Abstract
MicroRNAs (miRNAs) have been demonstrated to play a central role in the initiation and development in many types of cancers including colorectal carcinoma (CRC). The purpose of current study was to investigate the clinical significance of serum miR-140-5p in CRC. Real-time PCR was performed to compare the expression level of miR-140-5p in the serum samples from 63 CRC patients, 20 patients with adenomas and 20 healthy controls. Then the clinical significance of serum miR-140-5p was further determined. Serum miR-140-5p was downregulated in the serum samples derived from CRC patients compared to patients with adenomas patients and controls (P<0.01). In addition, its expression level was able to discriminate CRC from adenomas as well as healthy controls with high accuracy, and was also significantly associated with various clinicopathological parameters including TNM stage (P = 0.0013) and lymph node metastasis (P = 0.0323). Serum miR-140-5p was remarkably upregulated in the patients who received surgery (P<0.01). Patients with lower serum miR-140-5p expression suffered poorer 5-year overall survival (P = 0.0034). In conclusion, our study revealed that serum miR-140-5p might function as a tumor suppressor in CRC, which might serve as a novel prognostic biomarker and therapeutic target.
Keywords: Serum miR-140-5p, colorectal carcinoma, prognosis
Introduction
Colorectal carcinoma (CRC) is a common malignancy around the world, and remains a leading cause of cancer-related death in the developing countries [1]. Radiotherapy, chemotherapy and surgery are standard treatments for CRC and great strides have been made in the past few decades. However, the 5-year overall survival rate for the patients in the advanced stage remains very low [2]. Thus identification of novel biomarkers that contribute to the early detection of CRC is very important.
MicroRNAs (miRNAs), first discovered in Caenorhabditis elegans, are a class of naturally occurring, highly conserved, small non-coding RNA molecules with 21-25 nucleotides in length [3]. miRNAs regulate gene expression post-transcriptionally by binding to the 3’-UTR (untranslated region) and promoting repression of translation or degradation of the mRNA [4]. Numerous studies have shown that miRNAs play a critical role in most important biological events, including survival, proliferation and differentiation [5]. Deregulation of miRNA expression may result in activation of oncogenes or inactivation of tumor suppressor genes, which lead to the initiation and development of cancer [6]. In addition, miRNA is highly stable in the biofluids [7]. Therefore, identification of serum miRNA expression level in cancer is an effective strategy that might help early detection and diagnosis. For instance, the expression level of serum miR-21 was significantly overexpressed in patients with CRC compared with the normal controls. Moreover, upregulation of serum miR-21 was associated with tumor size, distant metastasis, and poor survival, indicating that serum miR-21 might serve as a promising biomarker for predicting the clinical outcome of CRC [8]. Vychytilova-Faltejskova et al identified a panel of noninvasive biomarkers including miR-23a-3p, miR-27a-3p, miR-142-5p and miR-376c-3p and this 4-miRNA signature showed high accuracy in discriminating CRC from healthy controls [9].
Aberrant expression of miR-140-5p has been demonstrated to play an important role in many types of cancers including CRC [10-13]. However, the expression levels of miR-140-5p in the serum samples of patients with CRC as well as its potential clinical significance remain poorly known. Therefore, the purpose of current study was to elucidate the prognostic value of serum miR-140-5p in CRC.
Materials and methods
Study population
This study was approved by the Research Ethics Committee at Hebei Cangzhou Central Hospital. Written informed consent to participate in the study was obtained from all subjects and their relatives. The serum samples were obtained from patients with CRC or colorectal adenomas and healthy volunteers who received treatment from Hebei Cangzhou Central Hospital. All the patients with CRC or adenomas were pathologically confirmed and CRC patients were classified according to the 2009 Union for International Cancer Control (UICC) Classification. The clinical information of the patient cohort was summarized in Table 1.
Table 1.
