Abstract
Long non-coding RNAs (lncRNA) have been shown to serve critical roles in human cancers development, including epithelial ovarian cancer (EOC). Here, we identified a novel lncRNA SNHG1, which was markedly upregulated in human EOC tissues and cell lines. High SNHG1 expression was associated with aggressive clinical features and poor prognosis of EOC patients. Moreover, the downregulation of SNHG1 remarkably inhibited the EOC cells proliferation, migration and invasion, suppressed S-phase entry in vitro, and repressed tumor growth in vivo. In contrast, overexpression of SNHG1 could promote the aggressive behaviors of EOC cells. Furthermore, through western blot, we found that SNHG1 enhanced the expression of several downstream genes in Wnt/β-catenin pathway. Our findings demonstrated that the dysregulation of SNHG1 is implicated in EOC tumorigenesis and progression through regulating Wnt/β-catenin pathway.
Keywords: Epithelial ovarian cancer, SNHG1, prognosis, cell cycle, Wnt/β-catenin
Introduction
Ovarian cancer represents approximately 3% of all gynecologic malignancies in women and is currently the most deadly tumor [1,2]. Epithelial ovarian cancer (EOC) accounts for 90% of all ovarian cancer cases [3]. Owing to high metastasis rate and lack of effective diagnostic techniques to identify early symptoms, the prognosis of most EOC patients remains unfavorable, and the 5-year overall survival rate is generally less than 50% [4,5]. The pathogenesis of EOC is a complicated biological process that involves both genetic and environmental factors [6]. Accordingly, a better understanding of the molecular mechanisms underlying EOC progression will lead to the development of better diagnostic approaches and more effective treatments for EOC.
Currently, large-scale genome-wide projects have confirmed that less than 3% of the human genome sequences can encode proteins, whereas the majority (~98%) is transcribed into a wide variety of conserved noncoding RNAs (ncRNAs) [7]. Long noncoding RNAs (lncRNAs) are important new members of the ncRNA family, which are greater than 200 nucleotides in length and lack protein-coding ability [8]. It has been well documented that lncRNAs participate in cellular proliferation, differentiation, and other biological events [9,10]. Notably, numerous studies have linked lncRNA dysregulation with human diseases, especially cancers [11,12]. LncRNAs have accordingly been highlighted as critical regulators of tumorigenesis and cancer progression, and numerous lncRNAs have been found to serve as oncogenes or tumor suppressors in various types of cancers. In recent years, several EOC-associated lncRNAs have been identified, and their biological properties and underlying molecular mechanisms have been determined, such as HOTAIR [13], HOST2 [14] and HOXA11-AS [15].
Among all the known cancer-related lncRNAs, small nucleolar RNA host gene 1 (SNHG1), localized in human chromosome 11q12.3 region, has been observed to be significantly upregulated in hepatocellular carcinoma [16] and nonsmall cell lung cancer [17]. However, the role of SNHG1 in EOC remains elusive. The purpose of our current study was to determine the expression of SNHG1 in EOC tissues and EOC cell lines. Furthermore, the clinicopathological and prognostic significance of SNHG1 levels in human EOC was also assessed. Next, the biological effects of SNHG1 on the phenotypes of EOC cells were analyzed through gain- and loss-of-function experiments.
Materials and methods
Human EOC tissue samples
Human EOC tissues (n=67) were collected from 67 patients who had received surgical treatment at Jinan Central Hospital (Shandong, China). The normal ovarian tissues (n=5) were obtained from five patients with perimenopausal hysteromyoma who received hysterectomy and prophylactic adnexectomy. Patients who underwent chemotherapy, radiotherapy, or targeted therapy prior to surgery were excluded. The follow-up periods were calculated from the date of surgery to death or the final follow-up. This study was carried out after approval by the Ethics Committee of Jinan Central Hospital and obtaining informed consent from all subjects. All tissue samples were immediately snap-frozen in liquid nitrogen, and stored at -80°C until further experiments.
Cell culture and transfection
Two ovarian cancer cell lines (HO-8910, OVCAR3) and a normal human ovarian epithelial cell line (HOEpiC), obtained from Chinese Type Culture Collection, Chinese Academy of Sciences, were cultured in RPMI 1640 medium (Gibco, Gaithersburg, MD, USA) containing 10% fetal bovine serum (FBS; HyClone, Logan, USA) and 100 U/ml penicillin/streptomycin in a humidified environment with 5% CO2 at 37°C.
