Abstract
Aim: Pulmonary tuberculosis (PTB) is an infectious disease with a high incidence worldwide. Previous genome-wide association studies have identified multiple susceptibility loci for pulmonary tuberculosis (PTB); however, validation of these findings is still needed. Methods: For this study, we recruited 300 subjects with PTB and 300 healthy subjects from a Tibetan population living in near or in Xi’an, China. Association analyses of single-nucleotide polymorphisms (SNPs) in TAP2 and SEC14L2 were performed with SPSS Statistics (version 17.0), SNPStats, Haploview (version 4.2), and SHEsis software. Results: We found a correction between one SNP (rs1061660) and PTB based on Chi-square or Fisher’s exact tests. In the allelic model analysis, the SNPs rs1061660 in SEC14L2 gene increased PTB 1.32-fold risk (OR = 1.32, CI = 1.05-1.66, P = 0.017). In the genetic model analysis, the rs3819721 in TAP2 gene was associated with increased 1.65-fold risk in the co-dominant model and 1.67-fold risk in the over-dominant model, respectively. For the rs1061660 in SEC14L2 gene, we found it was associated with a 1.49-fold increase the risk of PTB in the dominant model and a 1.37-fold increase the risk of PTB in the log-additive model, respectively. Conclusion: We found that two SNPs are associated with increased PTB risk in the Chinese Tibetan population.
Keywords: PTB, TAP2, SEC14L2, genetic polymorphisms, Tibetan population
Introduction
Pulmonary tuberculosis (PTB), as one of the most communicable diseases in humans, is caused by various strains of mycobacteria frequently Mycobacterium tuberculosis [1,2]. According to the World Health Organization (WHO) report, most of the estimated number of TB cases occurred in Asia (55%) and Africa (30%). PTB with 8 million new cases and 1.5 million deaths worldwide annually remains as a major global health problem [3]. The number of PTB patients in China is the second largest in the world, and China is ranked on a list of 22 high-burden countries. Though one third of human infected with Mycobacterium tuberculosis, only 10% of the infected persons develop the clinical disease [4]. Correlative evidence indicates that in addition to the environment, genetic factors play an important role in susceptibility to PTB [5-7].
Previous studies have identified genes that confer disease susceptibility by regulating the immune response [8,9]. we expect that identification of host genetic factors for PTB susceptibility might play a key role in PTB control worldwide. Genetic research has provided insight into tuberculosis, including the pathological and cytological bases of PTB. Several genes that influence PTB risk have been identified, including CCL2 [10], MIF [11], CD14 [12], TNF [13], IL18 [14], Vitamin D receptor gene [15], P2X7R [16], SP110 and PMP22 [17]. Most of genes contribute in immune response and their genetic polymorphisms may associate with PTB. Many studies showed that TAP2 and SEC14L2 gene are considered to be associated with a number of immune diseases, viral infection diseases, and even cancer [2,18-20]. Tuberculosis, as a kind of immune disease, its susceptibility may be associated with TAP2 and SEC14L2 gene.
Although genome-wide association studies (GWAS) found that some sites have relationships with PTB, but studies about the loci of TAP2 and SEC14L2 genes have little reported. Thus, in this study, we conducted a comprehensive association analysis between PTB and 6 susceptible SNPs in the TAP2 and SEC14L2 genes, to further clarify their potential roles in disease and reveal the association between common SNPs and PTB risk in the Tibetan Chinese population.
Materials and methods
Ethics statement
Our present study strictly observed the principles of the Declaration of Helsinki of the World Medical Association and was approved by the Ethics Committee of Tangdu Hospital affiliated with The Fourth Military Medical University in Xi’an. Informed consent forms were signed by all participants.
Study participants
Between October 2014 and September 2016, we selected 600 individuals in Tangdu Hospital and Xi’an Tuberculosis and Thoracic Tumor Hospital, including 300 PTB patients and 300 healthy controls. All PTB patients were ethnic of Tibetan and were newly diagnosed with consistent chest radiography and a positive sputum smear. Patients with human immunodeficiency virus (HIV), diabetes mellitus, or other tuberculosis diseases or who used immunosuppressive drugs were excluded. Individuals in the control group had no PTB history and no evidence of PTB in chest radiography or a positive sputum smear. We recruited subjects without consideration of age and gender.
