Abstract
Circulating microRNAs (miRNAs) have been demonstrated to be potential diagnostic and prognostic markers for many types of cancers including colorectal cancer (CRC). The aim of current study was to determine the clinical significance of serum miR-193b in CRC. The expression of serum miR-193b in CRC patients and healthy controls was detected by quantitative reverse transcription-polymerase chain reaction. Then the value of serum miR-193b as marker for CRC prognosis was evaluated. Our results showed that the expression levels of serum miR-193b were significantly reduced in CRC patients compared to healthy volunteers. Serum miR-193b discriminated CRC patients from healthy subjects with high accuracy. In addition, serum miR-193b levels were increased in CRC patients who received surgery treatment. Low serum miR-193b was associated with TNM stage, grade and lymph node metastasis. The overall/disease free survival time of patients with low serum miR-193b expression was significantly shorter compared to those with high serum miR-193b expression. Moreover, multivariate analysis showed that low serum miR-193b level predicted poor prognosis independently. These findings indicate that serum miR-193b serves as a promising novel prognostic biomarker for CRC.
Keywords: Biomarker, colorectal cancer, diagnosis, prognosis, serum
Introduction
Colorectal cancer (CRC) is one of the most commonly diagnosed cancer and a leading cause of cancer associated mortality around the world [1,2]. Regular screening and removal of polyps have significantly reduced the incidence of CRC. Metastasis is considered to be associated with poor clinical outcome of CRC. The prognosis of CRC patients diagnosed at the stage IV is dismal, with a five year overall survival rate of less than 10% [3]. Therefore, it is necessary to identify novel prognostic biomarkers that contribute to precise prediction of survival in CRC patients to determine optimal treatment modalities in clinical practice.
MicroRNAs (miRNAs) are a class of evolutionary conserved small non-coding RNA that negatively regulates gene expression by binding the 3’-untranslated region (UTR) of target mRNAs at the post-transcriptional level [4]. miRNAs plays important roles in various cellular processes including, but no limited to, proliferation, differentiation, apoptosis and survival [5]. Increasing studies have shown that circulating miRNAs are promising diagnostic and prognostic biomarkers for various types of cancers including CRC for the following reasons. Firstly, miRNAs might function as oncomiRs or tumor suppressor genes in cancer, thus deregulation of miRNAs might promote tumorigenesis [6]. Secondly, cancer cells actively secrete miRNAs into the circulation system [7]. Moreover, miRNAs are very stable in plasma or serum [8]. Zekri et al reported that the expression levels of miR-17, miR-19a, miR-20a and miR-223 were significantly elevated in patients with CRC and these four molecules might be used as diagnostic biomarkers for CRC [9]. Low levels of serum miR-21 were associated with higher risk of recurrence and mortality. In addition, reduced serum miR-21 was demonstrated to be an independent predictor of survival [10].
miR-193b is located on 16p13.12 and belongs to the miR-193b-365 cluster. Previous studies have shown that miR-193b is aberrantly expressed in many types of cancer such as pancreatic cancer, ovarian cancer and epidermal squamous cell carcinoma [11-13]. In this study, we aimed to investigate the expression pattern of serum miR-193b as well as its potential diagnostic and prognostic value in CRC.
Materials and methods
Patients
Serum samples were obtained from 90 CRC patients and 40 healthy volunteers who received treatment or physical examination in our hospital. Informed consent was obtained from each participant before serum samples collection. The study was approved by the Ethics Committee of the Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute. Inclusion criteria were a diagnosis of CRC based on histopathology. CRC patients with any other type of cancer or/and liver and kidney dysfunction were excluded from the current study. None of subjects had received preoperative radiation or chemotherapy before sample collection. The clinical stages were determined according to the criteria proposed by International Union Against Cancer (UICC) criteria. The demographic and clinicopathological data of all enrolled CRC patients was summarized in Table 1.
Table 1.