The full-length complementary DNA (cDNA) sequence of SNHG1, amplified by RT-PCR, was inserted into the pcDNA3.1 (GeneScript, Piscataway, NJ, USA) vectors (pcDNA-SNHG1). OVCAR3 cells were transfected with the aforementioned vectors by Lipofectamine 2000 (Invitrogen). HO-8910 cells were transfected with SNHG1-specific small interfering RNA (si-SNHG1) or a negative control (si-NC) (GenePharma, Shanghai, China). 48 h after transfection, the transfection efficiencies were evaluated by qRT-PCR.
RNA extraction and quantitative reverse transcription-PCR (qRT-PCR)
The total RNA of the tissue samples and cells was isolated using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized using a Reverse Transcription Kit (Takara, Dalian, China). Quantitative real-time RT-PCR was performed with the SYBR Premix EX TaqTM (Takala) using an ABI 7500 Real-Time PCR system (Applied Biosystems, Foster City, USA). The relative expression of individual genes was determined by 2-ΔΔCt methods [18], and GAPDH mRNA was used as an endogenous control. The following forward and reverse PCR primers were used in this study: SNHG1 forward: 5’-CCACCTTCTGTTCCCGTCAT-3’; SNHG1 reverse: 5’-GACAGCCAGTCCTCAAGGGA-3’; GAPDH forward: 5’-GACTCATGACCACAGTCCATGC-3’; GAPDH reverse: 3’-AGAGGCAGGGATGATGTTCTG-5’.
Western blot assay
Total protein was extracted from cells with RIPA buffer containing PMSF. Nuclear protein extracts were obtained using NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific, Rockford, IL, USA). The protein concentration was measured by the BCA Protein Assay Kit (Pierce, Illinois, USA). Equal quantities of each protein was subjected to SDS-PAGE and transferred to a PVDF membrane (Millipore, USA). Then, the membrane was probed with the corresponding primary antibodies at 4°C overnight, followed by incubation with secondary antibodies at room temperature for 1 h. The proteins were visualized using BandScan software (Bio-Rad, Hercules, CA, USA). β-actin or Lamin A was used as loading control.
MTT assay
Cell proliferation was measured after transfection for 24, 48 and 72 h by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cells (2×103/well) were plated into a 96-well plate. MTT solution (5 mg/mL) was added to each well. After 4 h, DMSO was added into each well for 30 min. The absorbance of each well was recorded at 490 nm and read on a microplate reader.
Transwell assay
Cell migration and invasion were performed with transwell assay using transwell chambers with 8-μm pores (Costar, Corning, NY, USA). For invasion assay, the upper side of the insert membrane was coated with Matrigel (BD Biosciences, New Jersey, USA). A number of 1×104 cells in 100 μl of serum-free medium were put into the top chamber. Also, the bottom chamber was filled with 600 μl of medium containing 10% FBS. After 24 h incubation, the cells that had migrated or invaded through the membrane were fixed in methanol and then stained with 1% crystal violet. From five random fields, the number of cells on the lower surface was counted under a light microscope and then photographed.
Flow-cytometric analysis
Cells for cell cycle analysis were stained with propidium iodide (PI) for 1 h at 4°C in the dark using the Cycle TEST PLUS DNA Reagent Kit (BD Biosciences, San Jose, CA, USA) and analyzed by a flow cytometer (FACScan®; BD Biosciences) with ModFit 3.0 software (BD Biosciences). The percentage of the cells in G0/G1, S, and G2/M phase were then counted and compared.
Animal experiments
Ten BALB/c athymic nude mice (male 4-5 weeks old), obtained from Shanghai Laboratory Animals Center (Shanghai, China), were randomized to two groups (n=5/group). A total of 2×106 HO-8910 cells transfected with si-SNHG1 or si-NC, were respectively injected into the dorsal scapular region of each mouse. The tumor size was assessed by a digital caliper every three days. After 30 days of monitoring, mice were killed by cervical dislocation and tumors were extracted for further analysis. Tumor volume was calculated as follows: Volume = length × width2/2. The protocol was approved by the Committee on the Ethics of Animal Experiments of Jinan Central Hospital.
