Abstract
Background: To explore the genetic diversity and drug resistance status of MTB in Xuzhou, China. Methods: A total of 325 clinical MTB strains were genotyped by spacer-oligonucleotide typing (spoligotyping) and mycobacterial interspersed repetitive unit variable number of tandem repeats (MIRU-VNTR). Phenotypic resistance was assessed by drug susceptibility testing (DST). Result: Based on the spoligotyping method, 325 MTB isolates were classified into 5 known genotypes and 12 unknown genotypes, and the largest branch comprised 268 strains belonging to the Beijing family. Based on the 15-loci VNTR typing method, 325 MTB isolates were divided into 35 clusters and 220 unique patterns. Compared to the low discriminatory power of spoligotyping genotyping (HGDI = 0.3444), 15-loci VNTR genotyping had a significantly higher discriminatory power for all strains (HGDI = 0.9980), particularly for the Beijing family strains (HGDI = 0.9892). When spoligotyping and 15-loci VNTR methods were used together, the discriminatory power increased to 0.9991. The Beijing family strain presented increased risks for developing multi-drug resistance TB (P < 0.05). Conclusion: The Beijing family isolates is the most prevalent strains in Xuzhou. Spoligotyping, in combination with 15-loci MIRU-VNTR, is useful for epidemiological analysis of MTB transmission in Xuzhou.
Keywords: Mycobacterium tuberculosis, genotype, spoligotyping, MIRU-VNTR
Introduction
Tuberculosis (TB) is a major global public health concern, particularly in developing countries. According to the World Health Organization (WHO), there were approximately 9.6 million new cases and 1.5 million fatal cases of TB in 2014 [1]. China ranks second among the 22 high burden countries, with approximately 0.98 million new cases in 2013 [1]. To reduce the global burden of TB, the use of effective drug treatments is considered to be the best approach. Moreover, due to the inappropriate use of anti-TB drugs in the world, approximately 0.48 million TB patients became multidrug-resistant in 2014. Therefore, it is critical to systematically monitor and track the transmission of drug-resistant TB strains to control MDR-TB infection [2,3].
Genotyping of Mycobacterium tuberculosis (MTB) isolates has significantly improved understanding of the epidemiology of TB, and allowed for better control of this disease [4]. Advances in molecular biotechniques can help to reveal the source of infection, trace the route of transmission, and determine risk factors [5] In order to monitor domestic spread, the spoligotyping method is the gold standard for MTB strain identification, particularly for Beijing family genotype strains [6,7]. Due to the low discriminatory power of spoligotyping, MIRU-VNTR is frequently used to fully explore the genetic diversity of MTB strains [8]. This method can determine the different numbers of MTB interspersed repetitive units with disparate VNTR loci, and is widely used for molecular typing.
Xuzhou, an important transportation hub located in eastern China, has remained a middle-prevalence area of TB. According to the 2010 National TB Epidemiology Survey in China, approximately 57,000 pulmonary TB cases occurred in Xuzhou from 2000 to 2010 [9]. However, information regarding the molecular epidemiology of TB in Xuzhou is scarce; therefore it is important to study the local molecular epidemiology of TB. In our study, we collected 325 clinical MTB strains in Xuzhou, and genotypes and drug resistance were analyzed by spoligotyping and MIRU-VNTR with 15 VNTR loci. This study not only provides valuable information for TB researchers, but also presents guidelines for the prevention and control of TB in Xuzhou.
Materials and methods
Study population and bacterial strains
A total of 325 clinical MTB strains were isolated from TB patients from Xuzhou Infectious Disease Hospital during 2014. Sputum smear examination by Ziehl-Neelsen staining and culture on Lowenstein-Jensen (L-J) medium were performed for all samples. The demographic data of all TB patients was collected, including age, sex, BCG vaccination status, drug treatment, and birth region. All patients enrolled in the study signed an informed consent form. The protocols performed in this study were approved by the Ethics Committee of Xuzhou Infectious Disease Hospital.