Relation between serum miR-193b expression level and the clinicopathologic characteristics of CRC patients
| Variables | Cases | Serum miR-193b levels | P | |
|---|---|---|---|---|
|
|
||||
| Low (n=43) | High (n=47) | |||
| Age | 0.7183 | |||
| <60 | 38 (42.22%) | 19 (21.11%) | 19 (21.11%) | |
| ≥60 | 52 (57.78%) | 24 (26.67%) | 28 (31.11%) | |
| Gender | 0.1021 | |||
| Male | 56 (62.22%) | 23 (25.56%) | 33 (36.67%) | |
| Female | 34 (37.78%) | 20 (22.22%) | 14 (15.56%) | |
| Tumor size (cm) | 0.0590 | |||
| <5 | 63 (70.00%) | 26 (28.89%) | 37 (41.11%) | |
| ≥5 | 27 (30.00%) | 17 (18.89%) | 10 (11.11%) | |
| Depth of invasion | 0.0730 | |||
| T1-T2 | 15 (16.67%) | 4 (4.44%) | 11 (12.22%) | |
| T3-T4 | 75 (83.33%) | 39 (43.33%) | 36 (40.00%) | |
| Lymph node metastasis | 0.0212 | |||
| Negative | 59 (65.56%) | 23 (25.56%) | 36 (40.00%) | |
| Positive | 31 (34.44%) | 20 (22.22%) | 11 (12.22%) | |
| Distant metastasis | 0.1063 | |||
| Negative | 82 (91.11%) | 37 (41.11%) | 45 (50.00%) | |
| Positive | 8 (8.89%) | 6 (6.67%) | 2 (2.22%) | |
| TNM stage | 0.0034 | |||
| I-II | 48 (53.33%) | 16 (17.78%) | 32 (35.56%) | |
| III-IV | 42 (46.67%) | 27 (30.00%) | 15 (16.67%) | |
| Grade | 0.0121 | |||
| I-II | 76 (84.44%) | 32 (35.56%) | 44 (48.89%) | |
| III | 14 (15.56%) | 11 (12.22%) | 3 (3.33%) | |
Serum sample harvesting
Whole blood sample was obtained from each participant and the processed with 1 h after collection. Briefly, the samples were centrifuged at 3000 g for 10 min at room temperature, followed by 10,000 g for 5 min. The serum samples were flash-frozen into liquid nitrogen and thereafter stored at -80°C.
RNA isolation and quantitative reverse transcription-polymerase chain reaction (RT-qPCR)
Total RNA was isolated from serum samples using miRNeasy Mini kits (Qiagen, Inc., Valencia, CA, USA), following the manufacturer’s protocol. The quality and quantity of isolated RNA was assessed using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE, USA). Reverse transcription of total RNA was performed using miScript reverse Transcription Kit (Qiagen). miScript SYBR Green PCR Kit (Qiagen) was used to amplify the first strand cDNA. qRT-PCR was carried out on an Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) with the following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. Fold-change was calculated using the delta Ct method. U6 small nuclear RNA was used as an internal control to normalize the amount of miR-193b. Each sample was run in triplicate.
Statistical analysis
All statistical analyses were performed by GraphPad Prism 6 software (GraphPad Software, Inc., San Diego, CA, USA). Mann-Whitney U test was used to compare the relative expression of serum miR-193b between CRC patients and healthy volunteers. The relationship between expression of serum miR-193b and clinicopathologic characteristics was examined using χ2 test. Receiver operating characteristic (ROC) curve was drawn to examine the effectiveness of serum miR-193b in distinguishing CRC from healthy volunteers. Survival curves were plotted using the Kaplan-Meier method and compared by log-rank test. Univariate and multivariate analyses were performed to identify the independent prognostic factors. A P-value less than 0.05 was considered as statistically significant.