SNP selection
We selected 6 SNPs with a minor allele frequency (MAF) above 5% in the HapMap Chinese Tibetan population. We selected these SNPs on the basis of their allele frequencies, location, and disease relevance through public HapMap databases (http://www.Hapmap.org/index.html.en).
Genotyping
Genomic DNA was extracted from peripheral blood samples using a genomic DNA purification kit (GoldMag, Xi’an, China). We used spectrometry (DU530 UV/VIS spectrophotometer, Beckman Instruments, Fullerton, CA) to measure the DNA concentration. The primers for amplification and extension reactions were designed with Sequenom MassARRAY Assay Design 3.0 Software (Sequenom, San Diego, CA) [21]. We used Sequenom MassARRAY RS1000 to perform the SNP genotyping with the agreement of the manufacturer [21], and we used Sequenom Typer 4.0 software for data management and analysis [21,22].
Statistical analysis
Microsoft Excel (Microsoft, Redmond, WA) and SPSS Statistics (version 17.0, SPSS, Chicago, IL) were used for statistical analyses. All p-values were two-tailed, and P ≤ 0.05 was considered to be statistically significant. SNP genotype frequencies in the case and control groups were calculated by Chi-square Test, and the Hardy-Weinberg equilibrium (HWE) was used to check the genotype frequency of the control group. Unconditional logistic regression analysis was used to examine the odds ratios (ORs) and 95% confidence intervals (CIs) in order to assess the association between SNPs and PTB [23]. Three models (dominant, recessive, log-additive) were used to test the association between SNPs and PTB [24]. Furthermore, Haploview (version 4.2, Broad Institute, Cambridge, MA) and SHEsis software (http://www.nhgg.org/analysis/) were used for checking the linkage disequilibrium structure.
Results
Characteristics of the participants
This study involved 600 subjects, including 300 patients (167 males and 133 females; age at diagnosis: 37.7±15.3 years) and 300 healthy controls (238 males, 61 females and 1 other; age: 22.8±1.6 years). There were statistical differences in age and sex distribution between the case and control groups (Table 1).
Table 1.
General characteristics the of this study population
| Variable | Cases (n = 300) | % | Controls (n = 300) | % | P-value |
|---|---|---|---|---|---|
| Sex | < 0.001 | ||||
| Male | 167 | 55.7% | 238 | 79.3% | |
| Female | 133 | 44.3% | 61 | 20.3% | |
| Age, yr (mean ± SD) | 37.7±15.3 | 22.8±1.6 | < 0.001 |
SNPs and the risk of PTB
Table 2 shows the basic information of candidate SNPs in our study, such as MAF, OR, 95% CI, position, HWE, chromosome position, alleles and the p-value of alleles. We used the chi-squared test to assess the influence of gene polymorphism of risk in the allele model, and found that rs1061660 was significantly associated with an increased risk of pulmonary tuberculosis (rs1061660, OR = 1.32, 95% CI = 1.05-1.659, P = 0.017). However, the other five SNPs (rs13501, rs241447, rs183585, rs3819721, rs1010324) had no significant association with PTB risk.
Table 2.
Allele frequencies in cases and controls and odds ratio estimates for PTB risk
| SNP | Gene (s) | Locus | Alleles A/B | MAF | HWE p value | OR | 95% CI | p-value | |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Case | Control | ||||||||
| rs13501 | TAP2 | 6p21.32 | A/G | 0.265 | 0.258 | 0.652 | 1.039 | 0.803-1.345 | 0.768 |
| rs241447 | TAP2 | 6p21.32 | A/G | 0.258 | 0.255 | 0.543 | 1.018 | 0.785-1.319 | 0.895 |
| rs183585 | TAP2 | 6p21.32 | C/T | 0.265 | 0.265 | 0.768 | 1.002 | 0.774-1.296 | 0.99 |
| rs3819721 | TAP2 | 6p21.32 | A/G | 0.233 | 0.192 | 1 | 1.284 | 0.972-1.684 | 0.078 |
| rs1010324 | SEC14L2 | 22q12.2 | A/G | 0.283 | 0.257 | 0.881 | 1.145 | 0.887-1.478 | 0.298 |
| rs1061660 | SEC14L2 | 22q12.2 | C/T | 0.475 | 0.407 | 0.122 | 1.32 | 1.05-1.659 | 0.017* |
SNP: single-nucleotide polymorphism; MAF: minor allele frequency; HWE: Hardy-Weinberg equilibrium; OR: odds ratio; CI: confidence interval.
