Abstract
It has been found that the abnormal expression of miR-23b is related to the poor prognosis or the weak ability of metastasis in colorectal cancer. Furthermore, high expression of PDE7A in tumors is known to be related to poor prognosis. The aim of this study was to explain whether miR-23b was involved in the invasion and metastasis of colon cancer by regulating PDE7A gene. The expression levels of miR-23b and PDE7A in colon tissues and colon cancer cell lines (SW620 and SW480 cells) were measured by real time PCR. The PDE7A protein level was determined by Western blotting. The relationship between miR-23b and PDE7A was verified by cell transfection and luciferase reporter assay. The effect of miR-23b or PDE7A level on cell invasion and migration was determined by Transwell assay. miR-23b was down-regulated when PDE7A was up-regulated in colon cancer tissues and cells. The result of dual luciferase reporter assay showed that PDE7A was a direct target gene of miR-23b. Overexpression of miR-23b not only reduced the expression of PDE7A in SW620 cells or SW480 cells, but also weakened the migration and invasion abilities of SW620 cells or SW480 cells. When SW620 cells or SW480 cells were transfected with si-PDE7A, the level of PDE7A increased and the migration and invasion abilities of cells diminished. In conclusion, miR-23b is lowly expressed in colon cancer tissues and cells, and miR-23b may play an inhibitory role in the development of colon cancer through down-regulating the expression of PDE7A.
Keywords: miR-23b, colon cancer, PDE7A, migration, invasion
Introduction
MiRNAs have been extensively studied as a short, noncoding single-stranded RNA that regulate the expression of target genes. The human miRNA gene has been found to account for about 1% to 3% of the human genome [1], and 30% to 60% of the human protein genes are regulated by miRNAs [2,3]. A single miRNA may bind up to 200 different functional targets, including transcription factors, receptors, vectors [4]. More than 186 miRNA genes have been localized in the region of tumor-associated chromosomal rearrangements [5], and some changes in specific miRNA expression genes have been occurred in some of the cancer-specific chromosomal fragility sites [6,7].
Many miRNA targets are proto-oncogenes or tumor suppressor genes [8]. The abnormal expression of miRNA is a common feature of tumorigenesis. HnRNPA1 connexin can bind to miR-18a to promote the expression of miR-18a and the development of tumor [9]. Bic (B-cell integration cluster) gene is one of the known oncogenes, and its encoded RNA can be further processed to form miR-155, miR-155 precursor RNA and bic mRNA are highly expressed in children Burkitt’s lymphoma [10]. Invasion and metastasis is one of the hallmarks of malignancy and the leading cause of death in cancer patients. Studies have confirmed that miRNAs are involved in the whole process of tumor metastasis. Upregulation of miR-29a expression can promote the development of EMT and lead to tumor metastasis [11]. MiR-9 can work directly on the CDH1 gene encoding E-cadherin and increase the invasion and metastasis of cells [12]. The miRNA expression profile is closely related to the embryonic origin of the tumor, and its expression is more tissue-specific, so miRNAs can be used as clinical markers for effective tumor diagnostic markers. Lu et al. proposed the use of whole-gene miRNA expression profiles in identifying poorly differentiated tumors [13], which are difficult to identify by imaging methods and mRNA-based diagnostic methods. In addition, miRNAs and their target genes can serve as effective drug targets for tumor treatment. The inhibition of oncogene microRNAs in mice with different organs injected with AMOs (an anti-microRNA oligonucleotide) leads to inhibitory action on tumor growth [14].
MiR-23b has been found to be lowly expressed in a variety of tumor tissues such as prostate cancer [15], oral squamous cell carcinoma [16], bladder cancer [17] and can act as a tumor suppressor miRNA in hepatocellular carcinoma [18], malignant glioma [19] and ovarian cancer [20] through inhibiting the growth, proliferation, migration and invasion of tumor cells. Similarly, miR-23b is also found to be highly expressed in gastric cancer and associated with poor prognosis of gastric cancer [21]. In order to further understand the development mechanism of colon cancer [22], this study examined the miR-23b expression in colon cancer tissue and colon cancer cell lines. In addition, miR-23b target gene was searched and verified by target gene prediction software and dual luciferase reporter assay, the function of miR-23b and its target gene in colon cancer cell line were analyzed too.
