Abstract
Chordoma is a rare, locally aggressive neoplasm of bone, usually with poor prognosis. The treatment for chordoma has been unsatisfactory for decades. MiRNAs were recently introduced into this field and provided new insights to the pathogenesis and pathophysiology of chordoma. However, molecular basis of chordoma remains ambiguous up to now. This research aims to discover novel miRNA molecules as potential biomarkers and therapeutic targets. We measured the expression of miRNA-1290 in chordoma tissues and fetal nucleus pulposus tissues by quantitative real-time PCR. Further, we analyzed its association with the clinical features as well as the prognosis of patients. The expression of miRNA-1290 in chordoma samples was significantly lower than fetal nucleus pulposus samples (P=0.026). Low expression of miRNA-1290 contributed to tumor invasion into surrounding muscle (P=0.013), while no obvious significance was identified between miRNA-1290 expression and patients’ age, gender, tumor location and size (P>0.05). Log-rank test showed that low-level miRNA-1290 expression had a prominent impact on the patients’ RFS (P=0.004). Conclusively, miRNA-1290 might be a valuable prognostic biomarker and efficient therapeutic target for sacral chordoma.
Keywords: Chordoma, nucleus pulposus, microRNA, invasion, recurrence, prognosis
Introduction
Chordoma is a primary malignant bone tumor arising from the embryonic notochordal remnants, which often occurs at axial skeleton, including sacral region, spinal and skull base [1]. The overall age-adjusted incidence rate of chordoma is 0.089 per 100,000 annually [2], while the sacral chordoma occurs in 0.03 out of every 100,000 people per year [3]. The sex distribution reflects an obvious differentiation that it is approximately 2 times more common in male than female [4]. The survival rate of 5 years is 65% and it drops to 35% when it comes to a decade [5]. Chordoma is highly recurrent, with an appalling rate ranging from 44% to 78% [6-10], making itself prone to malignancy in terms of clinical progression and outcome. Surgical resection is recommended as the most effective treatment. However, it is quite difficult to receive complete surgical resection due to the complexity of anatomy. Besides, even though high-dose radiotherapy achieves reasonable disease control in most cases, there still lacks sufficient chemotherapy for subsequent adjuvant treatment, which contributes to the poor prognosis due to the high risk of recurrence [11]. Up to now, few biomarkers has been explored to identify the clinical features and characteristics of chordoma. Therefore, finding biomarkers for target therapy and prognosis indication is of great significance.
MicroRNAs are small noncoding RNAs of about 22 nucleotides in length. They were usually considered as noises in gene regulators. However, as the evidences have been accumulating, these microRNAs are now thought to function as antisense regulators of other RNAs [12]. Previous studies have shown that plenty of miRNAs have different expression in a variety of cancers and the irregular level of them closely correlates with tumor characteristics [13-15]. Yet, little has been studied about the abnormally expressed miRNAs in chordoma. Our previous research showed that the miRNA-1290 had great potential as a regulator in chordma microenvironment [16]. More importantly, no one has ever reported its association with invasion and recurrence of chordoma. Hence, in this article, we explored the differential expression of miRNA-1290 between sacral chordoma and nucleus pulposus and furtherly evaluated its correlation with clinical features as well as prognosis.
Materials and methods
Patients and samples
Our study enrolled 28 patients (20 males and 8 females) from Jan. 2005 to Jun. 2015 at the First Affiliated Hospital of Soochow University (Suzhou, China), with their average age of 56.4 years (range 18-84 years) when they were admitted to hospital. All the patients received primary surgical treatment and have received no adjuvant therapy preoperatively. Meanwhile, to settle the control group, 10 nucleus pulposus tissues were collected from aborted fetus at obstetric out-patient clinic of our hospital, with average age of 20.6 months (range 12-28 months). The samples were verified by means of histopathology (the representative histological figure of chordoma is shown as Figure 1) and preserved in liquid nitrogen. All the patients or families are informed of standard written consent. Our study protocol was approved by the Ethics Review Committee of the Institutional Review Board of the hospital.
Figure 1.