Genomic DNA extraction
All clinical isolates of MTB were grown on fresh L-J slants at 37°C for 4-6 weeks. Cells were resuspended in 400 μl TE buffer (pH 8.0), centrifuged at 12,000 rpm for 2 min, and the pellet was resuspended in 400 μl of TE buffer and heated in a water bath at 95°C for 30 min. The cellular debris was centrifuged at 12,000 rpm for 3 min, and DNA in the supernatant was stored at -20°C for PCR analysis.
Spoligotyping
Spoligotyping of TB strains was performed as previously described [10]. First, the direct repeat (DR) region was amplified with the primers DRa (5’-CCGAGAGGGGACGGAAAC-3’) and DRb (5’-GGTTTTGGGTCTGACGAC-3’). Then, the PCR products were hybridized to a set of 43 oligonucleotide probes corresponding to each spacer, which were covalently bound to a membrane. Chemiluminescent detection was performed using ECL detection liquid and ECL Hyperfilm (GE Healthcare Life Sciences, UK). The spoligotypes in binary format were then compared with those in the SITVIT WEB database [11].
MIRU-VNTR
In accordance with previous studies, 15-loci VNTR were selected for MLVA typing for our MTB genetic diversity study [12]. This set of loci included 5 loci of exact tandem repeats (ETRs): ETR-A, -B, -C, -D, and -E; 8 MIRU loci: MIRU-10, -16, -23, -26, -27, -39, and -40; 3 Mtub loci: Mtub21, 30, and 39. The primer sequences for PCR amplification of each locus are described in previous studies [12]. Each PCR reaction was performed in a final volume of 20 μl containing 50 ng DNA, 10 μl Taq Mixture (TaKaRa, Japan), and 1 μl (10 µM) primers. The amplification was carried out in a thermal cycler (Bio-Rad, USA) using the following parameters: an initial denaturation at 94°C for 5 min; followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 62°C for 30 s, and extension at 70°C for 45 s; and a final extension at 72°C for 10 min. A positive control (H37Rv strain genomic DNA) and a negative control (ddH2O) were included in each test. PCR amplicons were analyzed on 2% agarose gels, which were stained with ethidium bromide and imaged using the Image Lab System (Bio-Rad). By comparing PCR amplicon sizes to those obtained from H37Rv, we obtained the copy numbers of each VNTR loci. BioNumerics 5.0 was used for the phylogenetic and cluster analysis, and the discriminatory power of the VNTR loci was determined by the Hunter-Gaston discrimination index (HGDI).
Drug susceptibility testing
The L-J proportion method was used for the DST at the Clinical Laboratory of the Xuzhou Infectious Disease Hospital. The drug concentrations were 0.2 μg/ml for isoniazid (INH), 40 μg/ml for rifampicin (RFP), 2 μg/ml for ethambutol (EMB), and 4 μg/ml for streptomycin (SM). H37Rv strain was used as a quality control, and DST results were analyzed after 4 weeks growth. When the growth rate was more than 1% compared to the control, the strain was determined to be resistant to the specific drug. Strains resistant to at least RIF and INH were defined as MDR-TB [13].
Data analysis
The statistical analysis was performed using GraphPad Prism 6.0. Chi-square for measuring the association between two categorical variables which is the Spoligotyping and MIRU-VNTR, and Fisher’s exact test analysis were run to identify significant differences. The genetic diversity was calculated using Hunter-Gaston diversity index. A two-sided P value of < 0.05 was considered to be statistically significant.
Results
Spoligotyping analysis
To demonstrate spoligotype diversity among MTB strains in Xuzhou, 325 strains were collected from different districts of the city during 2014. Spoligotyping clustering analysis indicated that 268 (82.46%) were clustered into the Beijing family, including typical Beijing genotype (263, 80.92%), and Beijing-like genotype (5, 1.54%), 29 (8.92%) were T family, 4 (1.23%) were U genotype, 3 (0.92%) were H3, and 3 (0.92%) were MANU2 genotype, and 18 strains were not identified in the SpolDB4.0 database and referred to 12 “Unknown” genotypes (Figure 1). These results support the hypothesis that Beijing strains are predominant in different regions of China. However, the spoligotyping method did not effectively distinguish genetic diversity among Beijing family strains.
Figure 1.