P < 0.05 indicates statistical significance.
Associations between genotype frequencies and PTB
As shown in Table 3, we found the genotype “C/T” of rs1061660 in SEC14L2 was associated with a 1.49-fold increase the risk of PTB in the co-dominant model (OR = 1.64, 95% CI = 1.14-2.38, P = 0.02), 1.14-fold increase the risk of PTB in the dominant model (OR = 1.14, 95% CI = 1.16-2.23, P = 0.005) and 1.31-fold increase the risk of PTB in the Log-additive model (OR = 1.31, 95% CI = 1.05-1.64, P = 0.019), respectively. However, when adjusted by sex and age (Table 4), the rs3819721 in TAP2 was associated with increased 1.67-fold risk of PTB in the over-dominant model (OR = 1.67, 95% CI = 1.07-2.62, P = 0.023). For rs1061660 in SEC14L2, we found it was associated with increased 1.49-fold risk of PTB in the dominant model (OR = 1.74, 95% CI = 1.10-2.75, P = 0.017) and a 1.37-fold increase the risk of PTB in the log-additive model (OR = 1.02, 95% CI = 1.01-1.86, P = 0.038), respectively.
Table 3.
Single SNP association with PTB (logistic regression, crude)
| SNP | Model | Genotype | Control | Case | OR (95% CI) | P-value | AIC | BIC |
|---|---|---|---|---|---|---|---|---|
| rs3819721 | Codominant | G/G | 196 (65.3%) | 176 (58.7%) | 1 | 0.21 | 834.6 | 847.8 |
| A/G | 93 (31%) | 108 (36%) | 1.29 (0.92-1.82) | |||||
| A/A | 11 (3.7%) | 16 (5.3%) | 1.62 (0.73-3.58) | |||||
| Dominant | G/G | 196 (65.3%) | 176 (58.7%) | 1 | 0.092 | 832.9 | 841.7 | |
| A/G-A/A | 104 (34.7%) | 124 (41.3%) | 1.33 (0.95-1.85) | |||||
| Recessive | G/G-A/G | 289 (96.3%) | 284 (94.7%) | 1 | 0.32 | 834.8 | 843.6 | |
| A/A | 11 (3.7%) | 16 (5.3%) | 1.48 (0.68-3.24) | |||||
| Overdominant | G/G-A/A | 207 (69%) | 192 (64%) | 1 | 0.19 | 834.1 | 842.9 | |
| A/G | 93 (31%) | 108 (36%) | 1.25 (0.89-1.76) | |||||
| Log-additive | --- | --- | --- | 1.28 (0.97-1.70) | 0.077 | 832.7 | 841.4 | |
| rs1061660 | Codominant | C/C | 112 (37.3%) | 80 (26.7%) | 1 | 0.02* | 829.9 | 843.1 |
| C/T | 132 (44%) | 155 (51.7%) | 1.64 (1.14-2.38) | |||||
| T/T | 56 (18.7%) | 65 (21.7%) | 1.63 (1.03-2.57) | |||||
| Dominant | C/C | 112 (37.3%) | 80 (26.7%) | 1 | 0.005* | 827.9 | 836.7 | |
| C/T-T/T | 188 (62.7%) | 220 (73.3%) | 1.64 (1.16-2.32) | |||||
| Recessive | C/C-C/T | 244 (81.3%) | 235 (78.3%) | 1 | 0.36 | 834.9 | 843.7 | |
| T/T | 56 (18.7%) | 65 (21.7%) | 1.21 (0.81-1.80) | |||||
| Overdominant | C/C-T/T | 168 (56%) | 145 (48.3%) | 1 | 0.06 | 832.2 | 841 | |
| C/T | 132 (44%) | 155 (51.7%) | 1.36 (0.99-1.88) | |||||
| Log-additive | --- | --- | --- | 1.31 (1.05-1.64) | 0.019* | 830.2 | 839 |
Abbreviations: OR, odds ratio; CI, confidence interval.
P < 0.05 indicates statistical significance.
Table 4.