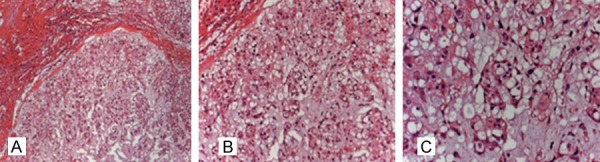
HE staining of chordoma tissue (A-C: ×100, ×200, ×300).
Total RNA extraction and quantitative real-time PCR
Total RNA was extracted from 0.1 cm3 frozen tissue with TRIzol reagent (TAKARA BIO INC., Shiga, JAPAN). cDNA was reverse transcribed from total RNA samples by RT primers of TaqMan MicroRNA Assays (Applied Biosystems, Foster City, USA) and MicroRNA reverse transcription Kit. TaqMan Small RNA Assay was equipped to operate the amplified reaction according to the manufacturer’s instructions (Applied Biosystems, Foster City, USA). U6 was used as internal control. The primers sequences were as follows: miR-1290, forward 5’-CAGTGCTGGATTTTTGGAT-3’, reverse 5’-TATGGTTGTTCACGACTCCTTCAC-3’; U6, forward 5’-ATTGGAACGATACAGAGAAGATT-3’, reverse 5’-GGAACGCTTCACGAATTTG-3’. Each operation was repeated for three times. The CT value for miRNA-1290 was normalized to U6 snRNA and the relative expression of miRNAs was calculated by the 2-ΔΔCt method. Patients were divided into two groups depending on the average of CT value.
Clinical data collection and follow-up
All the clinical data including age and gender of the patients as well as the size and location of tumors were collected. Largest diameter recorded in millimeters was used to represent the tumor size. As to tumor location, tumors located above S3 were regarded as one group while tumors located cephalad to S3 was considered as the other. Magnetic resonance imaging (MRI) was performed before surgery to assess tumor invasions into surrounding muscles. For all patients who have undergone surgery, plain radiographs, computed tomography scans (CT) and MRI are acquired every 3-6 months. Then assessment of which evaluated the tumor recurrence and postoperative life condition was made by imaging examination along with clinical manifestations. Recurrence free survival time (RFS) is defined as a time period dating from the primary surgery to tumor recurrence. All patients were followed up well for over 24 months. Disease free survival at the end of follow-up or lost to follow-up was regarded as censored date. Two typical cases were attached as Figures 2 and 3 displayed.
Figure 2.
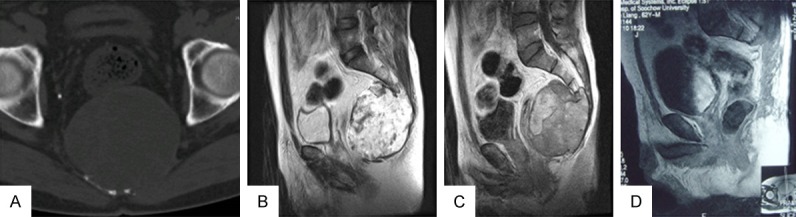
60-year-old male patient with high expression of miRNA-1290, preoperative CT and MRI showed no invasions into surrounding muscles (A-C); 3-year follow-up revealed no tumor recurrence (D).
Figure 3.
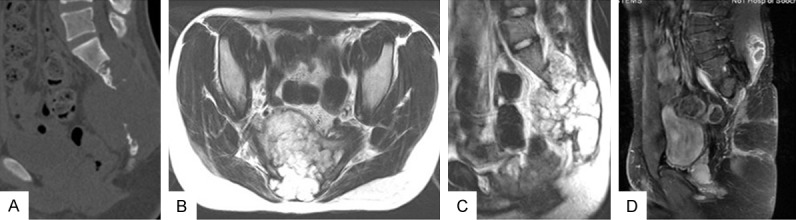
21-year-old male patient with down-regulated miRNA-1290, preoperative CT and MRI manifested obvious invasions (A-C); MRI took 5 years after the surgery showed that chordoma had recurred (D).