Statistical analysis
All statistical analyses were performed using SPSS 19.0 software (SPSS, Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad Software, LaJolla, CA, USA). Continuous data were compared using Student’s two-tailed t-test. For analysis on association of SNHG1 expression and clinicopathological variables of EOC patients, Chi-squared test was used. The overall survival (OS) of EOC patients were analyzed by the Kaplan-Meier method with the log-rank test. P values were calculated and those less than 0.05 were considered significant.
Results
SNHG1 is upregulated in EOC and associated with poor prognosis
To determine the expression pattern of SNHG1 in the human EOC tissues, we first analyzed its expression in a cohort of 67 EOC tissues and five nontumor tissues. As shown in Figure 1A, SNHG1 expression levels in tumor tissues of EOC patients were noticeably higher than those in normal tissues. Then we examined the expression of SNHG1 in two EOC cell lines, including HO-8910 and OVCAR3, and a normal human ovarian epithelial cell line HOEpiC, by qRT-PCR. The results showed that SNHG1 expression was obviously upregulated in EOC cells (Figure 1B).
Figure 1.
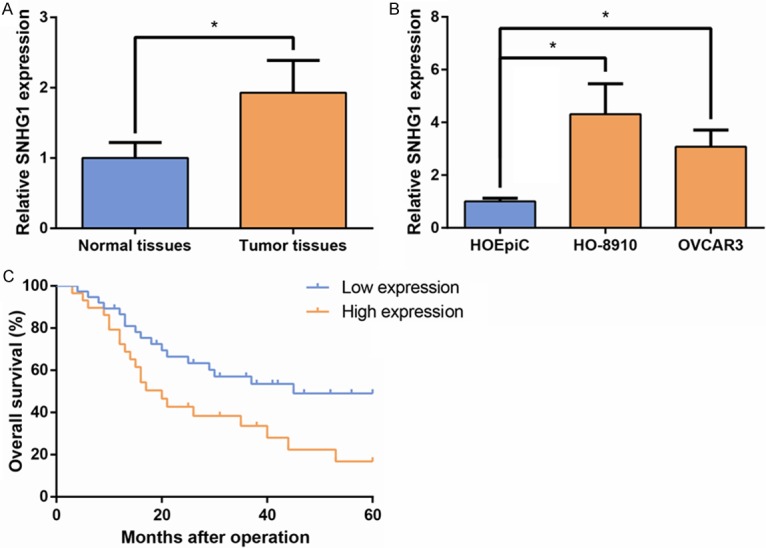
SNHG1 is upregulated in EOC and associated with poor prognosis. A. qRT-PCR analysis of SNHG1 expression in EOC tissues and normal ovarian tissues. B. qRT-PCR analysis of SNHG1 expression in the EOC cell lines HO-8910 and OVCAR3, and the normal human ovarian epithelial cell line (HOEpiC). SNHG1 level was normalized to the GAPDH level. Data are presented as mean ± SD. The experiments were all repeated at least three times. P value was assessed by Student’s t-test. *P<0.05. C. Kaplan-Meier analysis of overall survival (OS) in EOC patients according to SNHG1 expression. P value was assessed by log-rank test.
To assess whether SNHG1 expression was correlated with the clinicopathological factors and prognosis of EOC patients, according to relative SNHG1 expression in tumor tissues, the 67 EOC patients were classified into two groups: the high SNHG1 expression group (n=29); and the low SNHG1 expression group (n=38). As indicated in Table 1, high SNHG1 expression in gastric cancer was significantly correlated with advanced pathological grade (P=0.041) and advanced clinical stage (P=0.018). Moreover, Kaplan-Meier survival analysis revealed that patients with higher SNHG1 levels had shorter OS (P=0.02; Figure 1C) than those with lower SNHG1 levels.
Table 1.