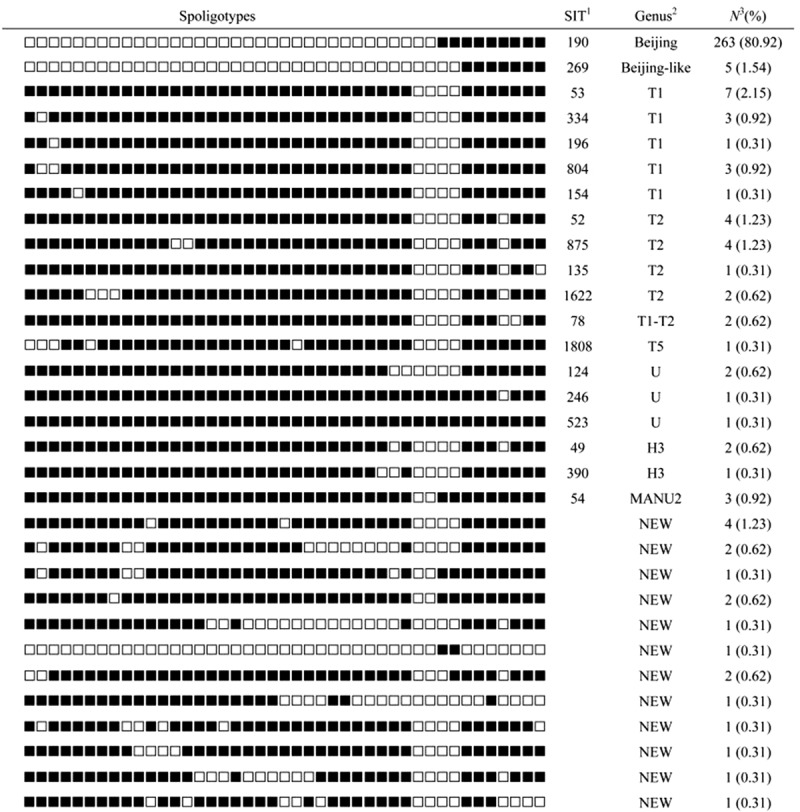
Spoligotypes of the MTB isolates (n = 325). 1SIT number from the SITVIT WEB database. SIT, spoligotype international type. 2Sppoligotype families as assigned in SITVIT WEB database. 3The number of the isolates with a common SIT.
MIRU-VNTR analysis of MTB strains
The 325 strains were divided into 35 clusters and 255 genotypes, including 220 unique patterns, by the MLVA with 15-loci VNTR (Figure 2 and Table S1). The number of clustered isolates was 105 and the clustering rate was 21.54%. To verify the discriminatory power of each locus and the cumulative discriminatory power of all 15 loci, we calculated the discriminatory power of each locus to obtain the HGDI discriminatory power (Table 1). Each of the 15 loci showed different discriminatory powers, which consisted of 5 high (> 0.6) discriminatory loci (Mtub21, MIRU26, MIRU10, ETRE, Mtub30), 7 moderate (0.3 to 0.6) discriminatory loci (MIRU39, MIRU16, ETRA, MIRU40, ETRD, Mtub39, MIRU27), and 3 poor (< 0.3) discriminatory loci (MIRU23, ETRB, ETRC), based on the HGDI values.
Figure 2.

Genotyping with spoligotyping and 15-loci MIRU-VNTR.
Table 1.