Statistical analysis
Statistical analysis was performed by SPSS 22.0 software (SPSS Inc., Chicago, IL). All the measurement data was is expressed by mean ± standard deviation (x̅ ± s). Shapiro-Wilk test was used to classify the measurement data into normal or abnormal distribution, then student’s t-test or Mann-Whitney U test will be furtherly adopted respectively, along with chi-square analysis to evaluate the correlation between miRNA-1290 expression and clinical features of patients. Kaplan-Meier survival curves and log-rank test were used to assess the association between miRNA-1290 expression and RFS. P<0.05 was considered statistically significant.
Results
The expression of miRNA-1290 in chordoma samples was significantly lower than fetal nucleus pulposus samples (P=0.026; Figure 4). The overall rate of tumor invasion was 60.7% (17/28). Notably, 77.8% (14/18) patients who had low miRNA-1290 expression occurred invasion while the rate in the opposite group was only 30% (3/10). Moreover, Chi-square analysis revealed that the low expression of miRNA-1290 contributes to tumor invasion (P=0.013; Table 1). In addition, no obvious significance was identified between miRNA-1290 expression and patients’ age, gender, tumor location and size.
Figure 4.
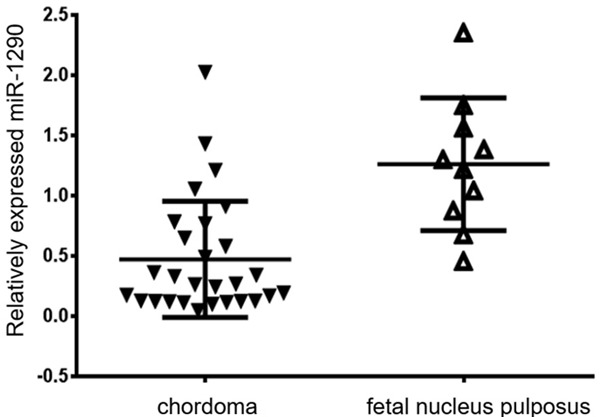
Relatively expressed miRNA-1290 in chordoma and fetal nucleus pluposus.
Table 1.
Association of miRNA-1290 expression with clinical parameters in sacral chordoma
| Parameters | miRNA-1290 expression | P value | |
|---|---|---|---|
|
| |||
| High (n=10) | Low (n=18) | ||
| Age (years) | |||
| <50 | 5 | 4 | 0.132 |
| ≥50 | 5 | 14 | |
| Gender | |||
| Male | 6 | 14 | 0.318 |
| Female | 4 | 4 | |
| Tumor location | |||
| Above S3 | 7 | 12 | 0.856 |
| S3 and below | 3 | 6 | |
| Tumor size (mm) | |||
| <90 | 5 | 9 | 1.000 |
| ≥90 | 5 | 9 | |
| Surrounding muscle invasion | |||
| Yes | 3 | 14 | 0.013* |
| No | 7 | 4 | |
| Recurrence | |||
| Yes | 2 | 14 | 0.003* |
| No | 8 | 4 | |
P<0.05.
Over an average span of 77.5±18.52 months (ranged from 25 to 139 months), satisfactory follow-up was achieved in all patients. The total recurrence rate was 57.1%, for local recurrence happened in 16 of the 28 patients. The median relapse time was 31.4±6.73 months. Within all the patients with low miRNA-1290 expression, 77.8% (14/18) had a tumor recurrence. However, only 20% (2/10) patients who expressed low miRNA-1290 developed local recurrence, which significantly lower than the former (P=0.03; Table 1). Kaplan-Meier survival curve and log-rank test showed that RFS (median 46.1±7.20 months) in low miRNA-1290 expression group was significantly shorter than RFS (median 115.9±14.61 months) in high miRNA-1290 expression group (F=8.101, P=0.004; Figure 5).
Figure 5.