Results
Serum miR-193b expression is reduced in CRC patients versus healthy volunteers
To evaluate the expression level of serum miR-193b, serum samples from 90 CRC patients and 40 healthy controls were used. The results of qRT-PCR showed that the expression levels of serum miR-193b were dramatically decreased in CRC patients compared to the healthy volunteers (**P<0.01) (Figure 1). Then ROC analysis was performed to examine the diagnostic performance of serum miR-193b, the results revealed that serum miR-193b was able to discriminate CRC patients from healthy volunteers with relative high accuracy (Sensitivity =76.67%, Specificity =80.00%, area under the curve (AUC) =0.853) (Figure 2).
Figure 1.
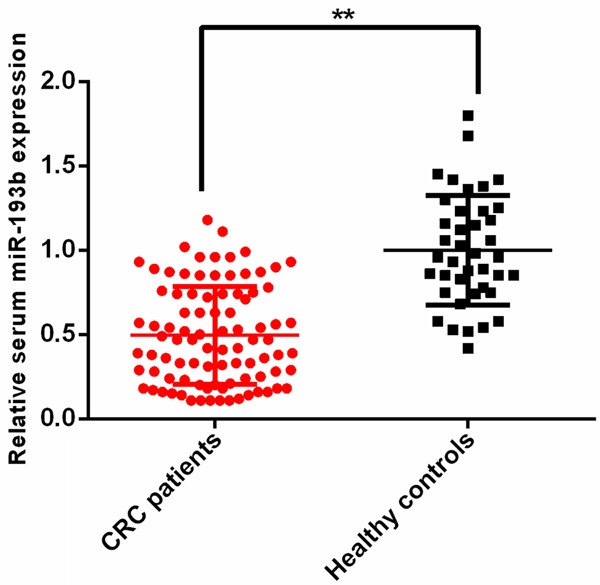
The expression levels of serum miR-193b in CRC patients and healthy controls.
Figure 2.
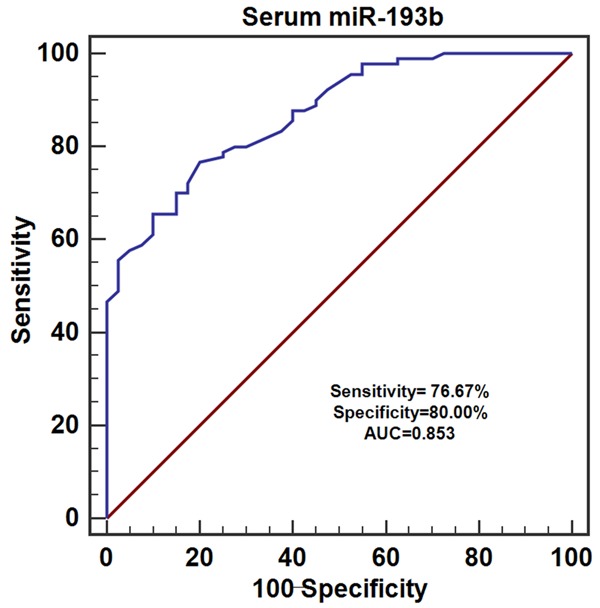
The diagnostic value of serum miR-193b in CRC.
Serum miR-193b levels was sensitive to surgery therapy
Seventy-five CRC patients received surgery therapy. We compared the expression levels of serum miR-193b before the operation and after surgery. Our results demonstrated that serum miR-193b levels were remarkably upregulated in CRC patients who received surgery treatment (**P<0.01) (Figure 3A). However, no significance in miR-193b levels was found in the other 15 stage IV CRC patients who did not receive surgery (P>0.05) (Figure 3B).
Figure 3.