Materials and methods
Clinical specimens
A total of 30 primary colon cancer tissues and its paired non-cancerous colon tissues were collected from the Renmin Hospital of Wuhan University. All patients had provided written consent and the study had been approved by the Renmin Hospital of Wuhan University Institutional Review Committee. All tissues have been histologically confirmed as colon adenocarcinoma. Tissue samples were collected, frozen quickly in liquid nitrogen, and stored at -80°C until further analysis.
Cell culture and transfection
The human colon cancer cell lines SW620 cells and SW480 cells were cultured at 37°C in a 5% CO2 atmosphere and maintained in DMEM containing 10% FBS and 2 mM L-glutamine (Invitrogen, CA, USA). Human miR-23b mimics and negative control oligonucleotide (NC) were designed and provided by GenePhram (Suzhou, China). Small interfering RNA of PDE7A (si-PDE7A) and negative control RNA (siRNA-NC) were synthesized and purified by Genifarma too. SW480 cells and SW620 cells were transfected with miR-23b mimics or NC, and si-PDE7A or siRNA-NC using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocol.
Dual luciferase reporter assay
Candidate target genes of miR-23b were identified by TargetScan, and we chose the PDE7A to perform the follow-up experiments because that PDE7A had been reported to play a role in migration and invasion of cancer cells [23]. To construct the PDE7A 3’UTR plasmid, the full-length 3’UTR of human PDE7A mRNA containing the putative miR-23b binding site was cloned into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, WI, USA). The putative miR-23b recognition site in PDE7A 3’UTR was mutated by site-directed mutagenesis. For dual luciferase assay, SW620 cells were transfected with 100 nM miR-23b mimics or negative control using Lipofectamine 2000. After 24 hours, 150 ng of pmir-GLO-PDE7A WT or pmir-GLO-PDE7A mut were transfected in these cells. After 48 hours, cell extracts were prepared and the detection of relative luciferase activity was performed using Dual-Luciferase® Reporter Assay System (Promega) according to the manufacturer’s protocol.
Cell migration and invasion assay
Transfection wells (BD Biosciences, NJ, USA) containing an uncoated or matrix adhesive coating containing 8 μm pores were used in cell migration or invasion assays. Cells transfected with 100 nM miR-149 mimics/NC or 100 nM siRNA-PDE7A/si-NC were seeded into the upper chamber of serum-free DMEM. After 24 hours the cells were fixed in 100% methanol and stained with crystal violet at room temperature for 30 minutes. The cells remaining on the filter side were removed with a cotton swab. The filter was then imaged under an inverted microscope and the number of migrating or invading cells was counted from these images.
QRT-PCR analysis
Total RNA was extracted from colon cancer cells or tumor tissues using TRIZOL reagent (Invitrogen). CDNA synthesis kit (Takara, Tokyo, Japan) was used for the synthesis of cDNA according to the manufacturer’s instructions. Quantitative RT-PCR was used to detect the expression levels of miR-23b and PDE7A mRNA, and was performed using the Light Cycler 480 detection system (Roche Diagnostics, IN, USA). U6 snRNA and GAPDH mRNA levels were detected and used for standardization. Primer sequences are listed in Table 1.
Table 1.
The sequences of primers
| Gene | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| MiR-23b | GAGCATCACATTGCCAGGG | GTGCAGGGTCCGAGGT |
| PDE7A | GGAAATAGTCTAGTAAGCTTAACC | GGCAGATGTGAGAATAAGCCTG |
| U6 | CGCTTCGGCAGCACATATAC | TTCACGAATTTGCGTGTCAT |
| GAPDH | TGAAGGTCGGAGTCAACGGATTTGGT | CATGTGGGCCATGAGGTCCACCAC |
Western blot analysis
Total protein was prepared from colon cancer tissues and colon cancer cells. The protein concentration was determined using a Bio-Rad protein assay system (Bio-Rad, CA, USA). Proteins were analyzed by SDS polyacrylamide gel electrophoresis. After electrophoresis, proteins were transferred onto NC membrane. The membrane containing protein was incubated with the appropriate antibody before scanning protein bands. The primary antibodies [anti-PDE7A antibody (ab154857), anti-GAPDH antibody (ab9485)] and horseradish peroxidase (HRP) conjugated secondary antibody [goat anti-Rabbit IgG H&L (HRP) (ab6721)] were obtained from Abcam and used according to the manufacturer’s instructions.