The repeat number of 15-loci VNTR for 325 MTB strains
| VNTR loci | HGDI score | No. of repeats in H37Rv | Repeat Numbers of VNTR-loci in the Isolates | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |||
| ETRA | 0.585 | 3 | 1 | 35 | 73 | 193 | 9 | 12 | 2 | ||
| ETRB | 0.229 | 3 | 38 | 283 | 4 | ||||||
| ETRC | 0.163 | 4 | 9 | 14 | 297 | 2 | 3 | ||||
| ETRD | 0.553 | 3 | 9 | 94 | 195 | 21 | 4 | 2 | |||
| ETRE | 0.665 | 3 | 2 | 16 | 63 | 152 | 91 | 1 | |||
| MIRU10 | 0.679 | 3 | 14 | 102 | 143 | 55 | 7 | 4 | |||
| MIRU16 | 0.591 | 2 | 11 | 193 | 61 | 45 | 15 | ||||
| MIRU23 | 0.242 | 6 | 1 | 7 | 281 | 34 | 2 | ||||
| MIRU26 | 0.726 | 3 | 2 | 4 | 11 | 8 | 21 | 53 | 143 | 72 | 11 |
| MIRU27 | 0.471 | 3 | 2 | 30 | 225 | 67 | 1 | ||||
| MIRU39 | 0.598 | 2 | 5 | 42 | 172 | 106 | |||||
| MIRU40 | 0.577 | 1 | 4 | 49 | 202 | 61 | 9 | ||||
| Mtub21 | 0.785 | 2 | 12 | 8 | 32 | 85 | 93 | 75 | 15 | 5 | |
| Mtub30 | 0.622 | 2 | 67 | 163 | 95 | ||||||
| Mtub39 | 0.477 | 5 | 27 | 19 | 229 | 43 | 7 | ||||
Comparison of spoligotyping and VNTR genotyping
Comparing the genotyping results of 325 isolates with spoligotyping and 15-loci VNTR indicated the discriminatory power of these two genotyping methods (Table 2). Compared to 15-loci VNTR, spoligotyping had a low discriminatory power (HGDI = 0.344), particularly when applied to Beijing family strains (HGDI = 0.0375). Moreover, among the 31 different types, spoligotyping identified 15 unique strains and 16 clusters grouped containing 310 strains, with a clustering rate of 90.46%. The 15-loci VNTR identified 220 unique strains and 35 clusters grouped containing 105 strains, with a clustering rate of 21.54%. Moreover, 15-loci VNTR genotyping had a significantly higher discriminatory power for all strains (HGDI = 0.9980), particularly for Beijing family strain genotyping (HGDI = 0.9892). When spoligotyping and 15-loci VNTR methods were combined, the discriminatory power increased to 0.9991, and the clustering rate was reduced to 18.77%.
Table 2.
Different discriminatory power of different typing methods used for 325 MTB strains
| Method | No. of clustered strains | No. of clusters | Cluster size | Clustering rate (%) | HGDI score | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| All strains | Beijing family | Other strains | |||||
| Spoligotyping | 310 | 16 | 2-263 | 90.46 | 0.3444 | 0.0375 | 0.9655 |
| 15-loci VNTR | 105 | 35 | 2-6 | 21.54 | 0.9980 | 0.9892 | 0.9963 |
| Spoligotyping and VNTR | 87 | 26 | 2-5 | 18.77 | 0.9991 | 0.9932 | 0.9989 |
Drug resistance
The DST results showed that 14.77% (48/325) were resistant to at least one of the four first-line drugs (Table 3). The proportions of single-drug resistance of RFP, INH, SM, and EMB were 1.54% (5/325), 3.08% (10/325), 2.77% (9/325), and 1.85% (6/325), respectively. Among all isolates, 5.54% (18/325) isolates were MDR-TB strains, including 2 (0.62%) strains resistant to all four first-line drugs. By comparing the distribution of drug resistance between Beijing and non-Beijing strains, we found that the Beijing family strains presented increased risks for developing MDR in Xuzhou (X2 = 4.053, P < 0.05).
Table 3.
First-line drug resistant frequency among 325 MTB strains
| Drugs | No. (%) of Isolates | |||
|---|---|---|---|---|
|
| ||||
| Beijing (n = 268) | Non-Beijing (n = 57) | X 2 | P | |
| Mono-resistance | ||||
| RFP | 4 (1.49) | 1 (1.75) | 0.02128 | > 0.05 |
| INH | 6 (2.24) | 4 (7.02) | 3.599 | > 0.05 |
| SM | 7 (2.61) | 2 (3.51) | 0.1404 | > 0.05 |
| EMB | 4 (1.49) | 2 (3.51) | 1.054 | > 0.05 |
| Total | 21(7.84) | 9 (15.79) | 3.549 | > 0.05 |
| MDR-TB | ||||
| RFP+INH | 6 (2.24) | 0 (0) | 1.300 | > 0.05 |
| RFP+INH+SM | 6 (2.24) | 0 (0) | 1.300 | > 0.05 |
| INH+SM+EMB | 4 (1.49) | 0 (0) | 0.8613 | > 0.05 |
| RFP+INH+SM+EMB | 2 (0.75) | 0 (0) | 0.4280 | > 0.05 |
| Total | 18 (6.72) | 0 (0) | 4.053 | < 0.05 |
Discussion
TB is a global public health issue, and the prevalence of TB is closely related to regional differences and population mobility. Systematic surveillance and tracking of transmission of TB strains have become critically important for infection control. Compared with traditional MTB typing, newer genotyping methods can quickly and accurately obtain information regarding outbreaks of MTB [4]. Moreover, certain MTB genotypes have been found to be related to the risk of transmission or anti-TB drug resistance. The number of MTB genotypes has increased throughout different regions of China [10,14-16]. However, these results do not represent the prevalence and epidemic trend of tuberculosis in Xuzhou, an important transportation hub in China.