Correlation of SNHG1 expression with clinicopathologic features in EOC patients
| Parameter | Total (n=67) | SNHG1 expression | P value | |
|---|---|---|---|---|
|
| ||||
| Low (n=38) | High (n=29) | |||
| Age (yrs) | 0.204 | |||
| <55 | 29 | 19 | 10 | |
| ≥55 | 38 | 19 | 19 | |
| Histological type | 0.784 | |||
| Serous | 45 | 25 | 20 | |
| Non-serous | 22 | 13 | 9 | |
| Pathological grade | 0.041 | |||
| 1-2 | 35 | 24 | 11 | |
| 3 | 32 | 14 | 18 | |
| Clinical stage | 0.018 | |||
| I-II | 27 | 20 | 7 | |
| III-IV | 40 | 18 | 22 | |
| Lymph nodes metastasis | 0.052 | |||
| No | 39 | 26 | 13 | |
| Yes | 28 | 12 | 16 | |
SNHG1 promotes EOC cell proliferation in vitro
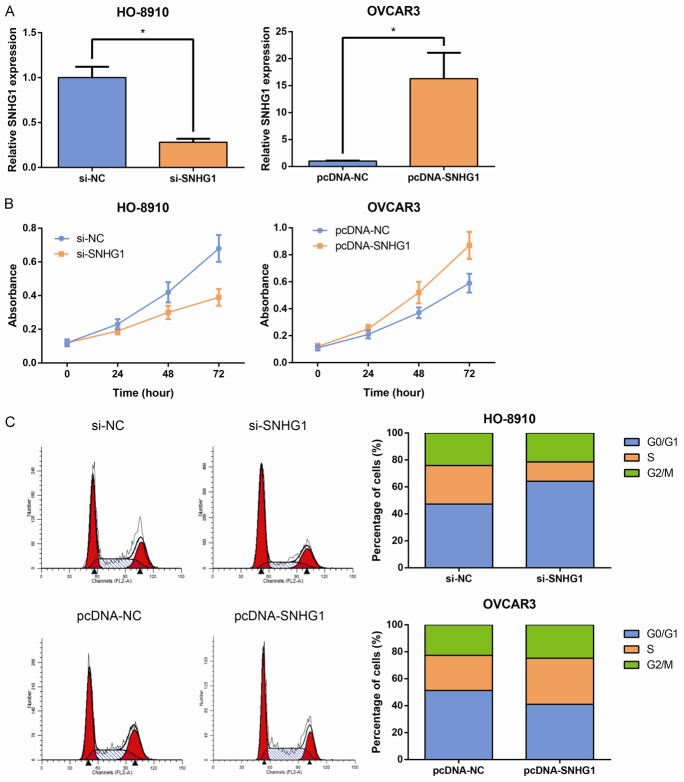
Next, we inhibited the endogenous expression of SNHG1 in HO-8910 cells by siRNA to further investigate the biological effects of SNHG1 on EOC. Moreover, SNHG1 was overexpressed in OVCAR3 cells by transfection of pcDNA-SNHG1 (Figure 2A). Curves of the growth in these experiments, as detected by MTT assays, showed that SNHG1 knockdown impaired HO-8910 cell growth, while SNHG1 overexpression promoted the proliferation of OVCAR3 cells (Figure 2B). To determine whether a change in cell cycle progression was involved in the SNHG1-mediated regulation of cell proliferation, flow cytometry analysis was subsequently performed. The results showed that silencing of SNHG1 increased the proportion of G0/G1 phase cells and reduced the proportion of S phase cells (Figure 2C). These findings indicate that SNHG1 could promote the cell cycle progression of EOC cells.
Figure 2.
SNHG1 promotes EOC cell proliferation in vitro. A. qRT-PCR analysis of SNHG1 expression in si-SNHG1-transfected HO-8910 and pcDNA-SNHG1-transfected OVCAR3 cells. B. In vitro proliferation of HO-8910 and OVCAR3 cells were assessed by MTT assay for 3 days. C. The cell cycle progression of HO-8910 and OVCAR3 cells was evaluated by flow cytometry by measuring the proportions of the cells in the G0/G1, S, and G2/M phases using PI staining. Data are presented as mean ± SD. The experiments were all repeated at least three times. P value was assessed by Student’s t-test. *P<0.05.
SNHG1 promotes EOC cell migration and invasion in vitro
To further determine whether SNHG1 is associated with the progression of EOC, we analyzed its effect on the migration and invasion of HO-8910 and OVCAR3 cells. By transwell assay, we found that the HO-8910 cell migration and invasion were significantly impaired after the knockdown of SNHG1 (Figure 3). Conversely, SNHG1 overexpression in OVCAR3 cells promoted cell migration and invasion. Taken together, these findings indicate that SNHG1 has important roles in EOC progression.
Figure 3.