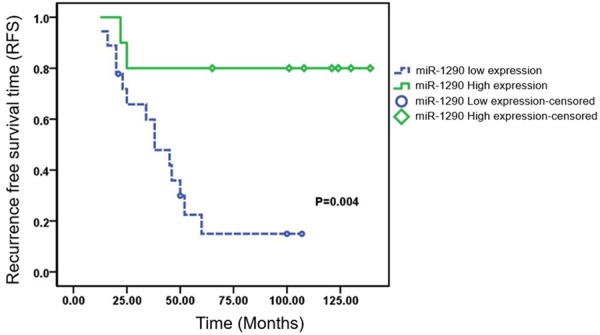
Recurrence free survival time according to the expression of miRNA-1290 in sacral chordoma.
Discussion
Sacral chordoma is a rare bone neoplasm characterized by strong invasiveness and high probability of recurrence [17]. The tumor is resistant to chemotherapy; consequently, surgery is the mainly recommended therapy [11]. However, despite the huge challenge for surgeons to perform complete resection, the mass still tends to recur even operates an en bloc resection [18]. This provokes us to discover relevant biomarkers associated with the clinical features of chordoma. Recently, miRNA has been well studied and there are countless inspiring findings that a variety of miRNAs may play an important role in tumor pathophysiology, including invasion, recurrence, proliferation, metastasis [19-21]. Our previous study has demonstrated the different expression of miRNAs between chormdoma and nucleus pulposus with miRNA microarray profiling and revealed that miRNA-1290 is the most down-regulated miRNA within the chromosome 1 [16]. It is worth mentioning that Chromosome 1 gene deletion or amplification is the most common chromosome variation in chordoma [22-24]. In this study, the different expression of miRNA-1290 was confirmed and clinical features as well as prognosis of chordoma were analyzed.
Chordoma shares favorable homology with fetal nucleus pulposus due to the fact that chordoma originates from notochord remnants, which shows great advantages over adjacent normal tissues. We detected the expression of miRNA-1290 in chordoma tissues and nucleus pulposus tissues. The result demonstrated that the expression of miRNA-1290 in chordoma was significantly lower than in fetal nucleus pulposus (P=0.026). Consistent with our result, Endo et al. reported that miRNA-1290 was down-regulated in ERhigh Ki67low breast cancer cohorts [25]. There were also findings reporting that miRNA-1290 was significantly varied in some sort of tumors [26,27]. All the above indicate that miRNA-1290 plays a crucial role in the molecular mechanism of the occurrence and development of chordoma.
Recently, many researchers have revealed that miRNAs function as pivotal regulators in chordoma cellular events. Duan et al. found that restoration of miR-1 expression suppresses migratory and invasive activities of chordoma cells [28] and verified that by further experiment showing that miR-1 reduces cell population growth and proliferation in chordoma cells in vitro [29]. Eiji Osaka’ team reported that miR-1 reduces cell proliferation, as well as migratory and invasive activities of chordoma cells [30]. Coincidently, our result suggests that dysregulation of miRNA-1290 significantly correlates with the tumor invasion into surrounding tissues (P=0.013), rather than other clinical features ranging from age, gender, tumor size to tumor location. What’s more, evidence has shown miRNA-1290 occupies a vital position impacting on tumor cell proliferation and metastasis, reprogramming, even anticancer reaction [27,31,32]. Taken together, we deem that abnormally expressed miRNA-1290 may be of great value in predicting the aggressiveness and malignancy of chordoma.
As one of the most troublesome features of chordoma, local recurrence is still a remaining problem to be solved [17,18], and always count against the prognosis of patients. Thus far, there is only one article explored the relationship between miRNA-1290 and tumor recurrence. Within colorectal cancer cohorts, miRNA-1290 was found have great potential to be a predictive biomarker of tumor recurrence and the Cox’s proportional hazard regression model was employed to demonstrate that serum miRNA-1290 could be an independent risk factor for early recurrence [33]. Current study found that differential miRNA-1290 was associated with local recurrence (P=0.03). In addition, Kaplan-Meier survival curve and log-rank test revealed that RFS was significantly shorter in patients with low-level miRNA-1290 (P=0.004). Besides the colorectal cancer, Mo et al. also draw a conclusion that disordered miRNA-1290 was closely related with the prognosis of non-small cell lung cancer patients [26,33]. Also, similar phenomenon has been observed in other miRNAs involved in chordoma. Duan et al. reported that miRNA-1 expression levels were related to the outcome clinical prognosis for patients [29]. The miR-140-3p and miR-1237-3p were reported to have strong relationships with the prognosis and were identified as an independent predictor for short recurrence-free survival of spinal chordoma patients [34,35]. As a result, miRNA-1290 may be proved to become a crucial prognostic indicator for sacral chordoma.