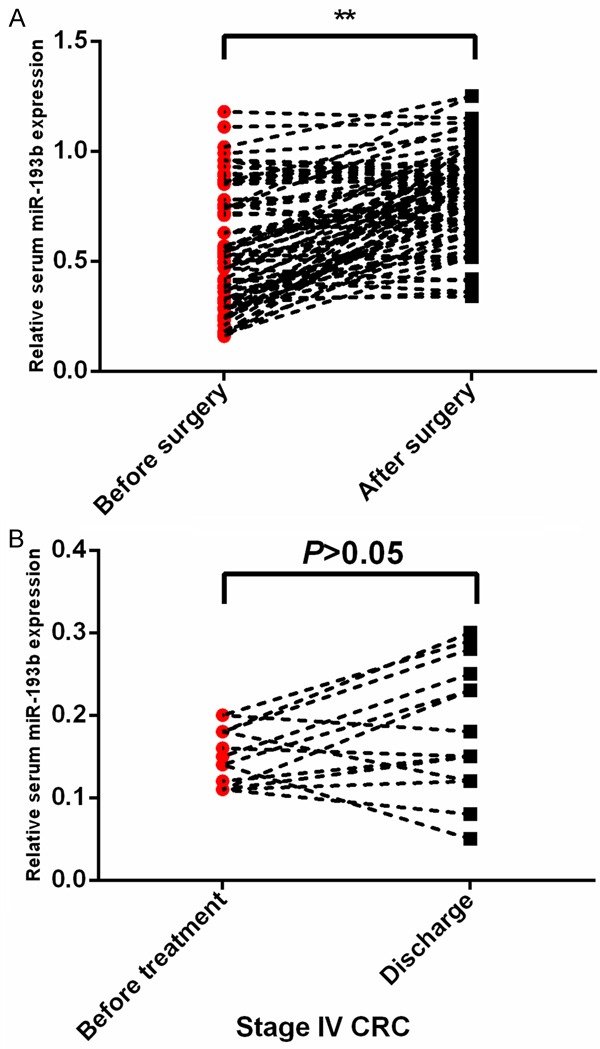
Serum miR-193b levels were increased in CRC patients who received surgery treatment.
Association between serum miR-193b levels and clinicopathological parameters of CRC
Chi-square test was used to reveal the correlation between serum miR-193b levels and clinicopathological parameters of CRC. The results showed that serum miR-193b was associated with lymph node metastasis (P=0.0212), TNM stage (P=0.0034), and grade (P=0.0121). While it was not correlated with age, gender, depth of invasion, tumor diameter, distant metastasis (Table 1).
Association of serum miR-193b levels with survival in patients with CRC
The Kaplan-Meier survival curve and the log-rank test were applied to explore whether serum miR-193b levels were correlated with survival. As anticipated, patients with lower levels of serum miR-193b had significantly worse five year overall survival (P=0.0026; log-rank test). Moreover, patients in the low serum miR-193b group also suffered a shorter five year disease free survival (P=0.0002; log-rank test) (Figure 4A, 4B).
Figure 4.
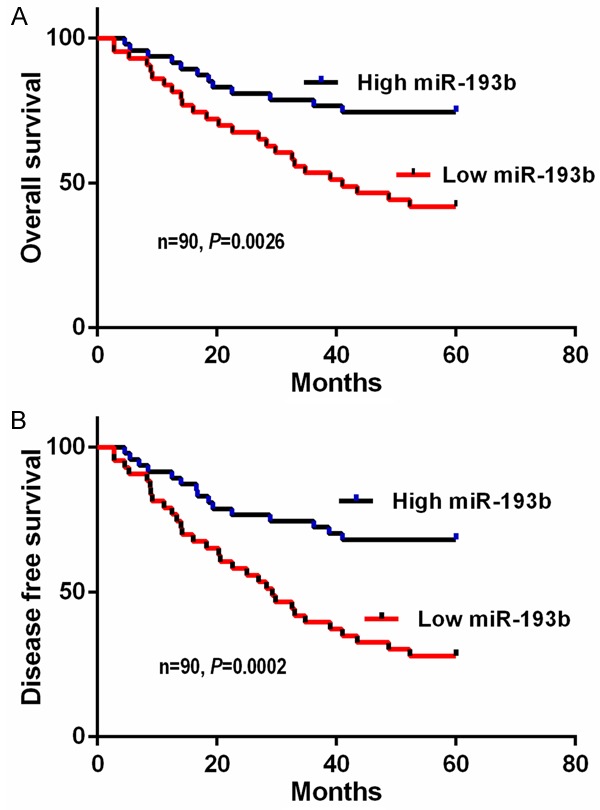
Kaplan-Meier survival analysis in CRC patients based upon serum miR-193b levels.