The association between serum miR-140-5p expression level and the clinical features of CRC
| Parameters | Case | Serum miR-140-5p | P | |
|---|---|---|---|---|
|
|
||||
| Low | High | |||
| Age | 0.9232 | |||
| <60 | 34 | 18 | 16 | |
| ≥60 | 29 | 15 | 14 | |
| Gender | 0.1634 | |||
| Male | 32 | 14 | 18 | |
| Female | 31 | 19 | 12 | |
| Tumor size | 0.5602 | |||
| <5 | 27 | 13 | 14 | |
| ≥5 | 36 | 20 | 16 | |
| TNM Stage | 0.0013 | |||
| I/II | 35 | 12 | 23 | |
| III/IV | 28 | 21 | 7 | |
| Lymph node metastasis | 0.0323 | |||
| No | 42 | 18 | 24 | |
| Yes | 21 | 15 | 6 | |
| Distant metastasis | 0.1105 | |||
| No | 57 | 28 | 29 | |
| Yes | 6 | 5 | 1 | |
| Histological grade | 0.2071 | |||
| Well/moderate | 39 | 18 | 21 | |
| Poor | 24 | 15 | 9 | |
Serum preparation
In each case, at least 3 mL peripheral blood sample was collected and then separated by centrifugation. Briefly, the samples were first centrifuged at 1,200 g for 10 min at 4°C, and then the supernatant was transferred to a clean centrifuge tube and undergo a second centrifugation at 16,000 g for 10 min at 4°C. Supernatant serum was then stored at -80°C for future use.
Real-time PCR
The mirVanaTM PARISTM kit (Applied Biosystems Life Technologies, Foster City, CA, USA) was used to extract the total RNA from serum samples according to the manufacturer’s instruction. The first strand cDNA was synthesized with TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). The cDNA was amplified and quantified using FastStart SYBR Green Master (Roche Applied Science, Indianapolis, IN, USA). The expression level of miR-140-5p in the serum samples was detected using an ABI PRISM 7500 Sequence Detection System (Applied Biosystems). The changes in miR-140-5p expression were determined using the 2-ΔΔCT method with the U6 small nuclear RNA (U6) as the reference gene.
Statistical analysis
All the data was expressed as mean ± standard deviation. Statistical analyses were performed using SPSS Software version 18.0 (SPSS Inc., Chicago, IL) and GraphPad Prism version 6.0 (GraphPad Software Inc., La Jolla, CA). The nonparametric Mann-Whitney U and Kruskal-Wallis H test were used to compare the expression level of serum miR-140-5p among the CRC patients, patients with adenomas and the healthy controls. The efficacy of serum miR-140-5p for disease diagnosis was evaluated by the area under receiver operating characteristic (ROC) curve (AUC). The relationship between serum miR-140-5p expression level and clinical parameters of CRC was evaluated using Chi-squared test. The survival curves were plotted using the Kaplan-Meier method, with differences assessed by the log-rank test. P<0.05 was considered to be statistical significance.
Results
Serum miR-140-5p was downregulated in patients with adenoma and CRC
The real-time PCR results showed that the expression level of miR-140-5p was significantly decreased in the serum samples derived from patients with CRC or adenomas in comparison with healthy volunteers (P<0.01) (Figure 1). In addition, serum miR-140-5p level was remarkably enhanced in those patients (patients with CRC or adenomas) who received surgery treatment, indicating serum miR-140-5p might be used to monitor the therapeutic responses (P<0.01). However, serum miR-140-5p changed little in the patients who received non-surgery treatment (NST) (P>0.05) (Figure 2).
Figure 1.
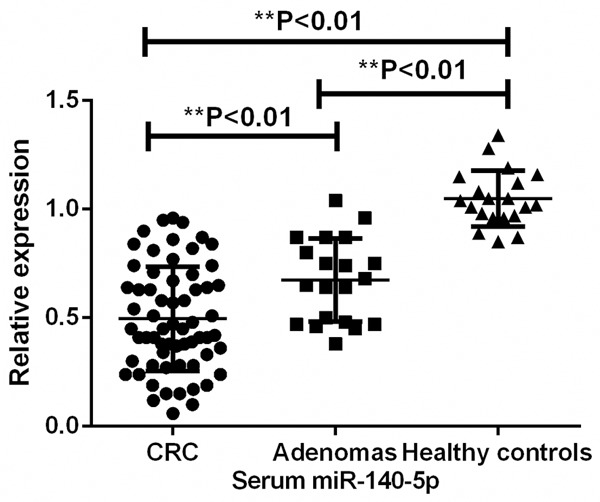
Expression level of serum miR-140-5p among CRC patients, adenomas patients and healthy controls.
Figure 2.