Notably, some articles concluded that the miRNA-1290 expression was up-regulated in tumor microenvironment while our result showed the opposite change [26,36]. Still, consensuses are made that deregulated miRNA-1290 expression could promote tumor invasion and recurrence and significantly associated with the prognosis, which indicates that miRNA-1290 may function as not only oncogene but also tumor suppressor to alter tumor characteristics. Supported by discovery proposing that down-regulated miRNA-1290 was linked with preferable subtype of the breast cancer [25], further studies are well-deserved to clarify the molecular mechanism of miRNA-1290 in chordoma genesis and progression.
In conclusion, examined by quantitative real-time PCR, low expression of miRNA-1290 was found within sacral chordoma tissues, and was significantly correlated with the invasion and recurrence of chordoma. Our result suggests that miRNA-1290 might become a valuable biomarker in predicting prognosis within sacral chordoma patients. Furthermore, it might serve as an effective therapeutic target for sacral chordoma in the near future.
Acknowledgements
The present study was funded by the Jiangsu Provincial Special Program of Medical Science (grant no. BL2012004), the Natural Science Foundation of the Colleges and Universities in Jiangsu Province (grant no. 16KJD320004) and the Suzhou Civic ‘Science and Education Guardian’ Youth Science and Technology Project (grant no. KJXW2014009).
Disclosure of conflict of interest
None.
References
- 1.Papagelopoulos PJ, Mavrogenis AF, Galanis EC, Savvidou OD, Boscainos PJ, Katonis PG, Sim FH. Chordoma of the spine: clinicopathological features, diagnosis, and treatment. Orthopedics. 2004;27:1256–63. doi: 10.3928/0147-7447-20041201-14. [DOI] [PubMed] [Google Scholar]
- 2.Chambers KJ, Lin DT, Meier J, Remenschneider A, Herr M, Gray ST. Incidence and survival patterns of cranial chordoma in the United States. Laryngoscope. 2014;124:1097–102. doi: 10.1002/lary.24420. [DOI] [PubMed] [Google Scholar]
- 3.Yu E, Koffer PP, DiPetrillo TA, Kinsella TJ. Incidence, treatment, and survival patterns for sacral chordoma in the United States, 1974-2011. Front Oncol. 2016;6:203. doi: 10.3389/fonc.2016.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maclean FM, Soo MY, Ng T. Chordoma: radiological-pathological correlation. Australas Radiol. 2005;49:261–8. doi: 10.1111/j.1440-1673.2005.01433.x. [DOI] [PubMed] [Google Scholar]
- 5.Yakkioui Y, van Overbeeke JJ, Santegoeds R, van Engeland M, Temel Y. Chordoma: the entity. Biochim Biophys Acta. 2014;1846:655–69. doi: 10.1016/j.bbcan.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 6.McPherson CM, Suki D, McCutcheon IE, Gokaslan ZL, Rhines LD, Mendel E. Metastatic disease from spinal chordoma: a 10-year experience. J Neurosurg Spine. 2006;5:277–80. doi: 10.3171/spi.2006.5.4.277. [DOI] [PubMed] [Google Scholar]
- 7.Hulen CA, Temple HT, Fox WP, Sama AA, Green BA, Eismont FJ. Oncologic and functional outcome following sacrectomy for sacral chordoma. J Bone Joint Surg Am. 2006;88:1532–39. doi: 10.2106/JBJS.D.02533. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs B, Dickey ID, Yaszemski MJ, Inwards CY, Sim FH. Operative management of sacral chordoma. J Bone Joint Surg Am. 2005;87:2211–6. doi: 10.2106/JBJS.D.02693. [DOI] [PubMed] [Google Scholar]