Statistical analysis
The concentration of the vector and the concentration of RNA and protein from each tissue or cell sample were detected three times and the mean value was used for subsequent experiments. Every treatment in the dual luciferase reporter assay was performed three times to achieve three values. Every treatment in Transwell assay was performed three times and every number of cell count was the mean value from 5 random sights. QRT-PCR was performed in triplicate. Data were analyzed by Student’s t-test using SPSS 16.0 statistical software (SPSS, NY, USA) to determine the statistical significance. The results were expressed as mean ± SD. P < 0.05, the results were considered statistically significant.
Results
Decreased expression of miR-23b and increased expression of PDE7A in colon cancer tissues and cells
The expression level of miR-23b in colon cancer tissues, adjacent non-cancerous tissues and colon cancer cell lines (SW620, SW480) were detected by qRT-PCR, and the results showed that miR-23b was down regulated in colon cancer tissues or colon cancer cell lines (SW620, SW480) compared with the adjacent non-cancerous tissues (P < 0.001) (Figure 1A). Meanwhile, the mRNA and protein levels of PDE7A in colon cancer tissues, adjacent non-cancerous tissues and colon cancer cell lines (SW620, SW480) were detected by qRT-PCR and western blot, respectively. As shown in Figure 1B and 1C, the mRNA level of PDE7A in colon cancer tissues or colon cancer cell lines (SW620, SW480) was significantly higher than that in adjacent non-cancerous tissues (P < 0.001) as well as the protein level of PDE7A. These results suggest the low expression of miR-23b and high expression of PDE7A in colon cancer tissues and colon cancer cell lines SW620 cells and SW480 cells.
Figure 1.
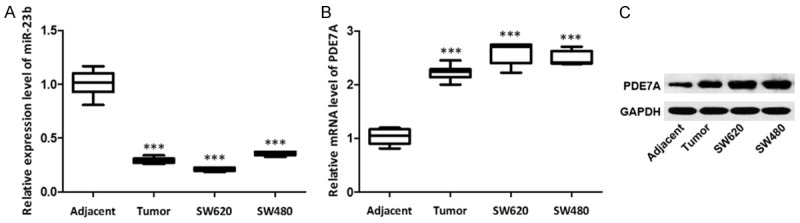
Down-regulation of miR-23b expression and up-regulation of PDE7A expression in colon cancer tissues and cells. A. miR-23b expression in colon cancer tissues and cells compared with adjacent tissues; B. PDE7A mRNA levels in colon cancer tissues and cells compared with adjacent tissues; C. PDE7A protein levels were significantly elevated in colon cancer tissues. ***P < 0.001, compared with adjacent tissues.
PDE7A is a direct target gene of miR-23b in colon cancer cells
To find the relationship between miR-23b and PDE7A in colon cancer cells, the miR-23b level was enhanced both in SW620 cells and SW480 cells by being transfected with miR-23b mimics (Figure 2A). Then the mRNA and protein levels of PDE7A in SW620 cells and SW480 cells were detected by qRT-PCR and western blot, the results showed that PDE7A mRNA level was obviously reduced in SW620 cells and SW480 cells with high miR-23b level (P < 0.01, P < 0.05) (Figure 2B). The protein level of PDE7A in SW620 cells and SW480 cells with high miR-23b level was also lower than that in SW620 and SW480 cells transfected with NC (Figure 2C). Furthermore, the result of dual luciferase reporter assay showed that miR-23b could directly target to the 3’-UTR of PDE7A mRNA and down-regulated the relative activity of luciferase (P < 0.05) (Figure 2D). These results suggest that PDE7A is a direct target gene of miR-23b and miR-23b can down-regulate the expression of PDE7A in SW620 cells and SW480 cells.
Figure 2.
