Abstract
Objective: This study focuses on the feasible molecular mechanism of the interaction between MALAT-1, CCR7 and related genes in oral squamous cell carcinoma, to find new target molecules that can block the lymph node metastasis. Methods: The expression of MALAT-1, miRNA-320s, SRSF1, YB-1 and CCR7 were detected in T3/T4-phase OSCC tissues of two groups with or without lymph node metastasis using real-time qPCR. CO-IP and western blot to test the interaction of RNAs (MALAT-1, miRNA-320s) with SRSF1 protein or YB-1 were evaluated by CO-IP, Western blot and real-time qPCR. The expression change of chemokine receptor CCR7 were investigated using CO-IP, Western blot and real-time qPCR after silencing miRNA-320d (one of the miRNA-320s family members) by transfection of miRNA mimics to explore related signaling pathway. Results: The expression levels of MALAT-1 SRSF1 and CCR7 in OSCC tissues with were differentially higher compared with those of samples without lymph node metastasis as well as para-carcinoma tissues, exclusive of miRNA-320d. Moreover, it is confirmed that the target RNA (MALAT-1, miRNA-320s) and SRSF1 protein can combine with each other, based on the statistically significant difference compared with negative control group (P<0.05). In addition, the expression of CCR7 was higher than the negative control group after silencing miRNA-320d. Conclusion: SRSF1 is likely to mediate the interactive relationship between MALAT-1 and miRNA-320d. CCR7 expression can be distinctly increased by silencing miRNA-320d. The effect of long-chain non-coding RNA MALAT-1 on chemokine receptor CCR7 and possibly further influence on lymph node metastasis of oral squamous cell carcinoma are revealed in molecular level to offer help for prevention and treatment of OSCC in future.
Keywords: Oral squamous cell carcinoma, long non-coding RNA, MALAT-1, human tongue carcinoma cell line (SCC9, SCC25), SRSF1, YB-1, hsa-miRNA-320s, chemokine receptor CCR7
Introduction
Oral squamous cell carcinoma (OSCC) is the most common type of oral malignant tumors, which is easier to induce lymphatic metastasis at early stage. Although the OSCC morbidity is still growing each year, but the complex reasons of this cancer need to be elucidate. This study focuses on the possible molecular mechanism of the interaction between long-chain non-coding RNA MALAT-1 and chemokine receptor CCR7 and its related genes in OSCC, aiming at finding out possible target spots to block the lymphatic metastasis [1].
Cell carcinogenesis and metastasis is a complex process, in which multifactor, multigene and complicated gene regulatory factors are involved. Recent studies indicate that long non-coding RNA (lncRNAs) plays an important role in cell development and metabolism as well as tumor genesis and development. No mutation exists in protein-coding genes of some tumors, nothing but expression of lncRNAs is abnormal, and several lncRNAs are enough for changing the transcription process [2]. Extensive molecular research revealed that unusually high expression levels of lncRNAs were detected in the various cancers in lung cancer [3], breast cancer [4], colorectal cancer [5], cervical cancer [6], hepatocellular carcinoma [7], kidney cancer [8] and so on [9]. The lncRNA transcripts (>200 nt in length) regulate gene expression in the levels of transcription, post-transcription, epigenetic modification and other aspects, and thus affect the biological functions of cells [10]. Metastasis associated lung adenocarcinoma transcript 1 (MALAT-1), which was first discovered in non-small cell lung cancer [11], is located on chromosome 11q13.1 and has highly evolutional conservation [12]. MALAT-1 is the pioneer related to tumor metastasis. It was found that MALAT-1 was highly expressed in the tissues of patients with non-small cell lung cancer and the prognosis was poor. Subsequent studies found that MALAT-1 also had high expression in a variety of malignant tumors including OSCC [13]. MALAT-1 can significantly promote the proliferation, metastasis and invasion of malignant tumor cells. MicroRNA (miRNA) is an endogenous small non-coding RNA with~22 nt in length in higher eukaryotes, which can mediate the regulation, translation or degradation of target mRNA by complementary binding [14]. Consequently, inhibition of miRNA maturation can accelerate cell transformation and tumorigenesis [15]. The abnormal expression of miRNA in tumor cells influences translation of the downstream mRNA and facilitates aberrant cell proliferation [16,17]. In the study of esophageal squamous cell carcinoma (ESCC), MALAT-1 was confirmed to restrain the proliferation, invasion and metastasis of tumor cells by regulating miRNA. MALAT-1 was synthesized by RNA polymerase II, whose expression product was located in the nuclear speckle [18], as well as the serine/arginine rich protein (SR protein) family involved in mRNA precursor processing [19]. In view of their co-localization relationship, the relevance between MALAT-1 and SF2/ASF of SR protein family was investigated through suppression of MALAT-1 transcription using 5,6-dichloro-1-b-D-ribobenzimidazole (DRB), which can reversibly inhibit RNA polymerase II. The results ensured that SF2/ASF was transferred from nuclear speckle to intranuclear and then repositioned after the MALAT-1 transcription was inhibited, while SF2/ASF would be resumed to the distribution of nuclear speckle distribution when the expression of MALAT-1 was restored by discontinuing DRB treatment. MALAT-1 can specifically recruit SR family members to participate in epigenetic regulation and cell-cycle control. The possible mechanism is MALAT-1 directly related to phosphorylation of SR protein. The localization of the nuclear speckle is the structural basis of MALAT-1 regulating expression, localization and activation of SR protein family members [20].