Single SNP association with PTB (logistic regression, adjusted by sex, age)
| SNP | Model | Genotype | Control | Case | OR (95% CI) | P-value | AIC | BIC |
|---|---|---|---|---|---|---|---|---|
| rs3819721 | Codominant | G/G | 196 (65.3%) | 176 (58.7%) | 1 | 0.0069* | 538.3 | 564.7 |
| A/G | 93 (31%) | 108 (36%) | 1.65 (1.05-2.59) | |||||
| A/A | 11 (3.7%) | 16 (5.3%) | 0.78 (0.25-2.40) | |||||
| Dominant | G/G | 196 (65.3%) | 176 (58.7%) | 1 | 0.056 | 538 | 560 | |
| A/G-A/A | 104 (34.7%) | 124 (41.3%) | 1.53 (0.99-2.37) | |||||
| Recessive | G/G-A/G | 289 (96.3%) | 284 (94.7%) | 1 | 0.45 | 541.1 | 563.1 | |
| A/A | 11 (3.7%) | 16 (5.3%) | 0.65 (0.22-1.98) | |||||
| Overdominant | G/G-A/A | 207 (69%) | 192 (64%) | 1 | 0.023* | 536.5 | 558.5 | |
| A/G | 93 (31%) | 108 (36%) | 1.67 (1.07-2.62) | |||||
| Log-additive | --- | --- | --- | 1.30 (0.90-1.88) | 0.17 | 539.8 | 561.7 | |
| rs1061660 | Codominant | C/C | 112 (37.3%) | 80 (26.7%) | 1 | 0.058 | 538 | 564.3 |
| C/T | 132 (44%) | 155 (51.7%) | 1.72 (1.06-2.79) | |||||
| T/T | 56 (18.7%) | 65 (21.7%) | 1.77 (0.96-3.29) | |||||
| Dominant | C/C | 112 (37.3%) | 80 (26.7%) | 1 | 0.017* | 536 | 558 | |
| C/T-T/T | 188 (62.7%) | 220 (73.3%) | 1.74 (1.10-2.75) | |||||
| Recessive | C/C-C/T | 244 (81.3%) | 235 (78.3%) | 1 | 0.38 | 540.9 | 562.9 | |
| T/T | 56 (18.7%) | 65 (21.7%) | 1.28 (0.74-2.19) | |||||
| Overdominant | C/C-T/T | 168 (56%) | 145 (48.3%) | 1 | 0.12 | 539.2 | 561.2 | |
| C/T | 132 (44%) | 155 (51.7%) | 1.40 (0.92-2.13) | |||||
| Log-additive | --- | --- | --- | 1.37 (1.02-1.86) | 0.038* | 537.3 | 559.3 |
Abbreviations: OR, odds ratio; CI, confidence interval.
P < 0.05 indicates statistical significance.
Associations between haplotype analyses and PTB risk
LD and haplotype analyses of the SNPs in the case and control samples were further studied. In order to assess the association between haplotypes and PTB risk, a Wald test was performed using an unconditional multivariate regression analysis. Although some candidate SNPs have showed strong linkage, but no positive result were observed (Figure 1). The result for the haplotype was not found to be associated with a risk of PTB, because the P value has no statistical difference (Table 5).
Figure 1.
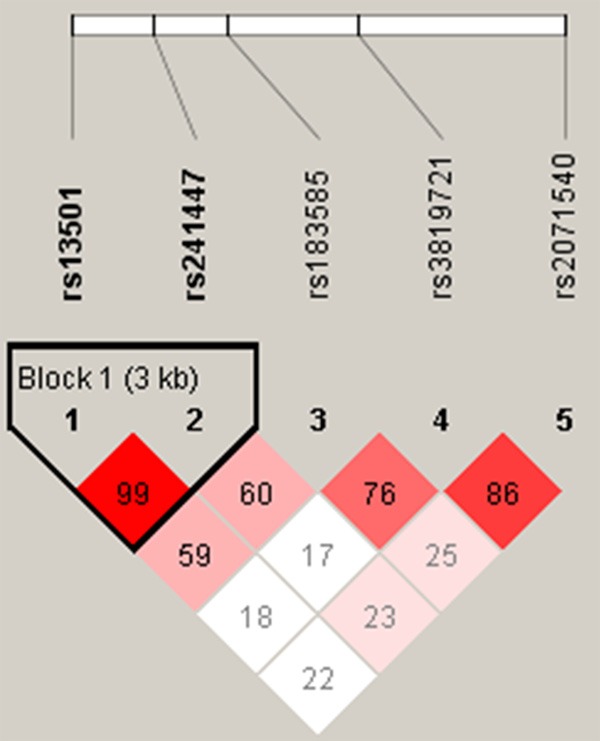
Linkage disequilibrium (LD) plots containing four SNPs from TAP2 gene.