To further evaluate the prognostic value of serum miR-193b, the univariate and multivariate Cox regression analyses were performed. The univariate Cox regression analysis showed that TNM stage, grade and serum miR-193b were the prognostic factors for the CRC patients. The multivariate Cox regression analysis revealed that serum miR-193b was an independent risk factor for CRC (HR=2.498, 95% CI=1.381-4.140, P=0.021) (Table 2).
Table 2.
Univariate and multivariate analyses of prognostic factors predicting the overall survival of CRC patients
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| HR | 95% CI | P | HR | 95% CI | P | |
| Serum miR-193b | ||||||
| Low vs high | 2.643 | 1.425-4.309 | 0.013 | 2.498 | 1.381-4.140 | 0.021 |
| Gender | ||||||
| Male vs female | 1.210 | 0.887-1.831 | 0.318 | |||
| Age | ||||||
| ≥60 vs <60 | 1.294 | 0.913-1.927 | 0.283 | |||
| Tumor size (cm) | ||||||
| ≥5 vs <5 | 1.519 | 0.944-2.415 | 0.192 | |||
| Depth of invasion | ||||||
| T3/T4 vs T1/T2 | 1.435 | 0.932-2.207 | 0.215 | |||
| LN metastasis | ||||||
| Yes vs no | 1.711 | 0.991-2.853 | 0.056 | |||
| Distant metastasis | ||||||
| Yes vs no | 1.683 | 0.984-2.720 | 0.072 | |||
| TNM stage | ||||||
| III-IV vs I-II | 3.120 | 1.941-5.752 | 0.006 | 3.418 | 1.861-6.346 | 0.005 |
| Grade | ||||||
| III vs I-II | 1.921 | 1.080-3.282 | 0.043 | 1.747 | 0.990-3.050 | 0.053 |
Discussion
Our results showed that serum miR-193b was significantly reduced in CRC patients and found to correctly discriminate CRC patients from healthy controls with an AUC value of 0.853. Serum miR-193b levels were remarkably elevated in CRC patients who received surgery treatment. We also investigated the association of serum miR-193b expression with clinicopathological features. Low serum miR-193b levels were correlated with multiple parameters including differentiation, TNM stage and lymph node metastasis. Survival analysis revealed that CRC patients with lower serum miR-193b expression had worse outcome. Moreover, multivariate analysis showed that serum miR-193b was an independent prognostic factor of overall survival.
These data indicate that miR-193b acts as a tumor suppressor in CRC and downregulation of miR-193b promotes its development. Serum miR-193b might serve as a potential prognostic biomarker for patients with CRC. Despite these findings, investigations at a large-scale are needed to validate the clinical significance of serum miR-193b in CRC. In addition, serum miR-193b levels are also found to increase in other diseases such as prediabetes, sepsis, chronic hepatitis B infection [14-16]. Thus these factors are required to be considered when interpreting the serum miR-193b levels in patients with CRC.