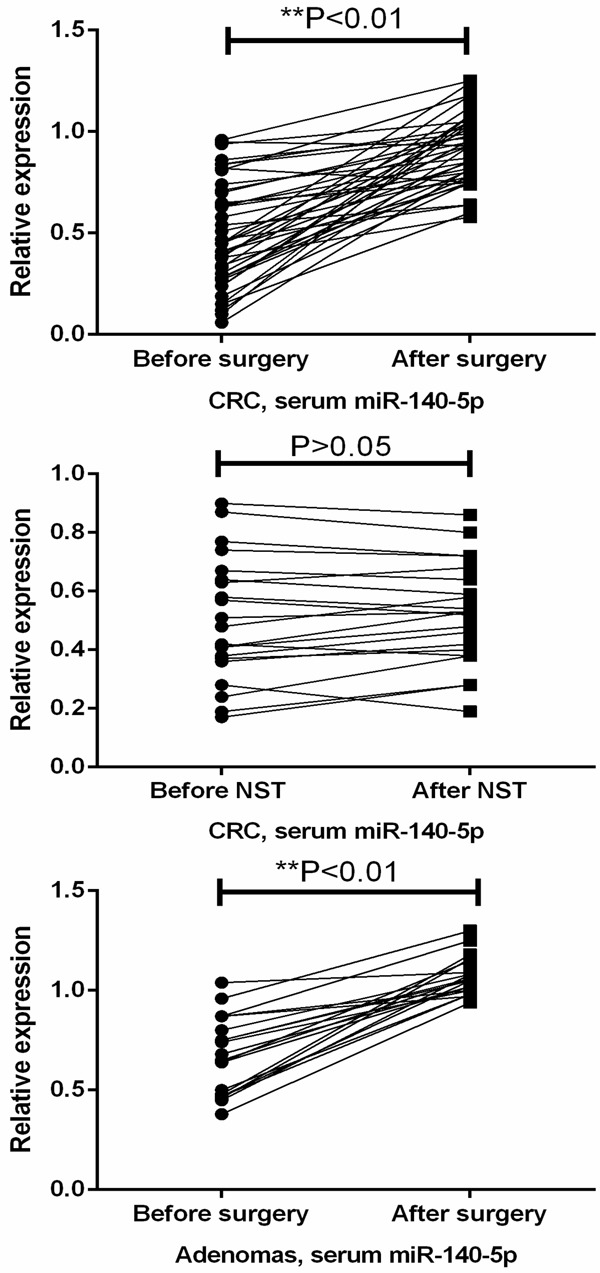
Serum miR-140-5p was increased follow surgery therapy.
Discrimination capacity of serum miR-140-5p for CRC
To determine whether the serum level of miR-140-5p had diagnostic values, the ROC curve was applied to analyze their diagnostic sensitivity and specificity. The optimal sensitivity and specificity of miR-140-5p were 86.21% and 88.45% in distinguishing CRC from normal controls (AUC = 0.915). The optimal sensitivity and specificity of miR-140-5p were 71.45% and 90.72% in distinguishing CRC from adenomas (AUC = 0.860) (Figure 3).
Figure 3.
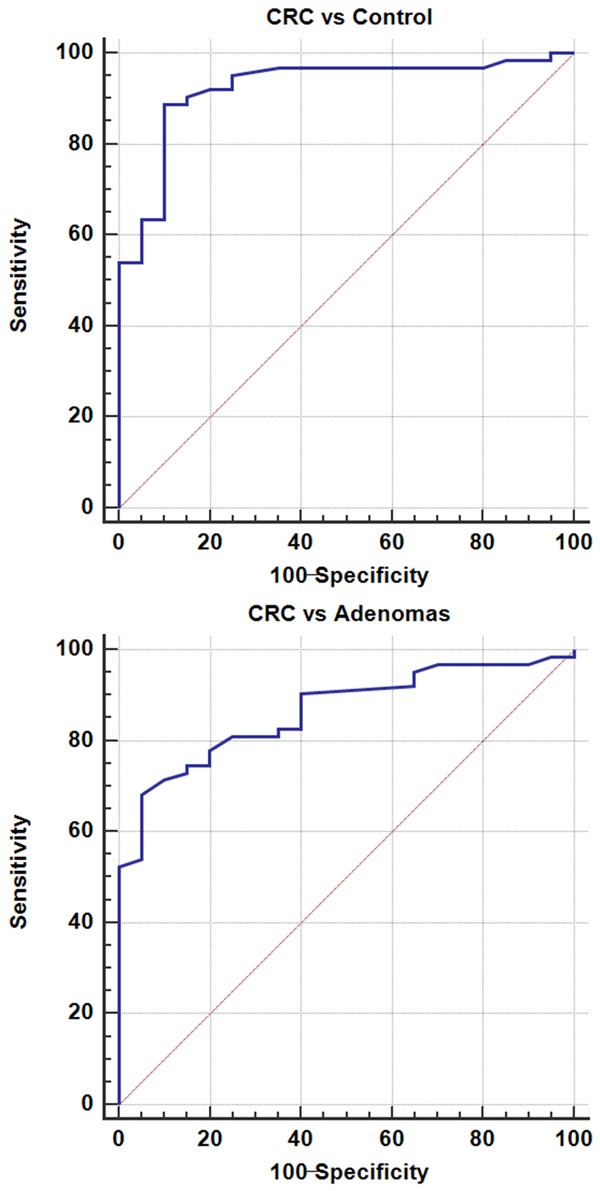
Diagnostic value of serum miR-140-5p in CRC.