Table 5.
Haplotype analysis results in this study
| Haplotypes | Crude analysis | Adjusted by Gender and Age | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| rs13501 | rs241447 | Freq | OR (95% CI) | P-value | OR (95% CI) | P a-value | |
| 1 | G | T | 0.7375 | 1 | --- | 1 | --- |
| 2 | A | C | 0.255 | 1.02 (0.79-1.33) | 0.86 | 1.03 (0.73-1.45) | 0.88 |
| Rare | * | * | 0.0075 | 1.80 (0.49-6.52) | 0.37 | 5.23 (0.90-30.24) | 0.065 |
Abbreviations: CI = confidence interval; OR = odds ratio; SNP = single nucleotide polymorphism.
P: Adjusted by gender and age.
Discussion
In this study, we investigated the association between TAP2 and SEC14L2 polymorphisms and PTB risk in a Tibetan Chinese population. We found two SNPs were significantly increased the PTB risk. Previous PTB studies investigated the function of host genetic factors and immunoreaction in M. tuberculosis infection [25]. In humans, macrophages are the main host cells for the intracellular replication pathway of M. tuberculosis. Macrophages also serve as the antigen-presenting cells during the reactivation of lymphocytes, and they function as a vital killer of mycobacteria [26]. In vivo and in vitro studies show that M. tuberculosis infection causes apoptosis in monocytes and macrophages [27]. and we previously found that apoptosis of these two cell types is a protective factor in human tuberculosis [28].
TAP2 gene, located in chromosome 6p21.32, plays an important role in antigen presentation on MHC class 1 molecule [29]. TAP2 gene polymorphisms can influence the antigen peptide selection and transport process and modify immune response regulation [30]. Polymorphic residues in TAP2 genes were identified which modify specificity of substrate transport [31]. Consequently, these genes are considered as candidate genes for susceptibility to a number of immune diseases, viral infection diseases, and even cancer [30,32-34]. Study has reported the associations between polymorphisms of TAP genes and susceptibility to PTB in a sample of Iranian [35]. In present study, we focused on the association of TAP2 gene polymorphisms with PTB susceptibility in a cohort of Tibetan Chinese population. We found a significant association between TAP2 rs3819721 A/G variant and PTB.
SEC14L2, as an immunohistochemical related-protein, regulating the immune response and mediate inflammatory cells, which is associated with immune disease and cancer, for example, SEC14L2 can promote the hepatitis C virus (HCV) [36] and prostate cancer [37]. SEC14L2, also known as c22orf6, supernatant protein factor 1 (SPF1), or tocopherol-associated protein 1 (TAP1), is a cytosolic lipid-binding protein family member [38,39], which is ubiquitously expressed in human tissues [40]. Study shows that TAP/SEC14L2 had a high expression in normal/benign breast, prostate, and liver tissues as compared to lung, colon, and kidney [20]. In our case-control study, we found that rs1061660, as the intronic SNP within the gene SEC14L2, were markedly associated with PTB risk according to both genotype and allele association analysis in a Tibetan Chinese population.
To our knowledge, our study is the first to investigate the association between polymorphic SNPs of TAP2 and SEC14L2 gene and PTB risk in a Tibetan Chinese population, which may provide new data to facilitate earlier diagnosis and promote early prevention and shed light on the new candidate genes and new ideas for the study of subsequent occurrence mechanism of PTB. Therefore, more studies should investigate these SNPs using more clinical data with bigger samples. In conclusion, our results show that TAP2 polymorphisms rs3819721 and SEC14L2 polymorphisms rs1061660 are associated with increased PTB risk in the Tibetan population.
Acknowledgements
This work was supported by National Natural Science Foundation of China (No. 81260240). We are grateful to the patients and control subjects for their participation in this study. We also thank the clinicians and hospital staff who obtained the blood samples and performed data collection for this study.