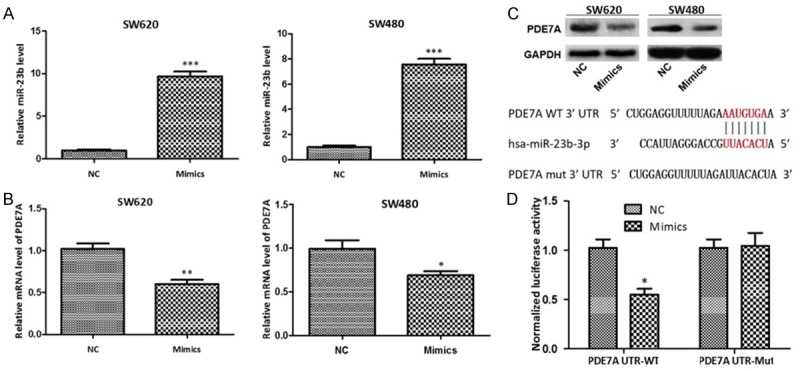
MiR-23b negatively regulates the PDE7A expression in colon cancer cells. MiR-23b mimics effectively increased the miR-23b level (A) and reduced the mRNA level (B) and protein level (C) of PDE7A in colon cancer cells; (D) The miR-23b targeting on the 3’UTR of PDE7A was predicted by the online tool (http://www.targetscan.org/) and verified by the luciferase reporter assay. *P < 0.05, **P < 0.01, ***P < 0.001, compared with negative control.
Overexpression of miR-23b weaken the migration and invasion ability of colon cancer cells
The effect of high level of miR-23b on the migration and invasion abilities of colon cancer cells was tested by Transwell assay, and the result showed that up-regulation of miR-23b reduced numbers of migrated SW620 cells (48 ± 3 vs. 101 ± 8, P < 0.01) and migrated SW480 cells (52 ± 5 vs. 100 ± 13, P < 0.05) (Figure 3A). Similarly, the aggressive cell numbers of SW620 cells or SW480 cells transfected with miR-23b mimics were dramatically reduced than that of SW620 and SW480 cells transfected with NC (45 ± 5 vs. 100 ± 10, P < 0.01; 38 ± 4 vs. 101 ± 12, P < 0.01) (Figure 3B). The above results indicate that high level of miR-23b can weak the migration and invasion abilities of SW620 cells and SW480 cells.
Figure 3.
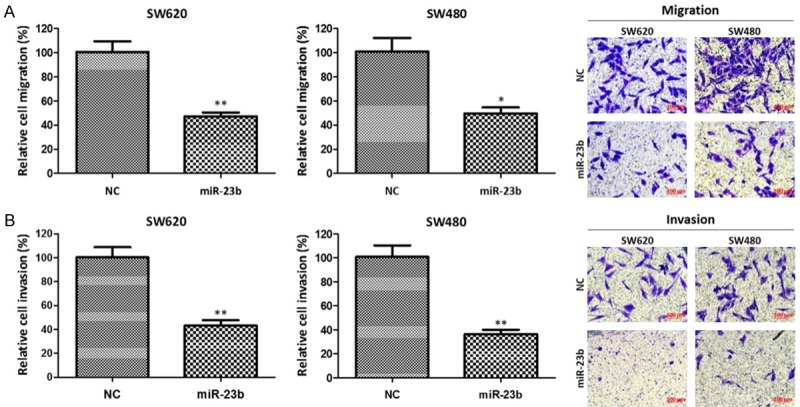
MiR-23b overexpression weakened migration and invasion of colon cancer cells. A. The effect of miR-23b overexpression on cell migration. B. The effect of miR-23b overexpression on cell invasion. *P < 0.05, **P < 0.01, compared with negative control.
Downregulation of PDE7A reduces the migration and invasion ability of colon cancer cells
The effects of si-PDE7A on mRNA level and protein level of PDE7A were tested by cell transfection, qRT-PCR and western blot. The results showed that PDE7A expression in SW620 cells and SW480 cells transfected with si-PDE7A-1 or si-PDE7A-2 was down-regulated both in mRNA level and protein level (P < 0.01, P < 0.05) (Figure 4A, 4B). In addition, the results of Transwell assay showed that migrated cell number was significantly reduced in SW620 cells or SW480 cells transfected with si-PDE7A-1 or si-PDE7A-2 than that in SW620 cells or SW480 cells transfected with NC (P < 0.01, P < 0.05) (Figure 4C); Invaded cells transfected with si-PDE7A-1 or si-PDE7A-2 were also decreased in SW620 cells or SW480 cells (P < 0.01, P < 0.05) (Figure 4D). These results suggest that si-PDE7A-1 and si-PDE7A-2 can down-regulate the PDE7A expression in SW620 cells or SW480 cells and weaken the migration and invasion of SW620 cells or SW480 cells.