Chemokine receptors and G protein-coupled receptors play diverse roles in immune defense by controlling cell migration and activating immune cells, as well as viral invasion, tumor growth and metastasis. Chemokine receptors play a crucial role in the pathogenesis of many diseases, including tumors [21]. The research on HIV infection shows that the mechanism of virus invasion into the host is escaping the host cell immune defense and enhancing its own viability with the help of protein encoded by self DNA similar to chemokine receptor [22]. Meanwhile, chemokine G protein-coupled receptor is one of the key carriers mediating HIV infection [23]. Therefore, blocking and inhibiting chemokine receptors can control HIV infection, and become the current target of anti-HIV drug targeting therapy.
The molecular regulation course of tumor cell migration to a specific tissue is similar to that of immune cell migration to the infection site. Therefore, chemokines and their receptors became the object of tumor research, which play an important role in the migration of immune cells. A large number of studies find that chemokines and their receptors are closely related to the occurrence, development, metastasis and prognosis of tumors [24]. For example, the biological axis consisted of chemokine CXCL12 and its receptor CXCR4 plays an important role in the growth and metastasis of several solid tumors, such as breast cancer [25]. CXCR4 is a highly conserved chemokine receptor in evolution, with a variety of effects on different immune cell lines [26]. Breast cancer cells with expression of chemokine receptors CXCR4 and CCR7 [27], can metastasize to target organs with high expression of their ligands CXCL12 and CCL2, including lung, liver and lymph nodes. Moreover, was used, the probability of breast cancer cell metastasis to mouse lungs can be reduced significantly after using anti-CXCR4 antibody [28]. Recent bioinformatics analysis of genomic data suggests that one of the target genes of chemokine receptor CCR7 may be hsa-miR-320a. However, certain evidence is not available yet. Previous studies of our group and other documents indicate reveal that chemokine receptor CCR7 plays an important role in lymphatic metastasis of OSCC.
The following hypothesis is proposed by the above mentioned: MALAT-1 regulates the processing of chemokine receptor CCR7 mRNA precursor miRNA-320s by activating SR protein or YB-1 protein to affect its expression in oral squamous cell carcinoma, and sequentially may influence the behavior of lymph node metastasis of OSCC.
This study contains three contents: 1) detection of expression of MALAT-1, miRNA-320s, SRSF1, YB-1 and CCR7 in OSCC samples using real-time fluorescence quantification; 2) correlation analysis between human tongue cancer cell lines SCC-9/SCC-25 and SRSF1 protein/YB-1 at the cellular level by co-immunoprecipitation, immunoblotting tests of related RNA (MALAT-1, miRNA-320s); 3) analysis of chemokine receptor CCR7 expression after silence of miRNA-320s by miRNA mimics transfection technology using co-immunoprecipitation, immunoblotting and real-time RT-PCR, and further investigation on possible interrelationships.
Materials and methods
Samples
OSCC carcinoma tissues and adjacent tissues were taken from patients in Department of Stomatology, Changhai Hospital. Patients with stage T3/T4 and without preoperative chemotherapy, radiotherapy and recurrence were selected, and their clinical information including age, sex, primary site, clinical stage and cervical lymph node metastasis were carefully recorded. The frozen sections were cut off from the excised tissues, and the identification of carcinoma tissues and adjacent tissues were performed using optical microscope (Olympus). The carcinoma tissues and adjacent tissues were divided into groups A and B numbered in sequence from 1 to 6, and the samples were transferred into sterile frozen tube and stored at -80°C for subsequent RNA extraction.
The human tongue squamous cell carcinoma cell lines, SCC-9 and SCC-25, were purchased from ATCC (American Type Culture Collection, US).
Quantification of OSCC-related gene expression
Total RNA was isolated from the carcinoma tissues and adjacent tissues using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the protocol. Because the molecular weight of miRNA-320 was small, which might exist in the cell nucleus, it was necessary to use strong lysis solution (Imprint RNA Immunoprecipitation Kit RIP-12Rxn, Sigma, louis, MO) to lyse the cells, and specific reverse transcription kit (TaqMan MicroRNA Reverse Transcription Kit and High-Capacity RNA-to-cDNA Kit, Thermo scientific, US). RNA concentration was measured spectrophotometrically on a NANO Drop 1000 spectrophotometer (Thermo scientific) and the integrity and purity of RNA was examined by running in 1.0% agarose gel. The first-strand synthesis was carried out based on Promega M-MLV RT Usage information (Takara, Tokyo, Japan). The cDNA mix was diluted to 1:10 and stored at -80°C for subsequent real-time quantitative PCR (qRT-PCR).