Spoligotyping is the gold standard for Beijing family identification, as it is simple and efficient. The current study spoligotyping showed 82.46% were clustered into the Beijing family, thus indicating the Beijing family is the most prevalent lineage of MTB strains in Xuzhou. The result is in concordance with previously published results. Compared with other Chinese cities, prevalence of the Beijing genotype was less than for Beijing (92%) [17], Shijiazhuang (91%) [18], but more than Hong Kong (70%) [19], Taibei (52%) [20], and Shanghai (77%) [21]. Although the Beijing family was the dominant genotypic family in the Xuzhou district, genotypic polymorphisms were also evident, such as the T1, T2, T1-T2, T5, U, H3, and MANU2 families. Notably, the T family genotype, which is prevalent in Africa, Central and South America, and Europe, was significantly higher in Xuzhou than in other districts [22]. The U types have previously been identified as high-incidence strains in the Middle East, and Central and Southern Asia [22]. This indicates that population mobility from these regions may have increased the prevalence of related genotypes in Xuzhou. In addition, we identified 12 novel MTB spoligotypes, indicating the complexity of the sources of strains in this region.
The discriminatory power of spoligotyping is low, and it cannot be utilized for complex analysis of the Beijing family strains. Genotyping with a higher discriminatory power is required to detect unnoticed transmission of M. tuberculosis among patients. Therefore, another molecular typing method based on MIRU-VNTR is used to complete the shortcoming of the spoligotyping.
In different regions, the disparate VNTR typing sets showed various efficiencies in MTB genotyping. The sets of common loci comprised of 12-loci, 15-loci, 24-loci, and others, and the discriminatory power of MIRU-VNTR is determined by the number of loci [23]. The selection of suitable sets of loci depends on the pattern of MTB strains in the investigated area. We selected the classic 15-loci VNTR method, based on available data for other Chinese cities [24,25], and found that the allelic diversity varied significantly at each VNTR locus. We identified 220 unique strains, and 35 clusters grouped containing 105 strains, and found a significantly higher discriminatory power for all strains (HGDI = 0.9980), particularly for genotyping of Beijing family strains (HGDI = 0.9892). Among the 15-loci VNTR, Mtub21 had the highest HGDI score of 0.785, similar to the Beijing results, however this locus was not optimal for genotyping in Tibet, Heilongjiang, and Taibei [26]. MIRU26, MIRU10, ETRE, and Mtub30 also revealed higher diversity than in other zones [26]. When the spoligotyping and 15-loci VNTR methods were used together, the discriminatory power increased to 0.9991. This result indicates that the combination of the two genotyping methods improves discriminatory power and reproducibility, and significantly contributes to the understanding of MTB epidemiology
The DST results showed that the multiple drug resistance rates were 5.54% in Xuzhou (Table 3). This was lower than the average level of China. This may be attributed to vigorous prevention and control methods used in Xuzhou, resulting in reduction of the spread of drug-resistant tuberculosis. According to published reports, the Beijing genotype is significantly associated with drug-resistance, and may be responsible for the emergence and spread of MDR-TB [12,27].
In order to identify any association of drug resistance with the Beijing genotype, we compared the distribution of drug resistance between Beijing and non-Beijing genotyping strains usieng the XXX methode and the result in Table 3 showed increased risk for developing MDR-TB in the Beijing family strains.