Association between serum miR-140-5p level and clinicopathological parameters of CRC
The median value of serum miR-140-5p was used as a cutoff point to divide the CRC patients into high and low serum miR-140-5p expression group. The Chi-square results showed that serum miR-140-5p level was associated with TNM stage (P = 0.0013) and lymph node metastasis (P = 0.0323). However, it was not correlated with age, gender tumor size, distant metastasis and histological grade (P>0.05) (Table 1).
Correlation between serum miR-140-5p level and overall survival of CRC patients
The survival analysis showed that the CRC patients in the low serum miR-140-5p expression group had a worse 5 year overall survival than those in the high serum miR-140-5p expression group (P = 0.0034) (Figure 4).
Figure 4.
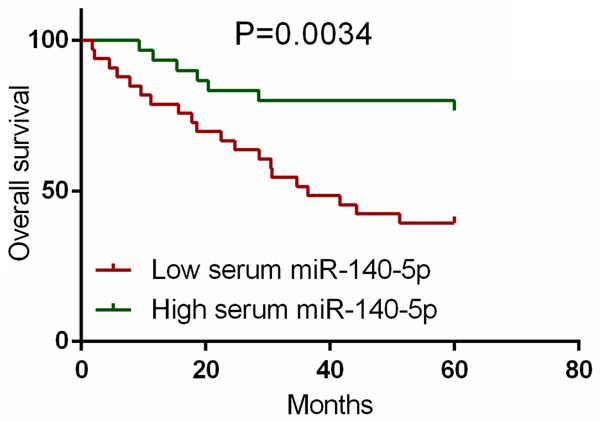
Association between serum miR-140-5p and overall survival of CRC.
Discussion
Our results showed that the expression level of serum miR-140-5p was significantly downregulated in patients with adenomas or CRC and it could discriminate CRC from adenomas as well as healthy controls with high accuracy. Serum miR-140-5p was remarkably upregulated in the patients who received surgery. In addition, its expression level was significantly associated with TNM stage and lymph node metastasis. Furthermore, the CRC with lower serum miR-140-5p expression had significantly shorter overall survival compared to those with higher serum miR-140-5p expression. Collectively, these data suggest that miR-140-5p plays a tumor suppressive role in the development of CRC and downregulation of miR-140-5p might promote the tumorigenesis of this malignancy.
Consistent with our findings, miR-140-5p expression was reduced in CRC tissue samples and it expression level was associated with poor prognosis of CRC. In addition, overexpression of miR-140-5p suppressed the proliferation, migration and invasion capacity of CRC cells both in vitro and in vivo, and opposite results was found when miR-140-5p was inhibited. Moreover, vascular endothelial growth factor A (VEGF-A) and smad2 were identified as the direct targets of miR-140-5p, indicating miR-140-5p might be crucial for the carcinogenesis of CRC [13,14]. Similarly, the expression level of miR-140-5p was decreased in hypopharyngeal squamous cell carcinoma and correlated with tumor stage and lymph node metastasis. Ectopic expression of miR-140-5p inhibited the migration and invasion capacity of cancer cells [15]. miR-140-5p was significantly decreased in hepatocellular carcinoma (HCC) and liver cancer cell lines. In addition, its expression level was correlated with various clinical features of HCC including multiple nodules, vein invasion, capsular formation, and differentiation, as well as overall and disease-free survival. The proliferation and metastasis capacity of liver cancer cells were remarkably inhibited when miR-140-5p was overexpressed, suggesting that miR-140-5p might function as a tumor suppressor in HCC [16]. Similar founding was also reported in a number of cancers such as head and neck cancer and ovarian cancer [17,18]. It seems that the tumor suppressive role of miR-140-5p might be independent of the tumor microenvironment and cancer type. To the best of knowledge, no study has reported that miR-140-5p can promote the development of cancer. Recently a study showed that the expression level of miR-140-5p was upregulated in non-small cell lung cancer (NSCLC) [19]. However, whether upregulation of miR-140-5p has clinical significance in NSCLC remains unknown.