- 9.Chen KW, Yang HL, Lu J, Liu JY, Chen XQ. Prognostic factors of sacral chordoma after surgical therapy: a study of 36 patients. Spinal Cord. 2010;48:166–71. doi: 10.1038/sc.2009.95. [DOI] [PubMed] [Google Scholar]
- 10.Berven S, Zurakowski D, Mankin HJ, Gebhardt MC, Springfield DS, Hornicek FJ. Clinical outcome in chordoma: utility of flow cytometry in DNA determination. Spine (Phila Pa 1976) 2002;27:374–9. doi: 10.1097/00007632-200202150-00010. [DOI] [PubMed] [Google Scholar]
- 11.Heery CR. Chordoma: the quest for better treatment options. Oncol Ther. 2016;4:35–51. doi: 10.1007/s40487-016-0016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yelamanchili SV, Morsey B, Harrison EB, Rennard DA, Emanuel K, Thapa I, Bastola DR, Fox HS. The evolutionary young miR-1290 favors mitotic exit and differentiation of human neural progenitors through altering the cell cycle proteins. Cell Death Dis. 2014;5:e982. doi: 10.1038/cddis.2013.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T, Xu H, Qi M, Yan S, Tian X. miRNA dysregulation and the risk of metastasis and invasion in papillary thyroid cancer: a systematic review and meta-analysis. Oncotarget. 2017 doi: 10.18632/oncotarget.16681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zedan AH, Blavnsfeldt SG, Hansen TF, Nielsen BS, Marcussen N, Pleckaitis M, Osther PJS, Sørensen FB. Heterogeneity of miRNA expression in localized prostate cancer with clinicopathological correlations. PLoS One. 2017;12:e0179113. doi: 10.1371/journal.pone.0179113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang T, Yang Z, Gao H. Advancements in the study of miRNA regulation during the cell cycle in human pituitary adenomas. J Neurooncol. 2017 doi: 10.1007/s11060-017-2518-5. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen K, Chen H, Zhang K, et al. MicroRNA profiling and bioinformatics analyses reveal the potential roles of miRNAs in chordoma. Oncol Letter. 2017 doi: 10.3892/ol.2017.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chugh R, Tawbi H, Lucas DR, Biermann JS, Schuetze SM, Baker LH. Chordoma: the nonsarcoma primary bone tumor. Oncologist. 2007;12:1344–50. doi: 10.1634/theoncologist.12-11-1344. [DOI] [PubMed] [Google Scholar]
- 18.Hanna SA, Aston WJ, Briggs TW, Cannon SR, Saifuddin A. Sacral chordoma: can local recurrence after sacrectomy be predicted? Clin Orthop Relat Res. 2008;466:2217–23. doi: 10.1007/s11999-008-0356-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmini G, Marini F, Brandi M. What is new in the miRNA world regarding osteosarcoma and chondrosarcoma? Molecules. 2017;22:417. doi: 10.3390/molecules22030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126:1283–90. doi: 10.1002/ijc.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riva P, Crosti F, Orzan F, Dalprà L, Mortini P, Parafioriti A, Pollo B, Fuhrman Conti AM, Miozzo M, Larizza L. Mapping of candidate region for chordoma development to 1p36.13 by LOH analysis. Int J Cancer. 2003;107:493–7. doi: 10.1002/ijc.11421. [DOI] [PubMed] [Google Scholar]
- 23.Scheil S, Brüderlein S, Liehr T, Starke H, Herms J, Schulte M, Möller P. Genome-wide analysis of sixteen chordomas by comparative genomic hybridization and cytogenetics of the first human chordoma cell line, U-CH1. Genes Chromosomes Cancer. 2001;32:203–11. doi: 10.1002/gcc.1184. [DOI] [PubMed] [Google Scholar]