The real-time quantitative PCR was performed with the SYBR Green on an ABI 7500 Real-Time Detection System (Applied Biosystems, USA) to investigate the expression of OSCC-related genes, including MALAT-1, SRSF1, YB-1 and CCR7. And qRT-PCR of miR-320s (miRNA320a/b/d/e) was performed using qRT-PCR kit (Takara, Dalian, China). All primers used in this assay were shown in Table 1. The qRT-PCR was carried out in a total volume of 20 μL containing SYBR Green Master Mix of Takara (Takara, Tokyo, Japan). The expression of MALAT-1, SRSF1, YB-1 and CCR7 genes was normalized to the expression of β-actin gene for each sample, while the expression of miRNA-320 genes was normalized to the expression of U6 gene for each sample. Because miRNA-320 has the precursor and mature miRNA, therefore, the expression of miRNA-320 was the average value of the expression of all the precursors and mature miRNAs. The relative expression of these genes was analyzed using the comparative CT (2-ΔΔCT) method [29].
Table 1.
Primers used in this study
| Gene name | Primer name | Sequence (5’-3’) |
|---|---|---|
| MALAT-1 | Forward | GAATTGCGTCATTTAAAGCCTAGTT |
| MALAT-1 | Reverse | GTTTCATCCTACCACTCCCAATTAAT |
| SRSF1 | Forward | CCTGTTCATCAGGAACGTCG |
| SRSF1 | Reverse | CGGCCACATACCCACTTTCTA |
| YB-1 | Forward | GGGGACAAGAAGGTCATCGC |
| YB-1 | Reverse | CGAAGGTACTTCCTGGGGTTA |
| CCR7 | Forward | TGAGGTCACGGACGATTACAT |
| CCR7 | Reverse | GTAGGCCCACGAAACAAATGAT |
| Has-miR-320a precursor | Forward | GCTTCGCTCCCCTCCGCCTTCTCTTCCCGGTTCTTCCCGGAGTCGGGAAAAG |
| Has-miR-320a precursor | Reverse | ACCTCATCCTTTTTCGCCCTCTCAACCCAGCTTTTCCCGACTCCGGGAAGAA |
| Has-miR-320a mature miRNA | Forward | AAAAGCTGGGTTGAGAGGGCGA |
| Has-miR-320a mature miRNA | Reverse | TCGCCCTCTCAACCCAGCTTTT |
| Hsa-miR-320b1 precursor | Forward | AATTAATCCCTCTCTTTCTAGTTCTTCCTAGAGTGAGGAAAAGCTGGGT |
| Hsa-miR-320b1 precursor | Reverse | AATTAATTAGTTAATTTGTTTGCCCTCTCAACCCAGCTTTTCCTCACTC |
| Hsa-miR-320b2 precursor | Forward | TGTTATTTTTTGTCTTCTACCTAAGAATTCTGTCTCTTAGGCTTTCTCTTCCCAGATTTCCCAAAGTTGGGAAAAGCTG |
| Hsa-miR-320b2 precursor | Reverse | TTCCCTATCTAATTATGTCAGAGACAGAATTCTTTTTTTTTCCTTTTGCCCTCTCAACCCAGCTTTTCCCAACTTTGGGA |
| Hsa-miR-320b mature miRNA | Forward | AAAAGCTGGGTTGAGAGGGCAA |
| Hsa-miR-320b mature miRNA | Reverse | TTGCCCTCTCAACCCAGCTTTT |
| Hsa-miR-320d1 precursor | Forward | TTCTCGTCCCAGTTCTTCCCAAAGTTGAGAAAAG |
| Hsa-miR-320d1 precursor | Reverse | TCCTCTCAACCCAGCTTTTCTCAACTTTGGGAAG |
| Hsa-miR-320d2 precursor | Forward | TTCTCTTCCCAGTTCTTCTTGGAGTCAGGAAAAG |
| Hsa-miR-320d2 precursor | Reverse | TCCTCTCAACCCAGCTTTTCCTGACTCCAAGAAG |
| Hsa-miR-320d mature miRNA | Forward | AAAAGCTGGGTTGAGAGGA |
| Hsa-miR-320d mature miRNA | Reverse | TCCTCTCAACCCAGCTTTT |
| Hsa-miR-320e precursor | Forward | GCCTTCTCTTCCCAGTTCTTCCTGGAGTCGGGGAAAAG |
| Hsa-miR-320e precursor | Reverse | ACCTTCTCAACCCAGCTTTTCCCCGACTCCAGGAAG |
| Hsa-miR-320e mature miRNA | Forward | AAAGCTGGGTTGAGAAGG |
| Hsa-miR-320e mature miRNA | Reverse | CCTTCTCAACCCAGCTTT |
Correlation of regulatory RNA (MALAT-1 and miRNA-320) and functional protein (SRSF1 and YB-1) in tongue cancer cell line
After the cell lines SCC-9 and SCC-25 recovered to the sixth generation, the cells with high expression of SRSF1 and YB-1 were selected as the experimental group and negative anti-rabbit antibody were selected as the control group. The interaction between two regulatory RNAs (MALAT-1 and miRNA-320) and two functional proteins (SRSF1 and YB-1) were tested using immunoprecipitation and Western blotting according to the previous study (anti-SF2 antibody and anti-YB-1 antibody, Abcam, US) [30]. The differential expression was verified using qRT-PCR.