Figure 4.
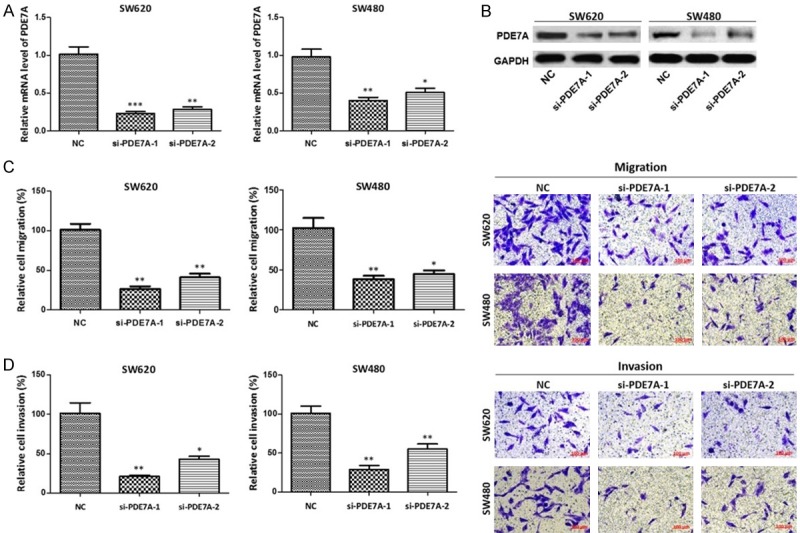
Knockdown of PDE7A weakened migration and invasion of colon cancer cells. A. The mRNA level of PDE7A in colon cancer cells transfected with si-PDE7A; B. The protein expression of PDE7A in colon cancer cells transfected with si-PDE7A; C. The effect of PDE7A knockdown on cell migration; D. The effect of PDE7A knockdown on cell invasion. *P < 0.05, **P < 0.01, compared with negative control.
Discussion
About 655,000 people die from colorectal cancer each year, and the incidence of colorectal cancer is on the rise. The 5-year survival rate for early colorectal cancer is about 90%, and this data drops to 15% in metastatic colorectal cancer [24]. Despite the development of many early screening and treatment programs in recent years, the survival rate of colorectal cancer has been not improved significantly over the past 20 years, and the introduction of invasive metastasis is the most common cause of treatment failure. The mechanism of tumor metastasis is complex, involving the invasion and adhesion of cancer cells, epithelial mesenchymal transition (EMT), extracellular matrix degradation, angiogenesis and microenvironmental chemotaxis. Which are affected by the abnormal activation or inactivation of many oncogenes or tumor suppressor genes and related signaling pathways. The activation or inactivation of oncogenes or tumor suppressor genes is regulated at transcriptional level, post-transcriptional and translational level, respectively. The miRNA regulates the target gene expression from the post-transcriptional level by completely or incompletely pairing with the 3’-UTR of target mRNA, causing the degradation or inhibited translation of target mRNA.
Studies have shown that miRNAs regulate the expression of at least 30% of the human protein-encoding genes [25] and play a greater role than protein-encoding genes [26]. In a variety of human tumors, miRNAs are characterized by abnormal expression due to gene mutation or gene fragility sites [27]. MiR-23b is currently known to be abnormally expressed in hepatocellular carcinoma cells, which reduce the proliferation and migration ability of hepatocellular carcinoma cells [18]. Our study find the low level of miR-23b in colon cancer tissues and cells, suggesting that miR-23b may play a tumor suppression role during the development of colon cancer. The colon cancer cells transfected with miR-23b mimics show high level of miR-23b and weaken migration or invasion ability, which indicate that miR-23b could exert the anti-tumor effect by inhibiting the migration and invasion of colon cancer cells.