Disclosure of conflict of interest
None.
References
- 1.Alqumber MA, Mandal RK, Haque S, Panda AK, Akhter N, Ali A. A genetic association study of CCL5 -28 C>G (rs2280788) polymorphism with risk of tuberculosis: a meta-analysis. PLoS One. 2013;8:e83422. doi: 10.1371/journal.pone.0083422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu MC, Huang CM, Wu MC, Wu JY, Tsai FJ. Association of TAP2 gene polymorphisms in Chinese patients with rheumatoid arthritis. Clin Rheumatol. 2004;23:35–39. doi: 10.1007/s10067-003-0769-3. [DOI] [PubMed] [Google Scholar]
- 3.Millet JP, Moreno A, Fina L, del Bano L, Orcau A, de Olalla PG, Cayla JA. Factors that influence current tuberculosis epidemiology. Eur Spine J. 2013;22(Suppl 4):539–548. doi: 10.1007/s00586-012-2334-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cobat A, Orlova M, Barrera LF, Schurr E. Host genomics and control of tuberculosis infection. Public Health Genomics. 2013;16:44–49. doi: 10.1159/000341499. [DOI] [PubMed] [Google Scholar]
- 5.Bahari G, Hashemi M, Taheri M, Naderi M, Eskandari-Nasab E, Atabaki M. Association of IRGM polymorphisms and susceptibility to pulmonary tuberculosis in Zahedan, Southeast Iran. ScientificWorldJournal. 2012;2012:950801. doi: 10.1100/2012/950801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashemi M, Eskandari-Nasab E, Moazeni-Roodi A, Naderi M, Sharifi-Mood B, Taheri M. Association of CTSZ rs34069356 and MC3R rs6127698 gene polymorphisms with pulmonary tuberculosis. Int J Tuberc Lung Dis. 2013;17:1224–1228. doi: 10.5588/ijtld.12.0762. [DOI] [PubMed] [Google Scholar]
- 7.Naderi M, Hashemi M, Pourmontaseri Z, Eskandari-Nasab E, Bahari G, Taheri M. TIRAP rs8177374 gene polymorphism increased the risk of pulmonary tuberculosis in Zahedan, southeast Iran. Asian Pac J Trop Med. 2014;7:451–455. doi: 10.1016/S1995-7645(14)60073-0. [DOI] [PubMed] [Google Scholar]
- 8.Lykouras D, Sampsonas F, Kaparianos A, Karkoulias K, Tsoukalas G, Spiropoulos K. Human genes in TB infection: their role in immune response. Monaldi Arch Chest Dis. 2008;69:24–31. doi: 10.4081/monaldi.2008.408. [DOI] [PubMed] [Google Scholar]
- 9.Britton WJ, Fernando SL, Saunders BM, Sluyter R, Wiley JS. The genetic control of susceptibility to mycobacterium tuberculosis. Novartis Found Symp. 2007;281:79–89. doi: 10.1002/9780470062128.ch8. discussion 89-92, 208-209. [DOI] [PubMed] [Google Scholar]
- 10.Feng WX, Mokrousov I, Wang BB, Nelson H, Jiao WW, Wang J, Sun L, Zhou SR, Xiao J, Gu Y, Wu XR, Ma X, Shen A. Tag SNP polymorphism of CCL2 and its role in clinical tuberculosis in Han Chinese pediatric population. PLoS One. 2011;6:e14652. doi: 10.1371/journal.pone.0014652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashemi M, Sharifi-Mood B, Rasouli A, Amininia S, Naderi M, Taheri M. Macrophage migration inhibitory factor -173 G/C polymorphism is associated with an increased risk of pulmonary tuberculosis in Zahedan, Southeast Iran. Excli J. 2015;14:117–122. doi: 10.17179/excli2014-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miao R, Ge H, Xu L, Xu F. CD14 -159C/T polymorphism contributes to the susceptibility to tuberculosis: evidence from pooled 1,700 cases and 1,816 controls. Mol Biol Rep. 2014;41:3481–3486. doi: 10.1007/s11033-014-3210-x. [DOI] [PubMed] [Google Scholar]
- 13.Mabunda N, Alvarado-Arnez LE, Vubil A, Mariamo A, Pacheco AG, Jani IV, Moraes MO. Gene polymorphisms in patients with pulmonary tuberculosis from Mozambique. Mol Biol Rep. 2015;42:71–76. doi: 10.1007/s11033-014-3741-1. [DOI] [PubMed] [Google Scholar]