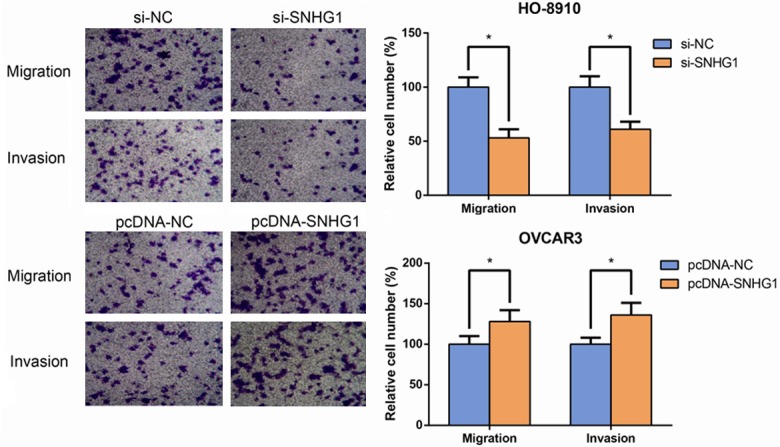
SNHG1 promotes EOC cell migration and invasion in vitro. The migratory and invasive abilities of HO-8910 and OVCAR3 cells were assessed using transwell assay. Data are presented as mean ± SD. The experiments were all repeated at least three times. P value was assessed by Student’s t-test. *P<0.05.
SNHG1 promotes tumorigenesis of EOC cells in vivo
To further investigate the tumorigenesis function of SNHG1 in vivo, we used nude mouse xenograft model. HO-8910 cells transfected with si-SNHG1 or si-NC were injected subcutaneous of ten nude mice, respectively. As exhibited in Figure 4A, tumor growth in the si-SNHG1 group was evidently slower than that in the si-NC group. 30 days later, the mice were sacrificed and the xenografts were collected. As expected, the si-SNHG1 group exhibited generally smaller tumors and displayed less weight compared to the si-NC group (Figure 4B). qRT-PCR analysis found that the levels of SNHG1 in tumor tissues formed from si-SNHG1-transfected cells were obviously lower than in tumors formed from si-NC-transfected cells (Figure 4C). These results indicate that SNHG1 down-regulation could inhibit tumorigenesis of EOC cells in vivo.
Figure 4.
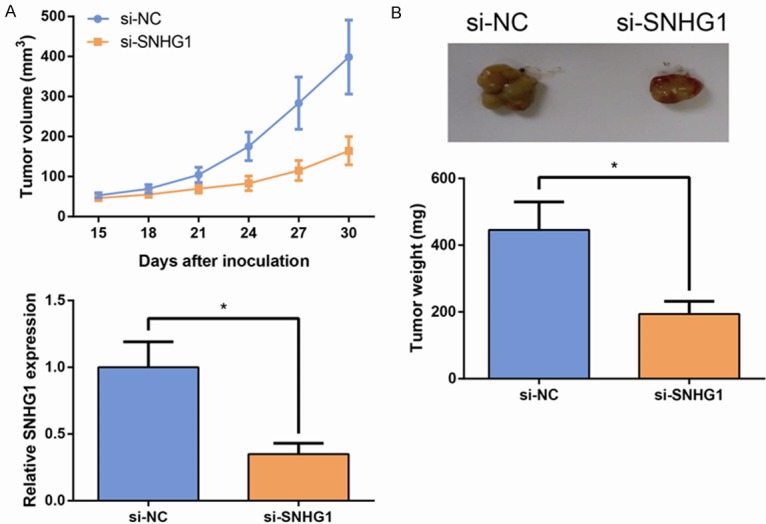
SNHG1 promotes tumorigenesis of EOC cells in vivo. A. Growth curves of tumor volumes in different groups of nude mice. B. Quantitative determination of the weight of the excised tumors. C. qRT-PCR analysis of SNHG1 expression levels in the excised tumor tissues. Data are presented as mean ± SD. The experiments were all repeated at least three times. P value was assessed by Student’s t-test. *P<0.05.
SNHG1 promotes Wnt/β-catenin signaling activation in EOC cells
As is known, Wnt/β-catenin signaling pathway plays a crucial role in the regulation of cell growth and migration. In the present study, using western blot assay, we detected the effect of SNHG1 on nuclear β-catenin expression and a few of the downstream genes of Wnt/β-catenin signaling pathway, including cyclin D1 and c-myc in EOC cells. The results showed that the expression of nuclear β-catenin, c-myc and cyclin D1 was significantly augmented when SNHG1 was overexpressed, whereas their levels were remarkably reduced in si-SNHG1-transfected cells (Figure 5). Thus, these data indicated that SNHG1 has an effect on the biological behavior of EOC cells by regulating Wnt/β-catenin signaling pathway.