Consistent with results of previous study, miR-193b was significantly reduced in CRC tissues compared with normal controls. In addition, low tissue miR-193b expression was associated with poorer overall survival. Ectopic expression of miR-193b suppressed the proliferation and invasive capacity of cancer cells and vice versa. Moreover, stathmin 1 was identified as a downstream target of miR-193b [17]. miR-193b has also been reported to function as a tumor suppressor in other types of cancer. For instance, it was common to observe that miR-193b was methylated in prostate cancer samples compared with benign prostate hyperplasia. Lower miR-193b levels were associated with more advanced pathological T stage, indicating miR-193b methylation might account for the initiation and progression of prostate cancer [18]. Similarly, miR-193b was remarkably reduced in breast cancer cell lines. Overexpression of miR-193b inhibited the proliferation, migration and invasion capacity of breast cancer cells. DNAJC13 and RAB22A was demonstrated to be the downstream targets [19].
However, miR-193b also plays an oncogenic role in some types of cancer. Serum miR-193b was significantly increased in patients with non-small cell lung cancer (NSCLC) compared with healthy volunteers, indicating that miR-193b might act as an oncomiR in NSCLC [20]. miR-193b expression was upregulated in both glioma tissues and cell lines, In addition, downregulation of miR-193b inhibited proliferation capacity of cancer cells by targeting Smad3 [21]. Similarly, miR-193b was overexpressed in head and neck squamous cell carcinomas (HNSCC) tissues and cell lines. Knockdown of miR-193b suppressed the proliferation, migration and invasion capacity in vitro as well as reduced tumor growth in vivo [22]. Therefore, the role of miR-193b is cancer type dependent.
In conclusion, we have observed for the first time the prognostic significance of serum miR-193b in CRC. Serum miR-193b might be a novel and promising biomarker for CRC. Future studies are required to interpret the underlying molecular mechanisms.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant nos.81672427).
Disclosure of conflict of interest
None.
References
- 1.Weitz J, Koch M, Debus J, Höhler T, Galle PR, Büchler MW. Colorectal cancer. Lancet. 2005;365:153–165. doi: 10.1016/S0140-6736(05)17706-X. [DOI] [PubMed] [Google Scholar]
- 2.Zhang B, Wang Q, Pan X. MicroRNAs and their regulatory roles inanimals and plants. J Cell Physiol. 2007;210:279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg RM, Rothenberg ML, Van Cutsem E, Benson AB 3rd, Blanke CD, Diasio RB, Grothey A, Lenz HJ, Meropol NJ, Ramanathan RK, Becerra CH, Wickham R, Armstrong D, Viele C. The continuum of care: a paradigm for the management of metastatic colorectal cancer. Oncologist. 2007;12:38–50. doi: 10.1634/theoncologist.12-1-38. [DOI] [PubMed] [Google Scholar]
- 4.Yang M, Li Y, Padgett RW. MicroRNAs: Small regulators with a big impact. Cytokine Growth Factor Rev. 2005;16:387–393. doi: 10.1016/j.cytogfr.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11:145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zekri AR, Youssef AS, Lotfy MM, Gabr R, Ahmed OS, Nassar A, Hussein N, Omran D, Medhat E, Eid S, Hussein MM, Ismail MY, Alenzi FQ, Bahnassy AA. Circulating serum miRNAs as diagnostic markers for colorectal cancer. PLoS One. 2016;11:e0154130. doi: 10.1371/journal.pone.0154130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menéndez P, Padilla D, Villarejo P, Palomino T, Nieto P, Menéndez JM, Rodríguez-Montes JA. Prognostic implications of serum microRNA-21 in colorectal cancer. J Surg Oncol. 2013;108:369–373. doi: 10.1002/jso.23415. [DOI] [PubMed] [Google Scholar]