- 14.Retraction note to: IL-18 genetic polymorphisms may contribute to the pathogenesis of tuberculosis among Asians: a meta-analysis of case-control studies. Mol Biol Rep. 2015;42:1473. doi: 10.1007/s11033-015-3900-z. [DOI] [PubMed] [Google Scholar]
- 15.Hu Q, Chen Z, Liang G, Mo F, Zhang H, Xu S, Wang Y, Kang L, Jin T. Vitamin D receptor gene associations with pulmonary tuberculosis in a Tibetan Chinese population. BMC Infect Dis. 2016;16:469. doi: 10.1186/s12879-016-1699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu X, Guo W, Ren G, He X, Hu Q, Zhang Y, Kang L, Yuan D, Jin T. P2X7R gene polymorphisms are associated with increased risk of pulmonary tuberculosis in the Tibetan Chinese population. Am J Trop Med Hyg. 2016;95:1016–1020. doi: 10.4269/ajtmh.16-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren G, You J, Gong X, Zhang X, Zhao L, Wei X, Jin T, Chen M. SP110 and PMP22 polymorphisms are associated with tuberculosis risk in a Chinese-Tibetan population. Oncotarget. 2016;7:66100–66108. doi: 10.18632/oncotarget.11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Correa PA, Molina JF, Pinto LF, Arcos-Burgos M, Herrera M, Anaya JM. TAP1 and TAP2 polymorphisms analysis in northwestern Colombian patients with systemic lupus erythematosus. Ann Rheum Dis. 2003;62:363–365. doi: 10.1136/ard.62.4.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozbas-Gerceker F, Bozman N, Gezici S, Pehlivan M, Yilmaz M, Pehlivan S, Oguzkan-Balci S. Association of TAP1 and TAP2 gene polymorphisms with hematological malignancies. Asian Pac J Cancer Prev. 2013;14:5213–5217. doi: 10.7314/apjcp.2013.14.9.5213. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Ni J, Hsu CL, Johnykutty S, Tang P, Ho YS, Lee CH, Yeh S. Reduced expression of tocopherol-associated protein (TAP/Sec14L2) in human breast cancer. Cancer Invest. 2009;27:971–977. doi: 10.3109/07357900802392659. [DOI] [PubMed] [Google Scholar]
- 21.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009 doi: 10.1002/0471142905.hg0212s60. Chapter 2: Unit 2.12. [DOI] [PubMed] [Google Scholar]
- 22.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, Wang M, Feng W, Zander T, MacConaill L, Lee JC, Nicoletti R, Hatton C, Goyette M, Girard L, Majmudar K, Ziaugra L, Wong KK, Gabriel S, Beroukhim R, Peyton M, Barretina J, Dutt A, Emery C, Greulich H, Shah K, Sasaki H, Gazdar A, Minna J, Armstrong SA, Mellinghoff IK, Hodi FS, Dranoff G, Mischel PS, Cloughesy TF, Nelson SF, Liau LM, Mertz K, Rubin MA, Moch H, Loda M, Catalona W, Fletcher J, Signoretti S, Kaye F, Anderson KC, Demetri GD, Dummer R, Wagner S, Herlyn M, Sellers WR, Meyerson M, Garraway LA. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 23.Bland JM, Altman DG. Statistics notes. The odds ratio. BMJ. 2000;320:1468. doi: 10.1136/bmj.320.7247.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 25.Kato-Maeda M, Bifani PJ, Kreiswirth BN, Small PM. The nature and consequence of genetic variability within mycobacterium tuberculosis. J Clin Invest. 2001;107:533–537. doi: 10.1172/JCI11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chackerian AA, Perera TV, Behar SM. Gamma interferon-producing CD4+ T lymphocytes in the lung correlate with resistance to infection with mycobacterium tuberculosis. Infect Immun. 2001;69:2666–2674. doi: 10.1128/IAI.69.4.2666-2674.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keane J, Balcewicz-Sablinska MK, Remold HG, Chupp GL, Meek BB, Fenton MJ, Kornfeld H. Infection by mycobacterium tuberculosis promotes human alveolar macrophage apoptosis. Infect Immun. 1997;65:298–304. doi: 10.1128/iai.65.1.298-304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mancino G, Placido R, Di Virgilio F. P2X7 receptors and apoptosis in tuberculosis infection. J Biol Regul Homeost Agents. 2001;15:286–293. [PubMed] [Google Scholar]