Correlation of miRNA-320 and functional protein CCR7 in tongue cancer cell line
The cells with silenced miRNA-320d transfected by miRNA-320d miRNA inhibitor (4464066, Thermo scientific, US) were defined as the experimental group, while the cells transfected by control miRNA (4464058, Thermo scientific, US) was selected as the control group. The interaction between miRNA-320 and CCR7 was tested using immunoprecipitation and Western blotting according to the previous study [31]. The differential expression was verified by qRT-PCR.
Statistical analysis
Statistical analysis of data was performed using SPSS 22.0 and Graphpad prism 7.0, and the quantitative data were given in terms of relative mRNA expression level as mean ± SD (N=3). Statistical significance was determined by paired t test. The P value less than 0.05 was considered statistically significant.
Results
Cell morphological characteristics of the sixth generation SCC-9 and SCC-25
When passaged to the sixth generation, the cell lines SCC-9 and SCC-25 were observed under TEM microscope. The cells showed epithelial like morphology, which were flattened and polygonal, and arranged like “paving stone”, and there was obvious atypia and more common mitotic with a small amount of megakaryocyte and multinucleated cells (Figure 1). Therefore, these cells can be identified as oral squamous cell carcinoma.
Figure 1.
The sixth generation cell lines of SCC9 (A) and SCC25 (B).
The differential expression of OSCC-related genes in carcinoma tissues and adjacent tissues
Three genes including MALAT-1 (Figure 2A), SRSF1 (Figure 2B) and CCR7 (Figure 2D) have the same expression trend in different groups, whose expression in carcinoma tissues with lymph node metastasis were significantly higher than carcinoma tissues without lymph node metastasis and adjacent tissues. The relative expression of MALAT-1 in carcinoma tissues with lymph node metastasis was 2.30 fold (P<0.01) and 11.04 fold (P<0.01) of that in carcinoma tissues without lymph node metastasis and adjacent tissues, respectively (Figure 2A). The relative expression of SRSF1 in carcinoma tissues with lymph node metastasis was 1.17 fold (P<0.01) and 1.74 fold (P<0.01) of that in carcinoma tissues without lymph node metastasis and adjacent tissues, respectively (Figure 2B). The relative expression of CCR7 in carcinoma tissues with lymph node metastasis was 1.85 fold (P<0.01) and 2.38 fold (P<0.01) of that in carcinoma tissues without lymph node metastasis and adjacent tissues, respectively (Figure 2D). However, the expression of YB-1 has no difference in carcinoma tissues with and without lymph node metastasis, while that in carcinoma tissues was significantly higher than adjacent tissues 1.28 fold, P<0.01) (Figure 2C).
Figure 2.
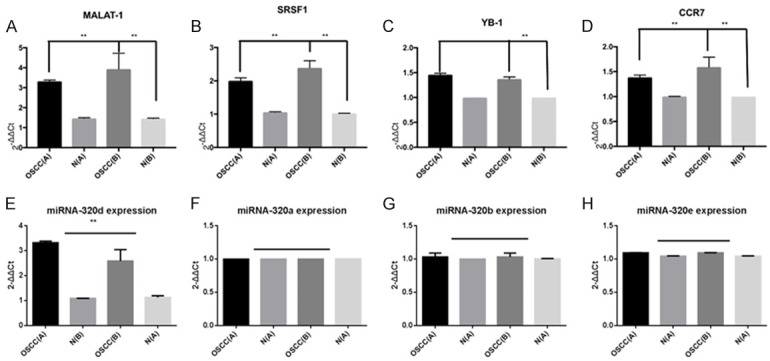
The differential expression of MALAT-1 (A), SRSF1 (B), YB-1 (C), CCR7 (D), has-miR-320a (E), has-miR-320b (F), has-miR-320c (G) and has-miR-320d (H) in carcinoma tissues and adjacent tissues. OSCC (A), OSCC carcinoma tissues with lymph node metastasis; N (A), adjacent tissues of OSCC (A); OSCC (B), OSCC carcinoma tissues without lymph node metastasis; N (B), adjacent tissues of OSCC (B).