PDE7A is a phosphodiesterase that regulates the cellular levels of cAMP and cGMP [28-30]. Low level of cAMP and high level of PDE were found in multiple cancer cells, whereas PDE7A was found to be overexpressed in lymphocytic leukemia and endometrial cancer [23,31], indicating that PDE7A may also be involved in the development of cancer. In this study, we find that low expression of miR-23b and overexpression of PDE7A in colon cancer tissues and cells, and when transfected with miR-23b mimics, colon cancer cells show high level of miR-23b and low level of PDE7A, indicating that the PDE7A level is affected by miR-23b level in colon cancer cells. PDE7A is a target of miR-23b, and miR-23b directly regulates PDE7A expression has been confirmed by dual luciferase reporter assay. In view of the role of miR-23b in colon cancer cells, PDE7A, which is negatively regulated by miR-23b, may act as an oncogene in colon cancer cells. And this is also confirmed by RNA interference test and Transwell assay.
In conclusion, our results suggest that miR-23b is lowly expressed in colon cancer tissues and cells, whereas PDE7A is highly expressed in colon cancer tissues and cells. Our further study shows that PDE7A is a target gene of miR-23b and the expression of PDE7A is negatively regulated by miR-23b. And increased miR-23b level or decreased PDE7A level both can suppress the migration and invasion ability of colon cancer cells. This suggests that miR-23b may function as a tumor suppressor in the development of colon cancer by regulating the expression of PDE7A. Our study contributes to revealing the development mechanisms of colon cancer and studying the treatment strategy for colon cancer.
Acknowledgements
This study was supported by National Natural Science Foundation of China (Grant No.: 81672387).
Disclosure of conflict of interest
None.
References
- 1.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao N, Lye KW, Barton MK. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell. 2004;7:653–662. doi: 10.1016/j.devcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell’Aquila ML, Alder H, Rassenti L, Kipps TJ, Bullrich F, Negrini M, Croce CM. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O’Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryazansky SS, Gvozdev VA. Small RNAs and cancerogenesis. Biochemistry (Mosc) 2008;73:514–527. doi: 10.1134/s0006297908050040. [DOI] [PubMed] [Google Scholar]
- 9.Koo JH, Lee HJ, Kim W, Kim SG. Endoplasmic reticulum stress in hepatic stellate cells promotes liver fibrosis via PERK-mediated degradation of HNRNPA1 and up-regulation of SMAD2. Gastroenterology. 2016;150:181–193. e188. doi: 10.1053/j.gastro.2015.09.039. [DOI] [PubMed] [Google Scholar]
- 10.van den Berg A, Kroesen BJ, Kooistra K, de Jong D, Briggs J, Blokzijl T, Jacobs S, Kluiver J, Diepstra A, Maggio E, Poppema S. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer. 2003;37:20–28. doi: 10.1002/gcc.10186. [DOI] [PubMed] [Google Scholar]
- 11.Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009;10:400–405. doi: 10.1038/embor.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, Westermann F, Speleman F, Vandesompele J, Weinberg RA. miR-9, a MYC/MYCN-activated microRNA, regulates Ecadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 14.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castillo RC, MacKenzie EJ, Archer KR, Bosse MJ, Webb LX, Group LS. Evidence of beneficial effect of physical therapy after lower-extremity trauma. Arch Phys Med Rehabil. 2008;89:1873–1879. doi: 10.1016/j.apmr.2008.01.032. [DOI] [PubMed] [Google Scholar]