Figure 5.
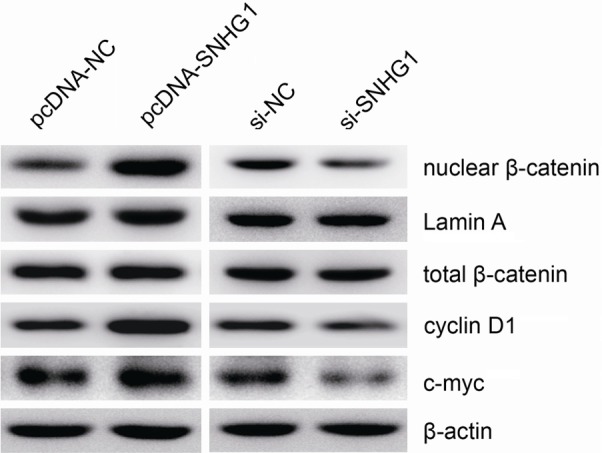
SNHG1 promotes Wnt/β-catenin signaling activation in EOC cells. Western blot analysis of nuclear β-catenin, cyclin D1 and c-myc levels in HO-8910 and OVCAR3 cells. β-actin or Lamin A was used as loading control. Data are presented as mean ± SD. The experiments were all repeated at least three times. P value was assessed by Student’s t-test. *P<0.05.
Discussion
In recent years, an increasing number of lncRNAs have been identified to be linked with human malignancies, including EOC. Thus, identification of EOC-associated lncRNAs and detection of their clinical significance and biological proprieties might provide promising therapeutic strategies and accelerate the development of lncRNA-based diagnostic methods for EOC.
SNHG1 has been found to have close relationships with multiple tumors. In hepatocellular carcinoma, SNHG1 was found to be significantly overexpressed compared to normal tissue [16,19]. Moreover, Cui et al. also reported lncRNA SNHG1 could promote the progression of non-small cell lung cancer by regulating miR-101-3p and SOX9 [20]. To our knowledge, the role and mechanism of SNHG1 in EOC have not been completely understood. In the present study, the analyses of clinical samples showed that SNHG1 was upregulated in EOC tissues and cells in comparison to the controls, and SNHG1 overexpression was closely associated with aggressive clinical characteristics and poor prognosis in EOC patients, revealing that SNHG1 could be a biomarker for EOC and might be a prognostic indicator for survival.
As high SNHG1 expression was correlated with an aggressive tumor phenotype in EOC, we speculated that SNHG1 might serve a pivotal role in tumor biology. After siRNA-mediated silencing of SNHG1, we found that decreased expression of SNHG1 significantly suppressed proliferation, cell cycle progression, migration and invasion, and caused cell cycle arrest at G1/G0 phase. Moreover, xenograft tumor models revealed that decreased SNHG1 expression evidently inhibited EOC tumorigenesis in vivo. This is in line with our clinical data. Taken together, the observations of this study indicated that SNHG1 might serve as an oncogene in EOC progression.
In normal physiological state, Wnt/β-catenin signaling pathway regulates downstream genes involved in multiple cellular processes, such as cell proliferation, differentiation, migration and cell death, of which β-catenin is one of the key downstream effectors [21]. It means that Wnt/β-catenin signaling should be kept in a normal level to exert normal physiological function. However, extensive findings have shown that Wnt/β-catenin pathway is constitutively activated in many human cancers, including EOC [22,23]. In the present study, we observed that knockdown SNHG1 expression markedly inhibited nuclear β-catenin, cyclin D1 and c-myc expression in EOC cells, indicating that in EOC, oncogenic function of SNHG1 was at least partly mediated by Wnt/β-catenin pathway.
In summary, this study provides the first link between SNHG1 expression and EOC development. We found that SNHG1 was overexpressed in EOC tissues and its upregulation may be correlated with unfavorable prognosis of EOC patients. The promotion of SNHG1 on EOC cell growth and tumorigenesis might partly through Wnt/β-catenin signaling. Accordingly, the present study may provide a novel perspective that SNHG1 functions as a non-coding oncogene in EOC tumorigenesis and a potential target for early diagnosis and treatment of EOC in the further. However, the other possible molecular mechanisms through which SNHG1 participates in EOC cell biological functions remains to be further explored.