The relative expression of miRNA-320d in carcinoma tissues with lymph node metastasis was significantly lower than that in carcinoma tissues without lymph node metastasis 0.78 fold, P<0.01) (Figure 2G), and both of them were significantly higher than that in adjacent tissues 3.09 fold, P<0.01; 2.29-fold, P<0.01;) (Figure 2G). There was no significant difference in the expression level of miRNA-320a (Figure 2E), miRNA-320b (Figure 2F) and miRNA-320e (Figure 2H) in different groups.
The difference of OSCC-related genes expression in carcinoma tissues and adjacent tissues (T/N)
The different expression of MALAT-1, YB-1, SRSF1, CCR7 and miR-320s in carcinoma tissues and adjacent tissues were shown by T/N value. The nonzero T/N value indicated that the gene expression could be detected in all 6 groups of samples. When the T/N value was greater than 1 indicating that the gene expression level in carcinoma tissues was higher than that in adjacent tissues, and when the T/N value was less than 1 indicating that the gene expression level in carcinoma tissues was lower than that in adjacent tissues. There were significantly higher expression of MALAT-1 (Figure 3A), SRSF1 (Figure 3B) and CCR7 (Figure 3C), and significantly lower expression of miRNA-320d (Figure 3G) in carcinoma tissues with lymph node metastasis group. No significant difference was observed in other genes.
Figure 3.
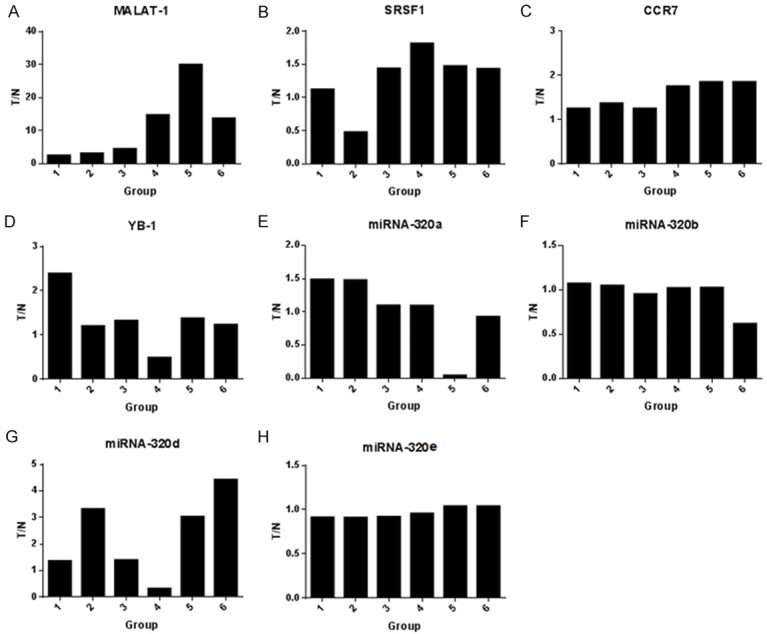
The T/N of MALAT-1 (A), SRSF1 (B), CCR7 (C) YB-1 (D) and miR-320s (E-H) in carcinoma tissues and adjacent tissues. 1-3 groups were carcinoma tissues without lymph node metastasis, and 4-6 groups were carcinoma tissues with lymph node metastasis.
Correlation of MALAT-1, SRF1, YB-1 and miRNA-320s
Expression of SRSF1 and YB-1 in cells SCC-9 and SCC-25
The cell lysis efficiency and the expression of SRSF1 and YB-1 in cells SCC-9 and SCC-25 were detected using Western Blot. The same express trend of reference GADPH indicated that the cleavage efficiency was similar, and the expression of SRSF1 and YB-1 in cells SCC9 and SCC25 were higher than that in the control groups (Figure 4).
Figure 4.
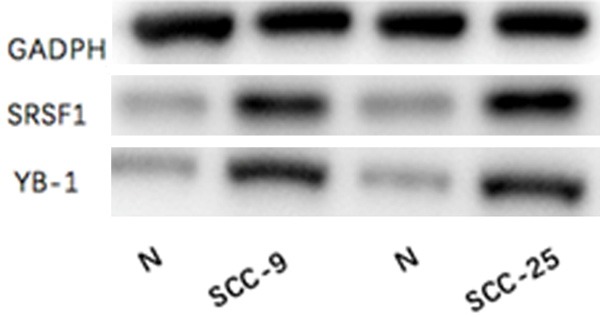
The expression of SRSF1 and YB-1 in cells SCC-9 and SCC-25.