The study demonstrates the Beijing family isolates is the most prevalent strains in Xuzhou. Furthermore, the Beijing family strain may be more virulent and associated with drug resistance.
The spoligotyping method is the gold standard for MTB strain identification, particularly for Beijing family genotype strains, nevertheless it has low. Therefore, 15-loci MIRU-VNTR, whic is useful for epidemiological analysis of MTB transmission in Xuzhou, is used to complete the shortcoming of the spoligotyping.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Ravi A, Singh Sunita D, Medical GR. Global tuberculosis report 2015, Global Tuberculosis Report 6. 2012 [Google Scholar]
- 2.Zignol M, Gemert WV, Falzon D, Sismanidis C, Glaziou P, Floyd K, Raviglione M. Surveillance of anti-tuberculosis drug resistance in the world: an updated analysis, 2007-2010. Bull World Health Organ. 2012;90:111–119. doi: 10.2471/BLT.11.092585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao Y, Xu S, Wang L, Chin DP, Wang S, Jiang G, Xia H, Zhou Y, Li Q, Ou X. National survey of drug-resistant tuberculosis in China. N Engl J Med. 2012;366:2161–2170. doi: 10.1056/NEJMoa1108789. [DOI] [PubMed] [Google Scholar]
- 4.Barnes PF, Cave MD. Molecular epidemiology of tuberculosis. N Engl J Med. 2003;349:1149–1156. doi: 10.1056/NEJMra021964. [DOI] [PubMed] [Google Scholar]
- 5.Roetzer A, Diel R, Kohl TA, Rückert C, Nübel U, Blom J, Wirth T, Jaenicke S, Schuback S, Rüsch-Gerdes S. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med. 2013;10:e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bifani PJ, Mathema B, Kurepina NE, Kreiswirth BN. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 2002;10:45–52. doi: 10.1016/s0966-842x(01)02277-6. [DOI] [PubMed] [Google Scholar]
- 7.Kamerbeek J, Schouls L, Kolk A, Van Agterveld M, Van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allix-Béguec C, Harmsen D, Weniger T, Supply P, Niemann S. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2008;46:2692–2699. doi: 10.1128/JCM.00540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Zhang H, Ruan Y, Chin DP, Xia Y, Cheng S, Chen M, Zhao Y, Jiang S, Du X, He G, Li J, Wang S, Chen W, Xu C, Huang F, Liu X, Wang Y. Tuberculosis prevalence in China, 1990-2010; a longitudinal analysis of national survey data. Lancet. 2014;383:2057–2064. doi: 10.1016/S0140-6736(13)62639-2. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Tian M, Wang X, Wei R, Xing Q, Ma T, Jiang X, Li W, Zhang Z, Xue Y. Genotypic diversity analysis of Mycobacterium tuberculosis strains collected from Beijing in 2009, using spoligotyping and VNTR typing. PLoS One. 2014;9:e106787. doi: 10.1371/journal.pone.0106787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demay C, Liens B, Burguière T, Hill V, Couvin D, Millet J, Mokrousov I, Sola C, Zozio T, Rastogi N. SITVITWEB--a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol. 2012;12:755–766. doi: 10.1016/j.meegid.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Pang H, Tong J, Liu HC, Du YG, Zhao XQ, Jiang Y, Wu XC, Yang JC, Wan Kl. Molecular characterization and drug-resistance of mycobacterium tuberculosis strains in Xuzhou, China. Biomed Environ Sci. 2014;27:960–964. doi: 10.3967/bes2014.136. [DOI] [PubMed] [Google Scholar]