- 11.Jin X, Sun Y, Yang H, Li J, Yu S, Chang X, Lu Z, Chen J. Deregulation of the miR-193b-KRAS axis contributes to impaired cell growth in pancreatic cancer. PLoS One. 2015;10:e0125515. doi: 10.1371/journal.pone.0125515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitra AK, Chiang CY, Tiwari P, Tomar S, Watters KM, Peter ME, Lengyel E. Microenvironmentinduced downregulation of miR-193b drives ovarian cancer metastasis. Oncogene. 2015;34:5923–5932. doi: 10.1038/onc.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gastaldi C, Bertero T, Xu N, Bourget-Ponzio I, Lebrigand K, Fourre S, Popa A, Cardot-Leccia N, Meneguzzi G, Sonkoly E, Pivarcsi A, Mari B, Barbry P, Ponzio G, Rezzonico R. miR-193b/365a cluster controls progression of epidermal squamous cell carcinoma. Carcinogenesis. 2014;35:1110–1120. doi: 10.1093/carcin/bgt490. [DOI] [PubMed] [Google Scholar]
- 14.Párrizas M, Brugnara L, Esteban Y, González-Franquesa A, Canivell S, Murillo S, Gordillo-Bastidas E, Cussó R, Cadefau JA, García-Roves PM, Servitja JM, Novials A. Circulating miR-192 and miR-193b are markers of prediabetes and are modulated by an exercise intervention. J Clin Endocrinol Metab. 2015;100:407–415. doi: 10.1210/jc.2014-2574. [DOI] [PubMed] [Google Scholar]
- 15.Wang HJ, Zhang PJ, Chen WJ, Feng D, Jia YH, Xie LX. Four serum microRNAs identified as diagnostic biomarkers of sepsis. J Trauma Acute Care Surg. 2012;73:850–854. doi: 10.1097/TA.0b013e31825a7560. [DOI] [PubMed] [Google Scholar]
- 16.Ninomiya M, Kondo Y, Kimura O, Funayama R, Nagashima T, Kogure T, Morosawa T, Tanaka Y, Nakayama K, Shimosegawa T. The expression of miR-125b-5p is increased in the serum of patients with chronic hepatitis B infection and inhibits the detection of hepatitis B virus surface antigen. J Viral Hepat. 2016;23:330–339. doi: 10.1111/jvh.12522. [DOI] [PubMed] [Google Scholar]
- 17.Guo F, Luo Y, Mu YF, Qin SL, Qi Y, Qiu YE, Zhong M. miR-193b directly targets STMN1 and inhibits the malignant phenotype in colorectal cancer. Am J Cancer Res. 2016;6:2463–2475. [PMC free article] [PubMed] [Google Scholar]
- 18.Kaukoniemi KM, Rauhala HE, Scaravilli M, Latonen L, Annala M, Vessella RL, Nykter M, Tammela TL, Visakorpi T. Epigenetically altered miR-193b targets cyclin D1 in prostate cancer. Cancer Med. 2015;4:1417–1425. doi: 10.1002/cam4.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Z, He M, Wang K, Sun G, Tang L, Xu Z. Tumor suppressive microRNA-193b promotes breast cancer progression via targeting DNAJC13 and RAB22A. Int J Clin Exp Pathol. 2014;7:7563–7570. [PMC free article] [PubMed] [Google Scholar]
- 20.Nadal E, Truini A, Nakata A, Lin J, Reddy RM, Chang AC, Ramnath N, Gotoh N, Beer DG, Chen G. A novel serum 4-microRNA signature for lung cancer detection. Sci Rep. 2015;5:12464. doi: 10.1038/srep12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong Q, Wang T, Lu P, Zhang R, Zou J, Yuan S. miR-193b promotes cell proliferation by targeting Smad3 in human glioma. J Neurosci Res. 2014;92:619–626. doi: 10.1002/jnr.23339. [DOI] [PubMed] [Google Scholar]
- 22.Lenarduzzi M, Hui AB, Alajez NM, Shi W, Williams J, Yue S, O’Sullivan B, Liu FF. MicroRNA-193b enhances tumor progression via down regulation of neurofibromin 1. PLoS One. 2013;8:e53765. doi: 10.1371/journal.pone.0053765. [DOI] [PMC free article] [PubMed] [Google Scholar]