- 16.Fukumoto I, Hanazawa T, Kinoshita T, Kikkawa N, Koshizuka K, Goto Y, Nishikawa R, Chiyomaru T, Enokida H, Nakagawa M, Okamoto Y, Seki N. MicroRNA expression signature of oral squamous cell carcinoma: functional role of microRNA-26a/b in the modulation of novel cancer pathways. Br J Cancer. 2015;112:891–900. doi: 10.1038/bjc.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itesako T, Seki N, Yoshino H, Chiyomaru T, Yamasaki T, Hidaka H, Yonezawa T, Nohata N, Kinoshita T, Nakagawa M, Enokida H. The microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. PLoS One. 2014;9:e84311. doi: 10.1371/journal.pone.0084311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salvi A, Sabelli C, Moncini S, Venturin M, Arici B, Riva P, Portolani N, Giulini SM, De Petro G, Barlati S. MicroRNA-23b mediates urokinase and c-met downmodulation and a decreased migration of human hepatocellular carcinoma cells. FEBS J. 2009;276:2966–2982. doi: 10.1111/j.1742-4658.2009.07014.x. [DOI] [PubMed] [Google Scholar]
- 19.Loftus JC, Ross JT, Paquette KM, Paulino VM, Nasser S, Yang Z, Kloss J, Kim S, Berens ME, Tran NL. miRNA expression profiling in migrating glioblastoma cells: regulation of cell migration and invasion by miR-23b via targeting of Pyk2. PLoS One. 2012;7:e39818. doi: 10.1371/journal.pone.0039818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, Liu Z, Chen L, Zhou L, Yao Y. MicroRNA-23b is an independent prognostic marker and suppresses ovarian cancer progression by targeting runt-related transcription factor-2. FEBS Lett. 2014;588:1608–1615. doi: 10.1016/j.febslet.2014.02.055. [DOI] [PubMed] [Google Scholar]
- 21.Zhuang K, Han K, Tang H, Yin X, Zhang J, Zhang X, Zhang L. Up-regulation of plasma miR-23b is associated with poor prognosis of gastric cancer. Med Sci Monit. 2016;22:356–361. doi: 10.12659/MSM.895428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kou CH, Zhou T, Han XL, Zhuang HJ, Qian HX. Downregulation of mir-23b in plasma is associated with poor prognosis in patients with colorectal cancer. Oncol Lett. 2016;12:4838–4844. doi: 10.3892/ol.2016.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto N, Nishikawa R, Chiyomaru T, Goto Y, Fukumoto I, Usui H, Mitsuhashi A, Enokida H, Nakagawa M, Shozu M, Seki N. The tumor-suppressive microRNA-1/133a cluster targets PDE7A and inhibits cancer cell migration and invasion in endometrial cancer. Int J Oncol. 2015;47:325–334. doi: 10.3892/ijo.2015.2986. [DOI] [PubMed] [Google Scholar]
- 24.Din FV, Theodoratou E, Farrington SM, Tenesa A, Barnetson RA, Cetnarskyj R, Stark L, Porteous ME, Campbell H, Dunlop MG. Effect of aspirin and NSAIDs on risk and survival from colorectal cancer. Gut. 2010;59:1670–1679. doi: 10.1136/gut.2009.203000. [DOI] [PubMed] [Google Scholar]
- 25.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 26.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Murray F, Zahno A, Kanter JR, Chou D, Suda R, Fenlon M, Rassenti L, Cottam H, Kipps TJ, Insel PA. Cyclic nucleotide phosphodiesterase profiling reveals increased expression of phosphodiesterase 7B in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2008;105:19532–19537. doi: 10.1073/pnas.0806152105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marko D, Romanakis K, Zankl H, Furstenberger G, Steinbauer B, Eisenbrand G. Induction of apoptosis by an inhibitor of cAMP-specific PDE in malignant murine carcinoma cells overexpressing PDE activity in comparison to their nonmalignant counterparts. Cell Biochem Biophys. 1998;28:75–101. doi: 10.1007/BF02737806. [DOI] [PubMed] [Google Scholar]
- 30.Savai R, Pullamsetti SS, Banat GA, Weissmann N, Ghofrani HA, Grimminger F, Schermuly RT. Targeting cancer with phosphodiesterase inhibitors. Expert Opin Investig Drugs. 2010;19:117–131. doi: 10.1517/13543780903485642. [DOI] [PubMed] [Google Scholar]
- 31.Dong H, Zitt C, Auriga C, Hatzelmann A, Epstein PM. Inhibition of PDE3, PDE4 and PDE7 potentiates glucocorticoid-induced apoptosis and overcomes glucocorticoid resistance in CEM T leukemic cells. Biochem Pharmacol. 2010;79:321–329. doi: 10.1016/j.bcp.2009.09.001. [DOI] [PubMed] [Google Scholar]