- 13.Shao Y, Yang D, Xu W, Lu W, Song H, Dai Y, Shen H, Wang J. Epidemiology of anti-tuberculosis drug resistance in a Chinese population: current situation and challenges ahead. BMC Public Health. 2011;11:110. doi: 10.1186/1471-2458-11-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Chen J, Shen X, Gui X, Mei J, DeRiemer K, Gao Q. Highly polymorphic variable-number tandem repeats loci for differentiating Beijing genotype strains of Mycobacterium tuberculosis in Shanghai, China. FEMS Microbiol Lett. 2008;282:22–31. doi: 10.1111/j.1574-6968.2008.01081.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Liu Y, Zhang CL, Ji BY, Zhang LZ, Shao YZ, Jiang SL, Suzuki Y, Nakajima C, Fan CL. Genotypes and characteristics of clustering and drug-susceptibility of Mycobacterium tuberculosis isolates in Heilongjiang Province, China. J Clin Microbiol. 2011;49:1354–62. doi: 10.1128/JCM.02274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Somerville W, Thibert L, Schwartzman K, Behr MA. Extraction of Mycobacterium tuberculosis DNA: a question of containment. J Clin Microbiol. 2005;43:2996–2997. doi: 10.1128/JCM.43.6.2996-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong H, Liu Z, Lv B, Zhang Y, Liu J, Zhao X, Liu J, Wan K. Spoligotypes of Mycobacterium tuberculosis from different provinces of China. J Clin Microbiol. 2010;48:4102–4106. doi: 10.1128/JCM.00549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Liu H, Gao H, Wang Y, Zhang Z, Wang H, Cao J, Zhang S, Wu X, Sun Q. Characterization of Mycobacterium tuberculosis isolates from Shijiazhuang, China: genotypes and drug resistance. Int J Clin Exp Pathol. 2016;9 1533-+ [Google Scholar]
- 19.Chan M, Borgdorff M, Yip C, De Haas P, Wong W, Kam K, Van Soolingen D. Seventy percent of the Mycobacterium tuberculosis isolates in Hong Kong represent the Beijing genotype. Epidemiol Infect. 2001;127:169–171. doi: 10.1017/s0950268801005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dou HY, Tseng FC, Lin CW, Chang JR, Sun JR, Tsai WS, Lee SY, Su IJ, Lu JJ. Molecular epidemiology and evolutionary genetics of Mycobacterium tuberculosis in Taipei. BMC Infect Dis. 2008;8:170. doi: 10.1186/1471-2334-8-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiao K, Yang C, Luo T, Mei J, Gao Q. The application of variable number of tandem repeats in the microevolution study of Mycobacterium tuberculosis strain of Beijing genotype in Chongming Island, Shanghai. Journal of Microbes and Infections. 2010;5:208–213. [Google Scholar]
- 22.Filliol I, Driscoll JR, Van Soolingen D, Kreiswirth BN, Kremer K, Valétudie G, Anh DD, Barlow R, Banerjee D, Bifani PJ. Global distribution of Mycobacterium tuberculosis spoligotypes. Emerg Infect Dis. 2002;8:1347–1350. doi: 10.3201/eid0811.020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ei PW, Aung WW, Lee JS, Choi GE, Chang CL. Molecular strain typing of mycobacterium tuberculosis: a review of frequently used methods. J Korean Med Sci. 2016;31:1673–1683. doi: 10.3346/jkms.2016.31.11.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiao WW, Mokrousov I, Sun GZ, Guo YJ, Vyazovaya A, Narvskaya O, Shen AD. Evaluation of new variable-number tandem-repeat systems for typing mycobacterium tuberculosis with Beijing genotype isolates from Beijing, China. J Clin Microbiol. 2008;46:1045–1049. doi: 10.1128/JCM.01869-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou A, Nawaz M, Xue X, Karakousis PC, Yao Y, Xu J. Molecular genotyping of Mycobacterium tuberculosis in Xi’an, China, using MIRU-VNTR typing. Int J Tuberc Lung Dis. 2011;15:517–522. doi: 10.5588/ijtld.10.0495. [DOI] [PubMed] [Google Scholar]
- 26.Qiao L, Yang D, Xu W, Wang J, Bing LV, Yan S, Song H, Li G, Dong H, Wan K. Molecular typing of mycobacterium tuberculosis isolates circulating in Jiangsu province, China. BMC Infect Dis. 2011;11:1–10. doi: 10.1186/1471-2334-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao WW, Mokrousov I, Sun GZ, Guo YJ, Vyazovaya A, Narvskaya O, Shen AD. Evaluation of new variable-number tandem-repeat systems for typing with Beijing genotype isolates from Beijing, China. J Clin Microbiol. 2008;46:1045–9. doi: 10.1128/JCM.01869-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