Acknowledgements
The work of our research was supported by National Natural Science Foundation of China (Grant number: 81471437).
Disclosure of conflict of interest
None.
References
- 1.Bast RC Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9:415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 3.Mutch DG, Prat J. 2014 FIGO staging for ovarian, fallopian tube and peritoneal cancer. Gynecol Oncol. 2014;133:401–404. doi: 10.1016/j.ygyno.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 4.Chornokur G, Amankwah EK, Schildkraut JM, Phelan CM. Global ovarian cancer health disparities. Gynecol Oncol. 2013;129:258–264. doi: 10.1016/j.ygyno.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minig L, Otano L, Diaz-Padilla I, Alvarez Gallego R, Patrono MG, Valero de Bernabe J. Therapeutic management of epithelial ovarian cancer during pregnancy. Clin Transl Oncol. 2013;15:259–264. doi: 10.1007/s12094-012-0963-3. [DOI] [PubMed] [Google Scholar]
- 6.Haruta S, Furukawa N, Yoshizawa Y, Tsunemi T, Nagai A, Kawaguchi R, Tanase Y, Yoshida S, Kobayashi H. Molecular genetics and epidemiology of epithelial ovarian cancer (review) Oncol Rep. 2011;26:1347–1356. doi: 10.3892/or.2011.1456. [DOI] [PubMed] [Google Scholar]
- 7.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 9.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 10.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calore F, Lovat F, Garofalo M. Non-coding RNAs and cancer. Int J Mol Sci. 2013;14:17085–17110. doi: 10.3390/ijms140817085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW, Jin HY, Zhang Y, Li Q, Hua KQ. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol. 2014;134:121–128. doi: 10.1016/j.ygyno.2014.03.556. [DOI] [PubMed] [Google Scholar]
- 14.Gao Y, Meng H, Liu S, Hu J, Zhang Y, Jiao T, Liu Y, Ou J, Wang D, Yao L, Hui N. LncRNAHOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum Mol Genet. 2015;24:841–852. doi: 10.1093/hmg/ddu502. [DOI] [PubMed] [Google Scholar]
- 15.Richards EJ, Permuth-Wey J, Li Y, Chen YA, Coppola D, Reid BM, Lin HY, Teer JK, Berchuck A, Birrer MJ, Lawrenson K, Monteiro AN, Schildkraut JM, Goode EL, Gayther SA, Sellers TA, Cheng JQ. A functional variant in HOXA11-AS, a novel long non-coding RNA, inhibits the oncogenic phenotype of epithelial ovarian cancer. Oncotarget. 2015;6:34745–34757. doi: 10.18632/oncotarget.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Zhou D, Ying M, Chen M, Chen P, Chen Z, Zhang F. Expression of long noncoding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) exacerbates hepatocellular carcinoma through suppressing miR-195. Med Sci Monit. 2016;22:4820–4829. doi: 10.12659/MSM.898574. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.You J, Fang N, Gu J, Zhang Y, Li X, Zu L, Zhou Q. Noncoding RNA small nucleolar RNA host gene 1 promote cell proliferation in nonsmall cell lung cancer. Indian J Cancer. 2014;51(Suppl 3):e99–e102. doi: 10.4103/0019-509X.154092. [DOI] [PubMed] [Google Scholar]
- 18.VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44:619–626. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- 19.Zhang M, Wang W, Li T, Yu X, Zhu Y, Ding F, Li D, Yang T. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother. 2016;80:73–79. doi: 10.1016/j.biopha.2016.02.036. [DOI] [PubMed] [Google Scholar]
- 20.Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/beta-catenin signaling pathway. Oncotarget. 2017;8:17785–17794. doi: 10.18632/oncotarget.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bodnar L, Stanczak A, Cierniak S, Smoter M, Cichowicz M, Kozlowski W, Szczylik C, Wieczorek M, Lamparska-Przybysz M. Wnt/beta-catenin pathway as a potential prognostic and predictive marker in patients with advanced ovarian cancer. J Ovarian Res. 2014;7:16. doi: 10.1186/1757-2215-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou M, Cheng Z, Shen H, He S, Li Y, Pan Y, Feng C, Chen X, Zhang Y, Lin M, Wang L, Ke Z. High expression of CTHRC1 promotes EMT of epithelial ovarian cancer (EOC) and is associated with poor prognosis. Oncotarget. 2015;6:35813–35829. doi: 10.18632/oncotarget.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]