- 29.Monaco JJ. A molecular model of MHC class-Irestricted antigen processing. Immunol Today. 1992;13:173–179. doi: 10.1016/0167-5699(92)90122-N. [DOI] [PubMed] [Google Scholar]
- 30.Sunder SR, Hanumanth SR, Gaddam S, Jonnalagada S, Valluri VL. Association of TAP 1 and 2 gene polymorphisms with human immunodeficiency virus-tuberculosis co-infection. Hum Immunol. 2011;72:908–911. doi: 10.1016/j.humimm.2011.07.304. [DOI] [PubMed] [Google Scholar]
- 31.Powis SH, Tonks S, Mockridge I, Kelly AP, Bodmer JG, Trowsdale J. Alleles and haplotypes of the MHC-encoded ABC transporters TAP1 and TAP2. Immunogenetics. 1993;37:373–380. doi: 10.1007/BF00216802. [DOI] [PubMed] [Google Scholar]
- 32.Ismail A, Bousaffara R, Kaziz J, Zili J, el Kamel A, Tahar Sfar M, Remadi S, Chouchane L. Polymorphism in transporter antigen peptides gene (TAP1) associated with atopy in Tunisians. J Allergy Clin Immunol. 1997;99:216–223. doi: 10.1016/s0091-6749(97)70099-x. [DOI] [PubMed] [Google Scholar]
- 33.Rajalingam R, Singal DP, Mehra NK. Transporter associated with antigen-processing (TAP) genes and susceptibility to tuberculoid leprosy and pulmonary tuberculosis. Tissue Antigens. 1997;49:168–172. doi: 10.1111/j.1399-0039.1997.tb02731.x. [DOI] [PubMed] [Google Scholar]
- 34.Cucca F, Congia M, Trowsdale J, Powis SH. Insulin-dependent diabetes mellitus and the major histocompatibility complex peptide transporters TAP1 and TAP2: no association in a population with a high disease incidence. Tissue Antigens. 1994;44:234–240. doi: 10.1111/j.1399-0039.1994.tb02388.x. [DOI] [PubMed] [Google Scholar]
- 35.Naderi M, Hashemi M, Amininia S. Association of TAP1 and TAP2 gene polymorphisms with susceptibility to pulmonary tuberculosis. Iran J Allergy Asthma Immunol. 2016;15:62–68. [PubMed] [Google Scholar]
- 36.Saeed M, Andreo U, Chung HY, Espiritu C, Branch AD, Silva JM, Rice CM. SEC14L2 enables pan-genotype HCV replication in cell culture. Nature. 2015;524:471–475. doi: 10.1038/nature14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zingg JM, Azzi A. Comment re: vitamin E transport gene variants and prostate cancer. Cancer Res. 2009;69:6756. doi: 10.1158/0008-5472.CAN-09-0535. author reply 6756. [DOI] [PubMed] [Google Scholar]
- 38.Allen-Baume V, Segui B, Cockcroft S. Current thoughts on the phosphatidylinositol transfer protein family. FEBS Lett. 2002;531:74–80. doi: 10.1016/s0014-5793(02)03412-9. [DOI] [PubMed] [Google Scholar]
- 39.Aravind L, Neuwald AF, Ponting CP. Sec14plike domains in NF1 and Dbl-like proteins indicate lipid regulation of Ras and Rho signaling. Curr Biol. 1999;9:R195–197. doi: 10.1016/s0960-9822(99)80127-4. [DOI] [PubMed] [Google Scholar]
- 40.Kempna P, Zingg JM, Ricciarelli R, Hierl M, Saxena S, Azzi A. Cloning of novel human SEC14p-like proteins: ligand binding and functional properties. Free Radic Biol Med. 2003;34:1458–1472. doi: 10.1016/s0891-5849(03)00173-4. [DOI] [PubMed] [Google Scholar]