One limitation of the current study is the relative sample size, large cohort studies should perform to further elucidate the clinical significance of miR-140-5p in CRC. In addition, whether combination of tissue and serum miR-140-5p contributes to higher diagnostic value remains further investigation. Moreover, future studies should reveal the molecular mechanisms of miR-140-5p downregulation that drive the progression of CRC.
In conclusion, serum miR-140-5p was decreased in patients with CRC and its downregulation was correlated with poor prognosis of this deadly disease, suggesting serum miR-140-5p might be a promising biomarker and could have potential therapeutic application in CRC.
Acknowledgements
This study was supported by the funding from Department of Oncology, Hebei Cangzhou Central Hospital.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 3.Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: synthesis, mechanism, function, andrecent clinical trials. Biochim Biophys Acta. 2010;1803:1231–1243. doi: 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 5.Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67:129–139. doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 6.Zhang W, Dahlberg JE, Tam W. MicroRNAs in tumorigenesis: a primer. Am J Pathol. 2007;171:728–738. doi: 10.2353/ajpath.2007.070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vychytilova-Faltejskova P, Radova L, Sachlova M, Kosarova Z, Slaba K, Fabian P, Grolich T, Prochazka V, Kala Z, Svoboda M, Kiss I, Vyzula R, Slaby O. Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer. Carcinogenesis. 2016;37:941–950. doi: 10.1093/carcin/bgw078. [DOI] [PubMed] [Google Scholar]
- 10.Güllü G, Peker I, Haholu A, Eren F, Küçükodaci Z, Güleç B, Baloglu H, Erzik C, Özer A, Akkiprik M. Clinical significance of miR-140-5p and miR-193b expression in patients with breast cancer and relationship to IGFBP5. Genet Mol Biol. 2015;38:21–29. doi: 10.1590/S1415-475738120140167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J, Zhang W, Tang H, Qian H, Yang J, Zhu Z, Ren P, Lu B. Septin 2 accelerates the progression of biliary tract cancer and is negatively regulated by mir-140-5p. Gene. 2016;589:20–26. doi: 10.1016/j.gene.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Su Y, Xiong J, Hu J, Wei X, Zhang X, Rao L. MicroRNA-140-5p targets insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) to suppress cervical cancer growth and metastasis. Oncotarget. 2016;7:68397–68411. doi: 10.18632/oncotarget.11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W, Zou C, Pan L, Xu Y, Qi W, Ma G, Hou Y, Jiang P. MicroRNA-140-5p inhibits the progression of colorectal cancer by targeting VEGFA. Cell Physiol Biochem. 2015;37:1123–1133. doi: 10.1159/000430237. [DOI] [PubMed] [Google Scholar]
- 14.Zhai H, Fesler A, Ba Y, Wu S, Ju J. Inhibition of colorectal cancer stem cell survival and invasive potential by hsa-miR-140-5p mediated suppression of Smad2 and autophagy. Oncotarget. 2015;6:19735–19746. doi: 10.18632/oncotarget.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jing P, Sa N, Liu X, Liu X, Xu W. MicroR-140-5p suppresses tumor cell migration and invasion by targeting ADAM10-mediated Notch1 signaling pathway in hypopharyngeal squamous cell carcinoma. Exp Mol Pathol. 2016;100:132–138. doi: 10.1016/j.yexmp.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Yang H, Fang F, Chang R, Yang L. MicroRNA-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor β receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology. 2013;58:205–217. doi: 10.1002/hep.26315. [DOI] [PubMed] [Google Scholar]
- 17.Kai Y, Peng W, Ling W, Jiebing H, Zhuan B. Reciprocal effects between microRNA-140-5p and ADAM10 suppress migration and invasion of human tongue cancer cells. Biochem Biophys Res Commun. 2014;448:308–314. doi: 10.1016/j.bbrc.2014.02.032. [DOI] [PubMed] [Google Scholar]
- 18.Lan H, Chen W, He G, Yang S. miR-140-5p inhibits ovarian cancer growth partially by repression of PDGFRA. Biomed Pharmacother. 2015;75:117–122. doi: 10.1016/j.biopha.2015.07.035. [DOI] [PubMed] [Google Scholar]
- 19.Hu L, Ai J, Long H, Liu W, Wang X, Zuo Y, Li Y, Wu Q, Deng Y. Integrative microRNA and gene profiling data analysis reveals novel biomarkers and mechanisms for lung cancer. Oncotarget. 2016;7:8441–8454. doi: 10.18632/oncotarget.7264. [DOI] [PMC free article] [PubMed] [Google Scholar]