Binding efficiency of RNA-protein-antibody complex
The binding efficiency of RNA-protein-antibody complex was detected using Western Blot after co-immunoprecipitation. The expression of reference GADPH was similar in cell lines. SF2-RNA protein complexes could be detected by anti-SF2 antibodies, indicating that SRSF1 and RNA could bind well. At the same time, we could see that the complex band of anti-YB-1 antibody and YB-1-RNA are not clear, similar to the negative control (Figure 5).
Figure 5.
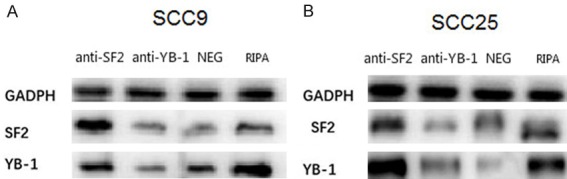
Co-Immunoprecipitation bands of RNA-protein-antibody complex. SF2-RIP, Co-Immunoprecipitation bands of SRSF1 protein; YB1-RIP, Co-Immunoprecipitation bands of YB1-RIP protein; NEG, Co-Immunoprecipitation bands of negative controlled rabbit antibody; RIPA, the original lysate.
Quantification of MALAT-1 in purified RIP products
Paired t-test was used for statistical analysis. The expression level of MALAT-1 in SCC9 (Figure 6A) and SCC25 (Figure 6B) purified RIP products was significantly higher than negative control samples (2.39-fold, P<0.01; 2.17 fold, P<0.01). This suggested that MALAT-1 was a component of the anti SRSF1-protein-RNA complex.
Figure 6.
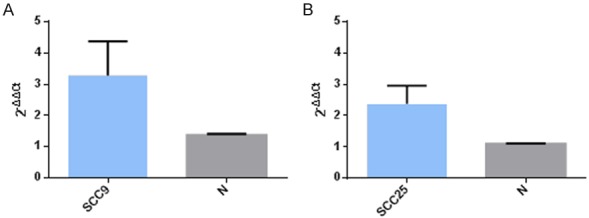
The expression of MALAT-1 after Co-Immunoprecipitation.
Quantification of miRNA-320d in purified RIP products
Paired t-test was used for statistical analysis. The expression level of miRNA-320d in SCC9 (Figure 7A) and SCC25 (Figure 7B) purified RIP products was significantly higher than that in normal tissues 1.43 fold, P<0.05; 1.34-fold, P<0.05). This suggested that miRNA-320d was a component of the anti SRSF1-protein-RNA complex.
Figure 7.
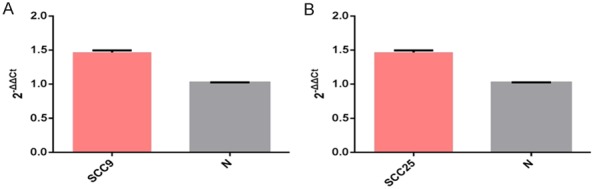
The expression of miRNA-320d after Co-Immunoprecipitation.
The interaction between miRNA-320d and CCR7
Cell line with high expression of miRNA-320d was selected and then its miRNA-320d was silenced by mimic miRNA transfection. Using CO-IP and western blotting, CCR7 expression changes were observed in miRNA-320d silent group and control. The relationship between miRNA-320d and CCR7 was checked by real-time qPCR.
Assessment of silence efficiency of miRNA-320d
After silencing miRNA-320d, bands of the experimental group were clearer than those of the control group, indicating excellent silence efficiency of miRNA-320d (Figure 8).
Figure 8.
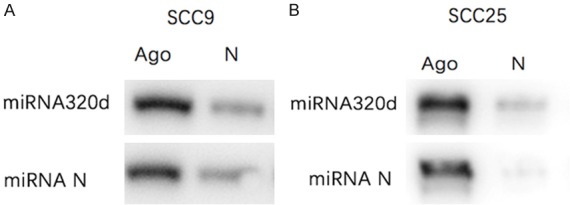
The silence efficiency of miRNA-320d.
Expression of CCR7 in miRNA-320d silent cells
The statistical analysis by paired t test, same as section 2.1, was used for comparison of CCR7 between SCC9/SCC25 experimental groups and that of the control (tscc9=4.84, P<0.01; tscc25=4.26, P<0.01). After logarithmic transformation of ΔCT values by 2-ΔΔCt, the expression level of CCR7 in experimental group was higher than that in control group, and the difference was statistically significant suggesting that CCR7 expression of was increased after silencing miRNA-320d (Figure 9).
Figure 9.
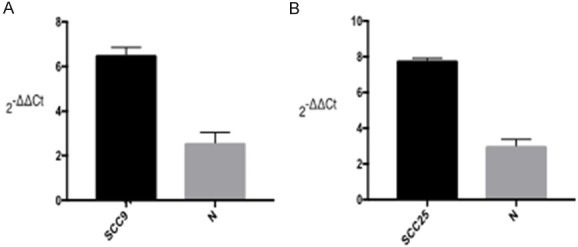
The differential expression of CCR7 in cells with and without miRNA-320.
