1002/gcc.22332. [DOI] [PubMed] [Google Scholar]
- 7.Zhang XL, Chen JH, Qin CK. MicroRNA-155 expression as a prognostic factor in patients with gallbladder carcinoma after surgical resection. Int J Clin Exp Med. 2015;8:21241–21246. [PMC free article] [PubMed] [Google Scholar]
- 8.Green D, Fraser WD, Dalmay T. Transfer RNA-derived small RNAs in the cancer transcriptome. Pflugers Arch. 2016;468:1041–7. doi: 10.1007/s00424-016-1822-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian LQ, Liu EQ, Zhu XD, Wang XG, Li J, Xu GM. MicroRNA-197 inhibits cell proliferation by targeting GAB2 in glioblastoma. Mol Med Rep. 2016;13:4279–4288. doi: 10.3892/mmr.2016.5076. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, Pang D, Wang C, Zhong S, Wang S. MicroRNA-134 modulates glioma cell U251 proliferation and invasion by targeting KRAS and suppressing the ERK pathway. Tumor Biol. 2016;37:11485–93. doi: 10.1007/s13277-016-5027-9. [DOI] [PubMed] [Google Scholar]
- 11.Hermansen SK, Nielsen BS, Aaberg-Jessen C, Kristensen BW. miR-21 is linked to glioma angiogenesis: a co-localization study. J Histochem Cytochem. 2016;64:138–48. doi: 10.1369/0022155415623515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paw I, Carpenter RC, Watabe K, Debinski W, Lo HW. Mechanisms regulating glioma invasion. Cancer Lett. 2015;362:1–7. doi: 10.1016/j.canlet.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herpin A, Lelong C, Favrel P. Transforming growth factor-β-related proteins: an ancestral and widespread superfamily of cytokines in metazoans. Dev Comp Immunol. 2004;28:461–485. doi: 10.1016/j.dci.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Mehra A, Wrana JL. TGF-β and the Smad signal transduction pathway. Biochem Cell Biol. 2002;80:605–622. doi: 10.1139/o02-161. [DOI] [PubMed] [Google Scholar]
- 15.Heldin CH, Miyazono K, Ten Dijke P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 16.Markowitz SD, Roberts AB. Tumor suppressor activity of the TGF-β pathway in human cancers. Cytokine Growth Factor Rev. 1996;7:93–102. doi: 10.1016/1359-6101(96)00001-9. [DOI] [PubMed] [Google Scholar]
- 17.Hua X, Miller ZA, Wu G, Shi Y, Lodish HF. Specificity in transforming growth factor β-induced transcription of the plasminogen activator inhibitor-1 gene: interactions of promoter DNA, transcription factor μE3, and Smad proteins. Proc Natl Acad Sci U S A. 1999;96:13130–13135. doi: 10.1073/pnas.96.23.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang YJ, Wang QY, Zhou CX, Yin QQ, He M, Yu XT, Cao DX, Chen GQ, He JR, Zhao Q. MiR-124 targets Slug to regulate epithelial-mesenchymal transition and metastasis of breast cancer. Carcinogenesis. 2013;34:713–22. doi: 10.1093/carcin/bgs383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia J, Wu Z, Yu C, He W, Zheng H, He Y, Jian W, Chen L, Zhang L, Li W. miR-124 inhibits cell proliferation in gastric cancer through downregulation of SPHK1. J Pathol. 2012;227:470–480. doi: 10.1002/path.4030. [DOI] [PubMed] [Google Scholar]
- 20.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 21.Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massagué J. TGFβ in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Chen E, Tang M, Yang X, Wang Y, Quan Z, Wu X, Luo C. The SMAD2/3 pathway is involved in hepaCAM-induced apoptosis by inhibiting the nuclear translocation of SMAD2/3 in bladder cancer cells. Tumor Biol. 2016:1–13. doi: 10.1007/s13277-016-4821-8. [DOI] [PubMed] [Google Scholar]
- 25.Qiu X, Cheng JC, Zhao J, Chang HM, Leung PC. Transforming growth factor-β stimulates human ovarian cancer cell migration by up-regulating connexin43 expression via Smad2/3 signaling. Cell Signal. 2015;27:1956–1962. doi: 10.1016/j.cellsig.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Zhao JL, Zhang L, Guo X, Wang JH, Zhou W, Liu M, Li X, Tang H. miR-212/132 downregulates SMAD2 expression to suppress the G1/S phase transition of the cell cycle and the epithelial to mesenchymal transition in cervical cancer cells. IUBMB Life. 2015;67:380–394. doi: 10.1002/iub.1381. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki R, Fukui T, Kishimoto M, Miyamoto S, Takahashi Y, Takeo M, Mitsuyama T, Sakaguchi Y, Uchida K, Nishio A. Smad2/3 linker phosphorylation is a possible marker of cancer stem cells and correlates with carcinogenesis in a mouse model of colitis-associated colorectal cancer. J Crohns Colitis. 2015;9:565–574. doi: 10.1093/ecco-jcc/jjv073. [DOI] [PubMed] [Google Scholar]