- 24.Longoni M, Orzan F, Stroppi M, Boari N, Mortini P, Riva P. Evaluation of 1p36 markers and clinical outcome in a skull base chordoma study. Neuro Oncol. 2008;10:52–60. doi: 10.1215/15228517-2007-048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endo Y, Toyama T, Takahashi S, Yoshimoto N, Iwasa M, Asano T, Fujii Y, Yamashita H. miR-1290 and its potential targets are associated with characteristics of estrogen receptor α-positive breast cancer. Endocr Relat Cancer. 2013;20:91–102. doi: 10.1530/ERC-12-0207. [DOI] [PubMed] [Google Scholar]
- 26.Mo D, Gu B, Gong X, Wu L, Wang H, Jiang Y, Zhang B, Zhang M, Zhang Y, Xu J, Pan S. miR-1290 is a potential prognostic biomarker in non-small cell lung cancer. J Thorac Dis. 2015;7:1570–9. doi: 10.3978/j.issn.2072-1439.2015.09.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, He XY, Zhang ZM, Li S, Ren LH, Cao RS, Feng YD, Ji YL, Zhao Y, Shi RH. MicroRNA-1290 promotes esophageal squamous cell carcinoma cell proliferation and metastasis. World J Gastroenterol. 2015;21:3245–55. doi: 10.3748/wjg.v21.i11.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan Z, Choy E, Nielsen GP, Rosenberg A, Iafrate J, Yang C, Schwab J, Mankin H, Xavier R, Hornicek FJ. Differential expression of microRNA (miRNA) in chordoma reveals a role for miRNA-1 in Met expression. J Orthop Res. 2010;28:746–52. doi: 10.1002/jor.21055. [DOI] [PubMed] [Google Scholar]
- 29.Duan Z, Shen J, Yang X, Yang P, Osaka E, Choy E, Cote G, Harmon D, Zhang Y, Nielsen GP, Spentzos D, Mankin H, Hornicek F. Prognostic significance of miRNA-1 (miR-1) expression in patients with chordoma. J Orthop Res. 2014;32:695–701. doi: 10.1002/jor.22589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osaka E, Yang X, Shen JK, Yang P, Feng Y, Mankin HJ, Hornicek FJ, Duan Z. MicroRNA-1 (miR-1) inhibits chordoma cell migration and invasion by targeting slug. J Orthop Res. 2014;32:1075–82. doi: 10.1002/jor.22632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J, Ji X, Zhu L, Jiang Q, Wen Z, Xu S, Shao W, Cai J, Du Q, Zhu Y, Mao J. Up-regulation of microRNA-1290 impairs cytokinesis and affects the reprogramming of colon cancer cells. Cancer Lett. 2013;329:155–63. doi: 10.1016/j.canlet.2012.10.038. [DOI] [PubMed] [Google Scholar]
- 32.Kim KB, Kim K, Bae S, Choi Y, Cha HJ, Kim SY, Lee JH, Jeon SH, Jung HJ, Ahn KJ, An IS, An S. MicroRNA-1290 promotes asiatic acid-induced apoptosis by decreasing BCL2 protein level in A549 non-small cell lung carcinoma cells. Oncol Rep. 2014;32:1029–36. doi: 10.3892/or.2014.3319. [DOI] [PubMed] [Google Scholar]
- 33.Imaoka H, Toiyama Y, Fujikawa H, Hiro J, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Mori T, Kato T, Toden S, Goel A, Kusunoki M. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann Oncol. 2016;27:1879–86. doi: 10.1093/annonc/mdw279. [DOI] [PubMed] [Google Scholar]
- 34.Zou MX, Huang W, Wang XB, Li J, Lv GH, Wang B, Deng YW. Reduced expression of miRNA-1237-3p associated with poor survival of spinal chordoma patients. Eur Spine J. 2015;24:1738–46. doi: 10.1007/s00586-015-3927-9. [DOI] [PubMed] [Google Scholar]
- 35.Zou MX, Huang W, Wang XB, Lv GH, Li J, Deng YW. Identification of miR-140-3p as a marker associated with poor prognosis in spinal chordoma. Int J Clin Exp Pathol. 2014;7:4877–85. [PMC free article] [PubMed] [Google Scholar]
- 36.Li A, Yu J, Kim H, Wolfgang CL, Canto MI, Hruban RH, Goggins M. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res. 2013;19:3600–10. doi: 10.1158/1078-0432.CCR-12-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]