Discussion
In the first part, in contrast to OSCC group with no metastasis, the higher expression of MALAT-1, SRSF1 [28] and CCR7, but the lower expression of miRNA-320d [32] in those samples with lymphatic metastasis were confirmed by real-time RT-PCR. This result is not only roughly consistent with our expectation, but also with the expression trend of these corresponding molecules in other cancers reported by recent research articles. In the second part, the interaction between MALAT-1, SRSF1 and miRNA-320d was investigated in the mature OSCC cell lines with the highest incidence rate, combined with co-immunoprecipitation method, immunoblotting assay and real-time RT-PCR. And their mutual combination and interaction in experimental group were determined by immuno-coloring WB band and quantitative statistical analysis in comparison with control group [33]. In the third part, the expression dynamics of the downstream chemokine receptor CCR7 [34] was measured in miRNA-320d silent group compared with that of control. This experiment used Ago 1/2/3 antibodies and the negative control mouse antibody to validate the silence of miRNA-320d by WB band. The real-time quantitative result showed that the expression of CCR7 in silent group was increased; indicating that miRNA-320d can affect the expression of CCR7.
The interaction between transmembrane proteins of OSCC cells and chemokine receptors may be a channel for long-chain non-coding RNA to regulate the biological behavior of chemokine receptors. The results of this study show that: (1) MALAT-1, SRSF1 and CCR7 have exceptionally high expression in samples of OSCC with lymphatic metastasis , and miRNA-320d expression is low in the same samples; (2) SRSF1 mediates mutual binding of MALAT-1 and miRNA-320d; (3) miRNA-320d can affect expression of the downstream chemokine CCR7. MALAT-1 interacts with miRNA-320d through SRSF1, where as gene-regulated silencing miRNA-320d can significantly promote the expression of CCR7, revealing the effect of MALAT-1 on chemokine receptor CCR7 in molecular level and possibly further effect on lymph node metastasis of OSCC. Unfortunately, because of small size sample and short cycle, no studies were carried out on the relative expression levels of MALAT-1, SRSF1, YB-1, miRNA-320d and CCR7 corresponding to TNM neoplasm staging in OSCC. Although the possible mutual effects among SRSF1, MiRNA-320d and CCR7 were found, but the specific mechanism is still unclear. In-depth study of the above studies may provide the theoretical basis for further targeted diagnosis and treatment of OSCC.
Disclosure of conflict of interest
None.
References
- 1.Obermajer N, Muthuswamy R, Odunsi K, Edwards RP, Kalinski P. PGE(2)-induced CXCL12 production and CXCR4 expression controls the accumulation of human MDSCs in ovarian cancer environment. Cancer Res. 2011;71:7463–7470. doi: 10.1158/0008-5472.CAN-11-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Ma YF, Liang T, Li CR, Li YJ, Jin S, Liu Y. Long non-coding RNA HNF1A-AS1 up-regulation in non-small cell lung cancer correlates to poor survival. Eur Rev Med Pharmacol Sci. 2016;20:4858–4863. [PubMed] [Google Scholar]
- 4.Liu Y, Sharma S, Watabe K. Roles of lncRNA in breast cancer. Front Biosci (Schol Ed) 2015;7:94–108. doi: 10.2741/s427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, Gu WL, Cai GX, Cai SJ. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7:3174–81. [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L, Bai HS, Deng Y, Fan L. High MALAT1 expression predicts a poor prognosis of cervical cancer and promotes cancer cell growth and invasion. Eur Rev Med Pharmacol Sci. 2015;19:3187–3193. [PubMed] [Google Scholar]
- 7.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–8. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 8.Lorenzen JM, Thum T. Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol. 2016;12:360–73. doi: 10.1038/nrneph.2016.51. [DOI] [PubMed] [Google Scholar]
- 9.Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W, Xu XK, Li JL, Kong KK, Li H, Chen C, He J, Wang F, Li P, Ge XS. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget. 2017;8:22783–22799. doi: 10.18632/oncotarget.15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Dan X, Li Y, Pan X, Zhang X, Kuang B, Ming Z, Li X, Wei X, Li G. The long noncoding RNA MALAT-1 is a novel biomarker in various cancers: a meta-analysis based on the GEO database and literature. J Cancer. 2016;7:991–1001. doi: 10.7150/jca.14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren Y, Wu Y, Mei M, Zhang L, Wang X. Long non coding RNA MALAT1 promotes tumor growth and metastasis by inducing epithelial-mesenchymal transition in oral squamous cell carcinoma. Sci Rep. 2015;5:15972. doi: 10.1038/srep15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhi F, Cao X, Xie X, Wang B, Dong W, Gu W, Ling Y, Wang R, Yang Y, Liu Y. Identification of circulating MicroRNAs as potential biomarkers for detecting acute myeloid leukemia. PLoS One. 2013;8:e56718. doi: 10.1371/journal.pone.0056718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Janiszewska J, Szaumkessel M, Kostrzewska-Poczekaj M, Bednarek K, Paczkowska J, Jackowska J, Grenman R, Szyfter K, Wierzbicka M, Giefing M. Global miRNA expression profiling identifies miR-1290 as novel potential oncomiR in laryngeal carcinoma. PLoS One. 2015;10:e0144924. doi: 10.1371/journal.pone.0144924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289–2294. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 19.Das S, Anczuków O, Akerman M, Krainer AR. Oncogenic splicing factor SRSF1 Is a critical transcriptional target of MYC. Cell Rep. 2012;1:110–7. doi: 10.1016/j.celrep.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schimanski CC, Schwald S, Simiantonaki N, Jayasinghe C, Gönner U, Wilsberg V, Junginger T, Berger MR, Galle PR, Moehler M. Effect of chemokine receptors CXCR4 and CCR7 on the metastatic behavior of human colorectal cancer. Clin Cancer Res. 2005;11:1743–1750. doi: 10.1158/1078-0432.CCR-04-1195. [DOI] [PubMed] [Google Scholar]
- 22.Rossi FW, Prevete N, Rivellese F, Lobasso A, Napolitano F, Granata F, Selleri C, Paulis AD. HIV-1 Nef promotes migration and chemokine synthesis of human basophils and mast cells through the interaction with CXCR4. Clin Mol Allergy. 2016;14:15. doi: 10.1186/s12948-016-0052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dolan MJ, Kulkarni H, Camargo JF, He W, Smith A, Anaya JM, Miura T, Hecht FM, Mamtani M, Pereyra F, Marconi V, Mangano A, Sen L, Bologna R, Clark RA, Anderson SA, Delmar J, O’Connell RJ, Lloyd A, Martin J, Ahuja SS, Agan BK, Walker BD, Deeks SG, Ahuja SK. Ccl3l1 and Ccr5 influence cell-mediated immunity and affect HIV-Aids pathogenesis via viral entry-independent mechanisms. Nat Immunol. 2007;8:1324–36. doi: 10.1038/ni1521. [DOI] [PubMed] [Google Scholar]
- 24.Zhang FR, Wang CL. The role of chemokine receptor CXCR7 in lung cancer. Clin Oncol Cancer Res. 2010;07:342–346. [Google Scholar]
- 25.Takayama Y, Aoki R, Uchida R, Tajima A, Aokiyoshida A. Role of CXC chemokine receptor type 4 as a lactoferrin receptor. Biochem Cell Biol. 2017;95:57–63. doi: 10.1139/bcb-2016-0039. [DOI] [PubMed] [Google Scholar]
- 26.Luker KE, Luker GD. Functions of CXCL12 and CXCR4 in breast cancer. Cancer Lett. 2006;238:30–41. doi: 10.1016/j.canlet.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 27.Sun X, Cheng G, Hao M, Zheng J, Zhou X, Zhang J, Taichman RS, Pienta KJ, Wang J. CXCL12/CXCR4/CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev. 2011;29:709–722. doi: 10.1007/s10555-010-9256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J. A splicing-independent function of SF2/ASF in MicroRNA processing. Mol Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adnan M, Morton G, Hadi S. Analysis of rpoS and bolA gene expression under various stress-induced environments in planktonic and biofilm phase using 2 (-Delta Delta CT) method. Mol Cell Biochem. 2011;357:275–282. doi: 10.1007/s11010-011-0898-y. [DOI] [PubMed] [Google Scholar]
- 30.Jedamzik B, Eckmann CR. Analysis of RNAprotein complexes by RNA coimmunoprecipitation and RT-PCR analysis from caenorhabditis elegans. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.prot5300. pdb.prot5300. [DOI] [PubMed] [Google Scholar]
- 31.Idris AI. Analysis of signalling pathways by western blotting and immunoprecipitation. Methods Mol Biol. 2012;816:223–232. doi: 10.1007/978-1-61779-415-5_15. [DOI] [PubMed] [Google Scholar]
- 32.Tian X, Chen Z, Shi S, Wang X, Wang W, Li N, Wang J. Clinical diagnostic implications of body fluid MiRNA in oral squamous cell carcinoma: a meta-analysis. Medicine. 2015;94:e1324. doi: 10.1097/MD.0000000000001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leucci E, Patella F, Waage J, Holmstrøm K, Lindow M, Bo P, Kauppinen S, Lund AH. MicroRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep. 2013;3:2535. doi: 10.1038/srep02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hui Y, Li Y, Jing Y, Feng JQ, Ding Y. miRNA-101 acts as a tumor suppressor in oral squamous cell carcinoma by targeting CX chemokine receptor 7. Am J Transl Res. 2016;8:4902–4911. [PMC free article] [PubMed] [Google Scholar]


